Abstract
Rolling circle replication is a mechanism for copying single-stranded genomes by means of double-stranded intermediates. A multifunctional replication inititiator protein (Rep) is indispensable for the precise initiation and termination of this process. Despite the ubiquitous presence and fundamental importance of rolling circle replication elements, structural information on their respective replication initiators is still missing. Here we present the solution NMR structure of the catalytic domain of Rep, the initiator protein of tomato yellow leaf curl virus. It is composed of a central five-stranded anti-parallel β-sheet, flanked by a small two-stranded β-sheet, a β-hairpin and two α-helices. Surprisingly, the structure reveals that the catalytic Rep domain is related to a large group of proteins that bind RNA or DNA. Identification of Rep as resembling the family of ribonucleoprotein/RNA-recognition motif fold proteins establishes a structure-based evolutionary link between RNA binding proteins, splicing factors, and replication initiators of prokaryotic and eukaryotic single-stranded DNA elements and mammalian DNA tumor viruses.
DNA replication is an intricate process largely regulated during early stages of initiation by specific proteins with multiple functionalities (1). Genetic entities evolutionarily as diverse as single-stranded self-complementary RNAs (2), some transposons (3), bacterial plasmids (4), bacteriophages (5), and single-stranded DNA (ssDNA) viruses of plants (6), birds (7), and mammals (8), multiply by rolling circle replication (RCR). RCR was first described ≈35 years ago (9) and became the paradigm for multiplication of autonomously replicating nucleic acid from archea to man, including the recently discovered and worldwide prevalent, yet quite elusive human TT virus (10). RCR is an asymmetric replication mechanism used in nature for copying nucleic acid information, in which leading- and lagging-strand synthesis is uncoupled. An abundance of detailed genetic and biochemical information is available for different systems (1). The replication initiator protein (Rep) of geminiviruses is a replicon-specific initiator enzyme and is an essential component of the replisome. Mechanistically, RCR initiation involves the binding of Rep protein to the replication origin, DNA nicking in a site- and strand-specific manner, thereby generating the 3′ primer for unidirectional DNA synthesis by host DNA polymerase, with Rep becoming covalently linked to the 5′ end (4, 11). After one round of polymerization, termination occurs on reencounter of another recognition site by Rep by means of cleavage of the newly synthesized strand and transfer of the 5′ Rep-linked DNA to the newly created 3′ hydroxyl. In this manner, a circular ssDNA molecule is created. In the case of coliphage ΦX174 protein A, two tyrosines were identified in the catalytic site that alternate in their cleavage and joining activities in a flip-flop type mechanism (12). Rep proteins of RCR plasmids possess only a single catalytically active tyrosine, and resolution is achieved via a water molecule, activated by a nearby carboxylate-bearing amino acid side chain (13). Thus, a circular, ssDNA molecule with simultaneous release of the protein occurs. For geminivirus Rep protein, the N-terminal region is crucial for origin recognition and DNA cleavage and nucleotidyl transfer (14, 15). A further variation of the RCR mechanism in eukaryotes is the “rolling-hairpin” replication of the linear parvovirus genome (16).
Here we present the three-dimensional (3D) solution NMR structure of the catalytic domain of the tomato yellow leaf curl virus (TYLCV) replication initiator protein. The comparative analysis of the present structure with other DNA and RNA binding proteins enabled us to discover a conserved architecture for a number of functionally diverse proteins. Based on this structural similarity, we suggest an evolutionary link between primordial single-stranded RNA binding proteins by means of prokaryotic and eukaryotic replication initiation proteins to mammalian DNA tumor viruses such as simian virus 40 (SV40) large Tumor antigen (T-ag).
Methods
Protein Expression.
The TYLCV Rep (GenBank accession no. CAA43466) catalytic domain comprising amino acids 1–136 was expressed with a N-terminal His-tag using plasmid pQE-32 (Qiagen, Courtaboeuf, France) and E. coli strain BL21. Uniformly 15N and/or 13C-labeled proteins were obtained by growth in minimal media containing 15NH4Cl and/or 13C-glucose as the sole nitrogen and carbon sources, respectively. The soluble fusion protein was purified by affinity chromatography, subjected to factor Xa protease digestion, and repurified by size-exclusion chromatography on a Superdex-75 column (Amersham Pharmacia). Purity, extent of labeling, and identity of the 15N-labeled Rep domain was assessed by mass spectrometry (13,730 Da expected mass, 13,716 Da observed mass) and N-terminal amino acid sequencing, establishing that the cleaved protein comprised residues 4–121. Samples for NMR contained 0.8–1.0 mM protein in 20 mM sodium phosphate buffer (pH 6.6), 0.1 M NaCl, 0.01% NaN3, and 1 mM DTT.
Binding Assays.
Activity assays were carried out essentially as described (17). Full-length wild-type Rep protein (1–359, C-terminal His-tagged), Rep1–136 (N-terminal His-tagged), and Rep4–121 were reacted with a 26-nt oligonucleotide (5′-CGTATAATATT*ACCGGATGGCCGCGC-3′) comprising the Rep recognition and cleavage (*) site, and covalent Rep-DNA adducts were monitored by SDS/PAGE. Reactions (10 μl) contained 0.1 μg wild-type Rep, 1.2 μg Rep1–136, or 1.0 μg Rep4–121 in 20 mM Tris⋅HCl buffer, pH 7.6/300 mM NaCl/5 mM MnCl2/1 mM DTT in the absence (−) or presence (≈2× and ≈8× molar excess) of oligonucleotide, respectively. Samples were incubated at 22°C for 30 min, followed by the addition of 2× SDS/PAGE sample buffer and heating for 5 min at >95°C before gel loading. For Western blot analysis, the gel was probed with a polyclonal rabbit anti-Rep serum and developed with alkaline phosphatase-conjugated goat anti-rabbit IgG by using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma) as substrate.
NMR Spectroscopy.
All NMR experiments were carried out at 25°C on Bruker 500, 600, 750 and 800 MHz spectrometers. 1H, 15H, and 13C backbone and side-chain resonances were assigned by 3D double- and triple-resonance NMR experiments (18). Interproton distance constraints were derived from 3D and four-dimensional 15N- and 13C-separated nuclear Overhauser enhancement (NOE) experiments. Torsion angle restraints were derived from backbone chemical shifts by using the program talos (19). Heteronuclear 3J couplings were measured by quantitative J-correlation spectroscopy (20).
Structure Calculations.
Structures were calculated from the experimental constraints in torsion angle space by using dyana (21). Upper-limit distance constraints of 2.7, 3.3, 5.0, and 5.5 Å (with appropriate corrections for methyl, aromatic, and nonstereospecifically assigned protons) were used, corresponding to strong-, medium-, weak-, and very-weak-intensity NOE crosspeaks, respectively. The experimental NMR constraints used for structure determination were as follows: 1,384 interproton distances, 251 torsion angles, and 84 3JHN couplings. The final structures exhibit no interproton distance or torsion angle violations >0.25 Å and >11°, respectively. The percentage of residues in the most favorable region of the Ramachandran map is 86%. Structural statistics were calculated with procheck (22) and dyana (21), and figures were generated with molmol (23).
Results and Discussion
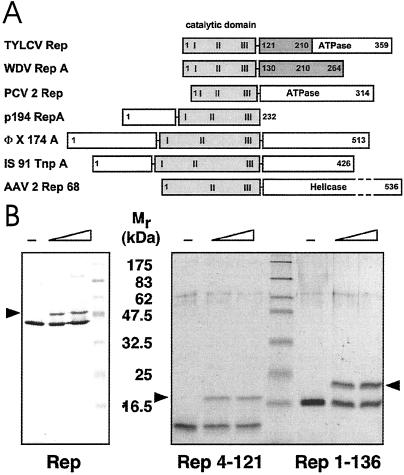
A comparison of selected Rep proteins from various sources illustrating their respective domain organisation is shown in Fig. 1A. Genetic and biochemical analyses of TYLCV Rep defined an N-terminal region spanning ≈130 aa as the catalytic domain (24, 25). Phosphodiester bond cleavage by Rep occurs via a nucleophilic attack by a tyrosine hydroxyl, Y103 in TYLCV Rep, resulting in a covalent DNA-5′ phosphotyrosyl Rep adduct (26). We prepared N-terminal Rep protein fragments and demonstrated that they were capable of forming covalent complexes with origin DNA (Fig. 1B). The final protein construct used for structure determination comprised residues 4–121 of TYLCV Rep.
Fig 1.
Domain organization and activity of Rep proteins. (A) Domain structure of TYLCV Rep, wheat dwarf virus (WDV) RepA, porcine circovirus 2 (PCV2) Rep, plasmid pC194 RepA, bacteriophage ΦX174 A, transposon IS91 TnpA, and AAV2 Rep68 proteins. The catalytic domains are displayed in gray, and the conserved sequence motifs (28) are labeled I, II, and III. The oligorimerization domains of TYLCV Rep and WDV RepA are shown in dark gray. (B) Activity assays of TYLCV Rep proteins. (Left) Western blot analysis of a 12% denaturing gel. (Right) A 15% gel after Coomassie staining. About 20–40% of Rep becomes covalently linked to the 5′ end of the 15-nt cleavage product (marked by arrow) under these conditions. The sizes of molecular weight markers are indicated between the panels.
Overall Structure.
The 3D structure of the catalytic Rep domain was determined by using heteronuclear multidimensional NMR spectroscopy (18). The structure of Rep4–121 is very well defined by the NMR data, which comprise about 1,850 total constraints, derived from 3D 15N- and 13C-edited NOE and heteronuclear correlation spectra. An ensemble of 30 NMR structures and ribbon representations of the regularized mean structure are shown in Fig. 2. Pertinent structural statistics are provided in Tables 1. Rep4–121 comprises nine β-strands and two α-helices, arranged in a central 5-stranded antiparallel β-sheet (β2, β3, β4, β8, and β9), decorated on the periphery by a small two-stranded β-sheet (β1, β5), a β-hairpin (β6, β7), and two α-helixes. The two strands in the minor sheet are extensions of strands β2 and β4 of the major sheet, solely separated by one or two residue bends (Fig. 2 B and C). One face of the central sheet is covered by the β1–β5 element, helix α1, strand β6, and the loops between β5 and β6 and β2 and α1. In contrast, the other side of the sheet is fairly exposed, only partially covered by helix α2, and its flanking, partially disordered loops (Fig. 2B).
Fig 2.
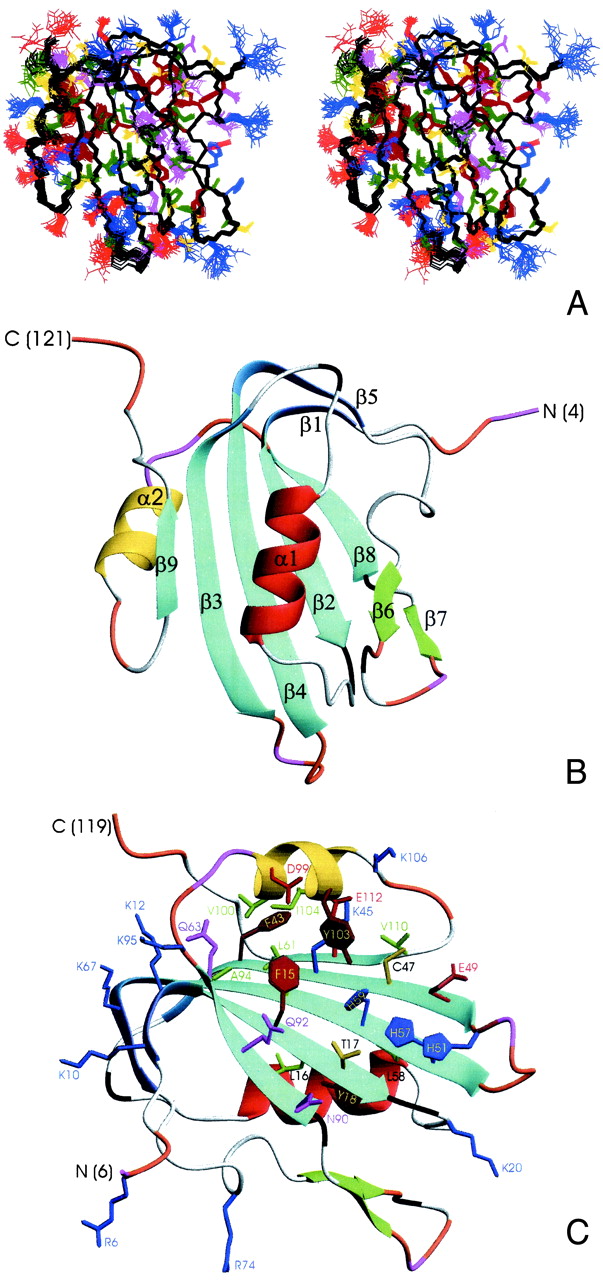
Three-dimensional structure of TYLCV Rep4–121. (A) Stereoview displaying best-fit superposition of the final ensemble (residues 6 to 119) of 30 conformers with the lowest dyana (21) target function (PDB ID ). The protein backbone (N, Cα, CO) is shown in black, and the side chains are colored according to residue type (YFW: brown; D,E: red; K,R,H: blue; A,V,L,I,P: green; T,S,C: yellow; N,Q: magenta). The coordinate precision for the protein backbone heavy atoms is 0.48 Å. (B and C) Ribbon representations of the TYLCV Rep4–121 regularized mean structure (PDB ID ). The central 5-stranded β-sheet is shown in blue, the small extension sheet in dark blue, the helix covering the β-sheet in red, the small 2-stranded sheet in green and loops in gray. The helix carrying the catalytic tyrosine is colored yellow. The strands and helices are numbered and the N and C termini labeled. Loop residues exhibiting substantial flexibility (low 15N heteronuclear NOE) or nondetected NH resonances are colored in orange and magenta, respectively. In C, selected amino acid side chains are displayed as well. They either belong to the conserved sequence motifs or occupy equivalent positions to those implicated in ss- or dsDNA/RNA binding of structurally related proteins (see Fig. 4).
Table 1.
NMR-derived constraints
| Total interproton | 1,384 |
| Intraresidue | 150 |
| Sequential (i − j = 1) | 330 |
| Short range (1 < i − j < 5) | 249 |
| Long range (i − j > 4) | 655 |
| Hydrogen bonds | 58 |
| Total dihedral angles | 251 |
| φ | 90 |
| ϕ | 94 |
| χ1 | 67 |
| Total coupling constants (3JHNHα) | 84 |
| Total number of constraints | 1,835 |
| Total number of constraints per residue | 16.4 |
Sequence comparison has defined a RCR-initiator protein superfamily and revealed conserved amino acid motifs (27). Some RCR initiators possess a C-terminal NTPase or helicase domain (Fig. 1A). Three amino acid motifs characterize the Rep catalytic domain: I, (FLTYP); II, (HxH); and III, (YxxxY) or (YxxK) (28) (Fig. 3). Motif III contains the active site tyrosine(s), motif II was postulated as a metal ion binding site, and no function was ascribed to motif I (27). Amino acids belonging to I, II, and III (Fig. 3) are displayed and highlighted in Fig. 2C. Residues of motif I reside on β2 with L16 and Y18 involved in the packing of the hydrophobic core. F15 and T17 reside on the solvent exposed side of the β-sheet, and are close to H57 and H59 of motif II on the adjacent strand (β4). Interestingly, a negatively charged side chain (E49) is also close to these histidines and constitutes a further ligand for potential divalent cation coordination. Whether and which metal is bound at this site cannot be ascertained at present, although it is known that Mn2+ or Mg2+ are essential for cleavage and strand transfer (17).
Fig 3.
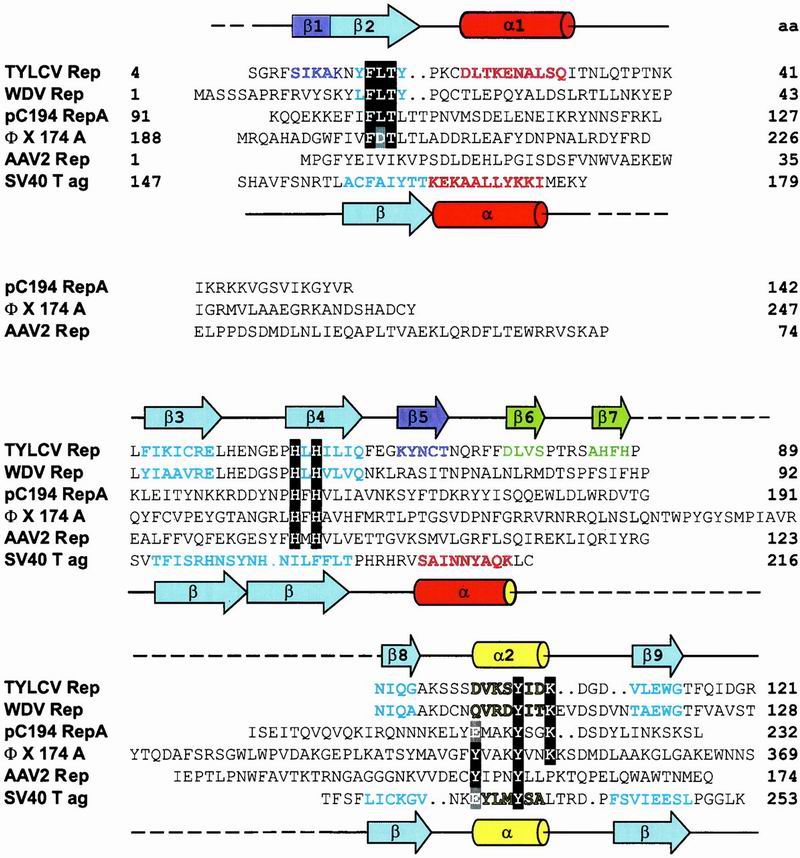
Structure-based sequence alignment of the catalytic domains of Rep proteins from TYLCV, WDV, pC194, ΦX, AAV2, and the DBD of SV40 T-ag. Amino acids of the motifs I, II, and III (28) are highlighted in black. The catalytic tyrosine(s) and equivalent residues in SV40 T-ag are highlighted in gray. Secondary structure elements present in the TYLCV Rep domain and SV40 T-ag 3D structures are indicated by cylinders (α-helices) and arrows (β-strands) above and below the sequence alignment, respectively. Amino acids in β-strands and α-helices of TYLCV Rep and SV40 T-ag, as well as predicted ones, are colored according to their location in the structure (see Fig. 2 B and C). Residue numbers are given at the end of each line. (GenBank accession nos: TYLCV Rep, CAA43466; WDV RepA, CAA57625; PCV2 Rep, AAC59462; pC194 RepA, NP_040435; IS91 TnpA, CAA34970; ΦX174 A, NP_040703; AAV2 Rep68, AAC03774; SV40 T-ag, P03070).
Nucleic Acid Binding Site and Catalytic Residue.
The catalytic tyrosine (Y103) resides on helix α2, pointing down onto the otherwise exposed side of the β-sheet. This finding suggests that this surface will be involved in ssDNA binding and catalysis. Tyrosines are frequently used by enzymes that cut and join DNA, such as recombinases (29) or type I topoisomerases (30), though they are in structurally different environments from Rep. As is evident from the present structure, Y103 is close to several potentially activating residues; either of the two histidines of motif II or C47 and E49 could be involved in catalytic activation and/or metal binding. Any involvement of C47, however, was ruled out because C47A substitution did not affect activity (unpublished results).
In ΦX174 a single, monomeric protein A is capable of substrate DNA cleavage and transesterification at initiation and termination of replication (31), whereas plasmid RCR initiators function as dimers or tetramers, that are modified in a second cleavage-transesterification step and cannot reinitiate replication (32). For geminivirus Rep proteins, oligomerization is known to occur, and mutations in the oligomerization domain have been shown to impact viral DNA replication in vivo (33). This finding allows for the attractive possibility that geminivirus Rep proteins could function in a “tyrosine only” mechanism, analogous to ΦX174 protein A, with termination of replication catalyzed again by Y103, now located on a different subunit in an oligomeric complex.
Based on our structure and the location of the catalytic tyrosine, we suggest that origin recognition by Rep results in partial melting of the DNA and binding of the conserved nonanucleotide sequence (6) as single-strand to the exposed surface of the β-sheet.
Similarity to Other Nucleic Acid Binding Proteins.
The present Rep structure reveals a surprising similarity in architecture to other nucleic acid binding proteins. dali (34) identifies more than 85 structurally similar proteins, despite very low sequence similarity with the Rep catalytic domain (identity <15%). Most of them are involved in DNA or RNA binding. RNA binding domains from U1A, PTB, U2AF, ribosomal protein S6, EF1, sex-lethal and nucleolin exhibit related structures, as do the DNA binding domains (DBD) of SV40 T-ag, E1 and E2 from papillomavirus, and Epstein–Barr nuclear antigen 1 (EBNA1). The structures of the DBDs of papillomavirus protein E1 and SV40 T-ag, the prototypic ribonucleoprotein (RNP)-fold protein U1A, and human polypyrimidine tract binding protein (PTB) are shown in Fig. 4. Comparison with Rep4–121 (Fig. 2C) illustrates the overall similarity in architecture, i.e., the central 4- or 5-stranded β-sheet covered on one face by two helices. In Rep, the second helix is absent and replaced by the β6–β7 hairpin, and their connection loops. Flanking the sheet are other secondary structure elements (shorter helices, loops, or a small 2-stranded sheet) that can be regarded as decorations or extra elements. Crucial amino acids involved in nucleic acid binding are also highlighted in Fig. 4. RNPs and RNA-recognition motifs (RRM) (35) have been extensively characterized, and several high-resolution structures of nucleic acid complexes are available (36–38). All of these proteins interact with single-stranded nucleic acids primarily through exposed hydrophobic residues of the central β-sheet, with bases stacking onto aromatic or hydrophobic side chains residing in the conserved RNP1 and RNP2 sequence motifs (39). The comparison of the location of these amino acids (Fig. 4; U1A) with those of motif I and motif II (Fig. 2C) is intriguing in that they appear to structurally coincide. In U1A, the single-stranded RNA binds on the open surface of the central sheet. This binding involves both RNP sequence motifs (in U1A, RNP1: R52GQAFVIF59, and RNP2: I12YINNL17). Direct mapping of these residues onto the Rep structure yields Y14-P19 in Rep β2, containing residues of motif I, and H57-F64, containing motif II, thus Y13 and F56 of U1A are equivalent to F15 and L61 of Rep, respectively. Given the fact that the nucleic acid binding face of U1A is similarly exposed to the one observed in the present Rep structure, we believe that ssDNA binding of Rep will most likely occur in a fashion related to that seen in these RNP complexes. For SV40 T-ag, the double stranded DNA (dsDNA) binding surface was identified by mutagenesis and NMR titration (40). It comprises the loop preceding β2 (residues S152, N153, R154, and T155) and the N-terminal end of the α-helix that has no counterpart in Rep (residues H203, R204, V205, S206, and A207 (Fig. 3, Fig. 4; T-ag). A similar area in the structure of papillomavirus E1 (Fig. 3; E1) was defined by mutagenesis (41), and very recently the atomic details of the contacts between E1 and dsDNA were determined from a x-ray structure of the complex (42). Equivalent regions in Rep are the β1 and β5 strands that form the minor or continuing sheet to β2 and β4 and the loop preceding α2 (Fig. 2C), suggesting that dsDNA binding by Rep could involve these elements. Based on the structural comparisons presented here and the functional similarity between Rep and SV40 T-ag (and E1) with respect to origin binding, we propose that Rep proteins recognize dsDNA with a cluster of positively charged residues protruding from the curved, extended sheet area (β1, β5). After local melting of the origin DNA, this would allow for easy positioning of the ssDNA on top of the exposed surface of the central β-sheet with Y103 poised to attack the phosphodiester bond. The cluster of histidine and glutamate residues could aid in positioning the nucleic acid, either directly or by metal coordination.
Fig 4.
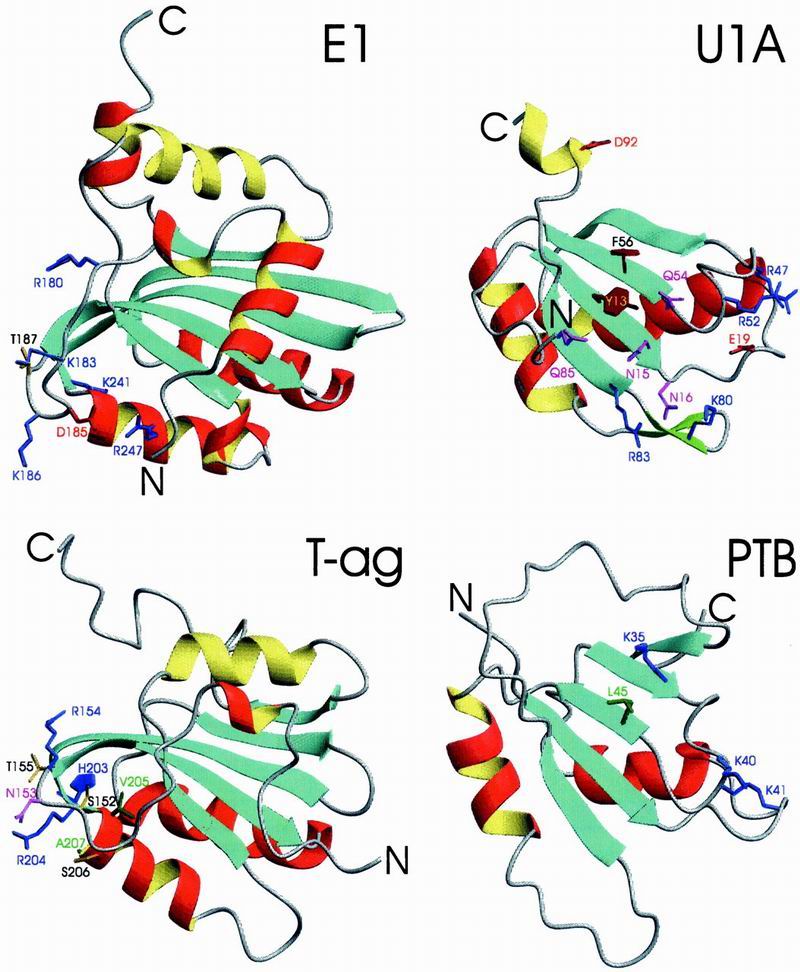
Ribbon representation of the DBDs of papillomavirus protein E1 and SV40 T-ag and RBDs of U1A and PTB. Helices conserved between Rep and D/RBDs are colored in red (α1) and yellow (α2), all other helices in red/yellow (outside/inside), strands in blue and loops and chain termini in gray. Side chains of amino acids implicated in nucleic acid recognition are displayed and colored identically to the color code given for Fig. 2C. PDB accession codes are E1, ; U1A, ; T-ag, ; and PTB, .
Evolutionary Implications.
The apparent similarity in the fold of the catalytic domain of RCR initiator proteins, members of the RNP/RRM family, and the DBD domain of small dsDNA viruses suggests a common root. As a group, all of these proteins act on nucleic acid sequences required to undergo transitions between double- and single-stranded forms, i.e., are linked to unwinding or hairpin formation. Double-strand recognition and catalysis are mediated by distinct structural elements “decorating” the primordial fold. Some proteins, e.g., SV40 T-ag and E1 have lost their catalytic activity, even if the original active center residues are still present (T-ag), albeit now in a different structural environment. This is easily appreciated from the structure-based sequence alignment for the catalytic domains of geminivirus Rep proteins, the origin DBD of SV40 T-ag, the plasmid pC194 RepA, adeno-associated virus (AAV)-2 Rep, and ΦX 174 protein A (Fig. 3). The development of initiator proteins from those of the ancient RCR elements to those found in the more sophisticated DNA tumor viruses mirrors the evolution of their host. Similarly, the recently discovered RC transposons in the Arabidopsis thaliana genome, termed Helitrons, have been suggested to represent the missing evolutionary link between prokaryotic RC elements and geminiviruses (43). Alternatively, they may have arisen from geminiviruses that were integrated into the genome of an early eukaryotic ancestor (44). They contain an 11-aa motif similar to the motif II (HxH) of the Rep proteins (see Fig. 3), followed by a conserved two-tyrosine-containing motif ≈100 aa further toward the C terminus that contains tyrosines and lysine with identical spacing to that observed in Rep (highlighted in black in Fig. 3). Thus, it could well be possible that the A. thaliana ATHEL1p exon encoded protein contains an N-terminal catalytic domain that is structurally similar to that of Rep and SV40 T-ag fused to a C-terminal helicase.
Table 2.
Structural quality
| Data set | Statistics | |
|---|---|---|
| Residual violations | ||
| No. of violations | ||
| Upper limits | 0 ± 1 (0; 3) | |
| Lower limits | 0 ± 0 (0; 1) | |
| van der Waals | 1 ± 1 (0; 2) | |
| Torisons | 0 ± 0 (0; 0) | |
| Couplings | 0 ± 0 (0; 0) | |
| Maximum violation | ||
| Upper limits | 0.18 ± 0.04 (0.11; 0.25) | |
| Lower limits | 0.13 ± 0.02 (0.11; 0.22) | |
| van der Waals | 0.23 ± 0.07 (0.14; 0.38) | |
| Torisons | 0.09 ± 0.03 (0.06; 0.19) | |
| Couplings | 0.22 ± 0.18 (0.06; 0.72) | |
| DYANA target function, Å2 | 1.34 ± 0.18 (1.00; 1.81) | |
| Ramachandran statistics‡ | ||
| 30-conformer ensemble | ||
| Residues in most favored regions | 85.9 | |
| Residues in additional favored regions | 11.6 | |
| Residues in generously allowed regions | 2.5 | |
| Residues in disallowed regions | 0.0 | |
| Regularized mean structure | ||
| Residues in most favored regions | 86.3 | |
| Residues in additional favored regions | 10.8 | |
| Residues in generously allowed regions | 2.9 | |
| Residues in disallowed regions | 0.0 |
Average values, standard deviation, and maximum and minimum values (in parentheses) for the 30-conformer ensemble. Upper limits, lower limits, and van der Waals are given in Å, torsion violations are given in radians, and coupling constant violations are given in Hz.
Number of distance constraint violations larger than 0.2 Å (upper limits, lower limits, and van der Waals), torisonal violations larger than 5 degrees, and coupling constant violations larger than 0.25 Hz.
‡ The 102 non-Gly, non-Pro residues in segment 7–118 of the 30 conformers and those of the regularized mean structure are considered separately. Values are given in percentages.
Table 3.
Coordinate precision
| All residues | Well defined residues | |
|---|---|---|
| N, Cα, C′ | 0.48 ± 0.18/0.54 ± 0.17 | 0.41 ± 0.13/0.44 ± 0.12 |
| (0.23; 0.75)/(0.20; 0.91) | (0.23; 0.66)/(0.20; 0.76) | |
| All heavy atoms | 0.89 ± 0.16/1.05 ± 0.14 | 0.81 ± 0.11/0.94 ± 0.10 |
| (0.67; 1.19)/(0.71; 1.35) | (0.59; 1.05)/(0.69; 1.19) |
Average rms difference between the 30 conformer ensemble and the regularized mean structure/pairwise rms difference between members of the ensemble. Data are given in Å.
Only 7 residues (95–99, 117, and 118) out of the total 112 in the 7–118 segment were excluded.
Acknowledgments
We thank D. Garrett and F. Delaglio for software, R. Tschudin and J. Baber for technical support, L. K. Pannell for mass spectrometry, and F. Bernardi for constructive comments on the manuscript. This work was supported in part by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to A.M.G.) and by the Ministerio de Ciencia y Tecnología (BIO2001-2287) (to R.C.-O.).
Abbreviations
RC, rolling circle
RCR, RC replication
Rep, replication initiator protein
dsDNA, double-stranded DNA
ssDNA, single-stranded DNA
NOE, nuclear Overhauser effect
RNP, ribonucleoprotein
RRM, RNA-recognition motif
TYLCV, tomato yellow leaf curl virus
WDV, wheat dwarf virus
AAV, adeno-associated virus
SV40, simian virus 40
3D, three-dimensional
T-ag, large T antigen
DBD, DNA-binding domain
Data deposition: Structural coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Kornberg A. & Baker, T. A., (1992) DNA Replication (Freeman, New York).
- 2.Been M. D. & Wickham, G. S. (1997) Eur. J. Biochem. 247 741-753. [DOI] [PubMed] [Google Scholar]
- 3.Mahillon J. & Chandler, M. (1998) Microbiol. Mol. Biol. Rev. 62 725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan S. A. (2000) Mol. Microbiol. 37 477-484. [DOI] [PubMed] [Google Scholar]
- 5.Baas P. D. & Jansz, H. S. (1988) Curr. Top. Microbiol. Immunol. 136 31-70. [DOI] [PubMed] [Google Scholar]
- 6.Laufs J., Jupin, I., David, C., Schumacher, S., Heyraud-Nitschke, F. & Gronenborn, B. (1995) Biochimie 77 765-773. [DOI] [PubMed] [Google Scholar]
- 7.Bassami M. R., Berryman, D., Wilcox, G. E. & Raidal, S. R. (1998) Virology 249 453-459. [DOI] [PubMed] [Google Scholar]
- 8.Allan G. M. & Ellis, J. A. (2000) J. Vet. Diagn. Invest. 12 3-14. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert W. & Dressler, D. (1968) Cold Spring Harbor Symp. Quant. Biol. 33 473-484. [DOI] [PubMed] [Google Scholar]
- 10.Bendinelli M., Pistello, M., Maggi, F., Fornai, C., Freer, G. & Vatteroni, M. L. (2001) Clin. Microbiol. Rev. 14 98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Mansfeld A. D., Baas, P. D. & Jansz, H. S. (1984) Adv. Exp. Med. Biol. 179 221-230. [DOI] [PubMed] [Google Scholar]
- 12.Van Mansfeld A. D., van Teeffelen, H. A., Baas, P. D. & Jansz, H. S. (1986) Nucleic Acids Res. 14 4229-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noirot-Gros M. F., Bidnenko, V. & Ehrlich, S. D. (1994) EMBO J. 13 4412-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi I. R. & Stenger, D. C. (1995) Virology 206 904-912. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez C. (2000) EMBO J. 19 792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore S. F. & Tattersall, P. (1996) in DNA Replication in Eukaryotic cells, ed. DePamphilis, M. L. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 799–813.
- 17.Laufs J., Traut, W., Heyraud, F., Matzeit, V., Rogers, S. G., Schell, J. & Gronenborn, B. (1995) Proc. Natl. Acad. Sci. USA 92 3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clore G. M. & Gronenborn, A. M. (1998) Trends Biotechnol. 16 22-34. [DOI] [PubMed] [Google Scholar]
- 19.Cornilescu G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13 289-302. [DOI] [PubMed] [Google Scholar]
- 20.Bax A., Vuister, G. W., Grzesiek, S., Delaglio, F., Wang, A. C., Tschudin, R. & Zhu, G. (1994) Methods Enzymol. 239 79-105. [DOI] [PubMed] [Google Scholar]
- 21.Güntert P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273 283-298. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. W. (1993) J. Appl. Crystallogr. 26 283-291. [Google Scholar]
- 23.Koradi R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graph. 14 51-55. [DOI] [PubMed] [Google Scholar]
- 24.Jupin I., Hericourt, F., Benz, B. & Gronenborn, B. (1995) FEBS Lett. 362 116-120. [DOI] [PubMed] [Google Scholar]
- 25.Heyraud-Nitschke F., Schumacher, S., Laufs, J., Schaefer, S., Schell, J. & Gronenborn, B. (1995) Nucleic Acids Res. 23 910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laufs J., Schumacher, S., Geisler, N., Jupin, I. & Gronenborn, B. (1995) FEBS Lett. 377 258-262. [DOI] [PubMed] [Google Scholar]
- 27.Koonin E. V. & Ilyina, T. V. (1993) Biosystems 30 241-268. [DOI] [PubMed] [Google Scholar]
- 28.Ilyina T. V. & Koonin, E. V. (1992) Nucleic Acids Res. 20 3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickman A. B., Waninger, S., Scocca, J. J. & Dyda, F. (1997) Cell 89 227-237. [DOI] [PubMed] [Google Scholar]
- 30.Krogh B. O. & Shuman, S. (2000) Mol. Cell 5 1035-1041. [DOI] [PubMed] [Google Scholar]
- 31.Hanai R. & Wang, J. C. (1993) J. Biol. Chem. 268 23830-23836. [PubMed] [Google Scholar]
- 32.Jin R., Fernandez-Beros, M. E. & Novick, R. P. (1997) EMBO J. 16 4456-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orozco B. M., Miller, A. B., Settlage, S. B. & Hanley-Bowdoin, L. (1997) J. Biol. Chem. 272 9840-9846. [DOI] [PubMed] [Google Scholar]
- 34.Holm L. & Sander, C. (1996) Science 273 595-603. [DOI] [PubMed] [Google Scholar]
- 35.Burd C. G. & Dreyfuss, G. (1994) Science 265 615-621. [DOI] [PubMed] [Google Scholar]
- 36.Musco G., Stier, G., Joseph, C., Castiglione Morelli, M. A., Nilges, M., Gibson, T. J. & Pastore, A. (1996) Cell 85 237-245. [DOI] [PubMed] [Google Scholar]
- 37.Allain F. H., Gubser, C. C., Howe, P. W., Nagai, K., Neuhaus, D. & Varani, G. (1996) Nature (London) 380 646-650. [DOI] [PubMed] [Google Scholar]
- 38.Ding J., Hayashi, M. K., Zhang, Y., Manche, L., Krainer, A. R. & Xu, R. M. (1999) Genes Dev. 13 1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Cañadillas J. M. & Varani, G. (2001) Curr. Opin. Struct. Biol. 11 53-58. [DOI] [PubMed] [Google Scholar]
- 40.Luo X., Sanford, D. G., Bullock, P. A. & Bachovchin, W. W. (1996) Nat. Struct. Biol. 3 1034-1039. [DOI] [PubMed] [Google Scholar]
- 41.Enemark E. J., Chen, G., Vaughn, D. E., Stenlund, A. & Joshua-Tor, L. (2000) Mol. Cell 6 149-158. [PubMed] [Google Scholar]
- 42.Enemark E. J., Stenlund, A. & Joshua-Tor, L. (2002) EMBO J. 21 1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapitonov V. V. & Jurka, J. (2001) Proc. Natl. Acad. Sci. USA 98 8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feschotte C. & Wessler, S. R. (2001) Proc. Natl. Acad. Sci. USA 98 8923-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]