Abstract
By primarily measuring changes in transcript and protein abundance, conventional genomics and proteomics methods may fail to detect significant posttranslational events that regulate protein activity and, ultimately, cell behavior. To address these limitations, activity-based proteomic technologies that measure dynamics in protein function on a global scale would be of particular value. Here, we describe the application of a chemical proteomics strategy to quantitatively compare enzyme activities across a panel of human breast and melanoma cancer cell lines. A global analysis of the activity, subcellular distribution, and glycosylation state for the serine hydrolase superfamily resulted in the identification of a cluster of proteases, lipases, and esterases that distinguished cancer lines based on tissue of origin. Strikingly, nearly all of these enzyme activities were down-regulated in the most invasive cancer lines examined, which instead up-regulated a distinct set of secreted and membrane-associated enzyme activities. These invasiveness-associated enzymes included urokinase, a secreted serine protease with a recognized role in tumor progression, and a membrane-associated hydrolase KIAA1363, for which no previous link to cancer had been made. Collectively, these results suggest that invasive cancer cells share discrete proteomic signatures that are more reflective of their biological phenotype than cellular heritage, highlighting that a common set of enzymes may support the progression of tumors from a variety of origins and thus represent attractive targets for the diagnosis and treatment of cancer.
In recent years, DNA microarrays have become a standard tool for the molecular analysis of cancer, providing global profiles of transcription that reflect the origin (1–3), stage of development (4), and drug sensitivity (5) of tumor cells. The ability to complement these genomic approaches with methods that analyze the proteome (6, 7) is crucial for the identification and functional characterization of proteins that support tumorigenesis. However, to date, the field of proteomics has had only a limited impact on cancer research, in large part because of the myriad technical challenges that accompany the analysis of complex protein samples (8). For example, conventional proteomics approaches that rely on two-dimensional gel electrophoresis encounter difficulty analyzing important fractions of the proteome, including membrane-associated (9) and low abundance proteins (10). Additionally, most proteomics technologies are restricted to detecting changes in protein abundance (11), and therefore, offer only an indirect readout of dynamics in protein activity. Numerous posttranslational forms of protein regulation, including those governed by protein–protein interactions, remain undetected. To address these limitations, we have developed a chemical proteomics strategy referred to as activity-based protein profiling (ABPP) that allows significant fractions of the enzyme proteome to be analyzed in an activity-dependent manner (12). This approach employs chemical probes that covalently label the active sites of enzyme superfamilies in a manner that provides a direct readout of changes in catalytic activity, distinguishing, for example, functional proteases from their inactive zymogens and/or endogenously inhibited forms (12–14). Moreover, by providing a covalent link between the labeled proteins and a chemical tag, ABPP permits the consolidated detection, isolation, and identification of active enzymes directly from complex proteomes (13).
Here we show that ABPP probes that target the serine hydrolase superfamily of enzymes generate molecular profiles that classify human breast and melanoma cancer cell lines into subtypes based on higher-order cellular properties, including tissue of origin and state of invasiveness.
Materials and Methods
Preparation of Human Cancer Cell Line Proteomes.
All cell lines, with the exception of MUM-2B and MUM-2C, are part of the NCI60 panel of cancer cell lines and were obtained from the National Cancer Institute's Developmental Therapeutics Program. The MUM-2B and MUM-2C lines were provided by Mary Hendrix. All cell lines were grown to 80% confluence in RPMI medium 1640 containing 10% FCS and then cultured in serum-free media for 48 h, after which conditioned media was collected on ice and the cells were harvested. Conditioned media samples were centrifuged at 2,400 × g for 5 min, and the protein content of the supernatant was precipitated with ammonium sulfate (80%), resuspended in 50 mM Tris⋅HCl, (pH 7.5; Buffer 1), and desalted over a PD-10 column (Amersham Pharmacia) to provide secreted proteome fractions. Cell pellets were sonicated and Dounce homogenized in Buffer 1 followed by centrifugation at 100,000 × g to provide soluble cellular proteome fractions (supernatant) and a membrane pellet. Membrane pellets were homogenized in Buffer 1 with 1% Triton X-100, rotated at 4°C for 1 h and then centrifuged at 100,000 × g to provide membrane proteome fractions (supernatant). A typical ratio of 8:2:1 was observed for the relative quantity of soluble/secreted/membrane protein isolated for each cell line.
Proteome Labeling and Quantification of Enzyme Activities.
Standard conditions for fluorophosphonate (FP)–proteome reactions were as follows: proteomes were adjusted to a final protein concentration of 1 mg/ml in Buffer 1 and treated with 1 or 4 μM (soluble/membrane and conditioned medium proteomes, respectively) rhodamine-coupled FP (15) for 1 h at room temperature. After labeling, a portion of each proteome sample was treated with PNGaseF (New England Biolabs) to provide deglycosylated proteomes. Where indicated, proteome samples were preincubated with recombinant plasminogen activator inhibitor (PAI)-1 (20 μg/ml; Calbiochem) for 30 min before the addition of FP–rhodamine. Reactions were quenched with one volume of standard 2× SDS/PAGE loading buffer (reducing), separated by SDS/PAGE (10–14% acrylamide), and visualized in-gel with a Hitachi FMBio IIe flatbed fluorescence scanner (MiraiBio) as described (15). Integrated band intensities (normalized for volume) were calculated for the labeled proteins. For each enzyme activity, 4–6 data points were generated from independent labeling reactions conducted on 2 or 3 independently prepared proteomic samples. These data points were averaged to provide the level of each enzyme activity in each cell line. The activity levels of each enzyme were compared across the cell lines by using the Tukey's honestly significant difference test, where P values <0.05 were considered statistically significant.
Isolation and Identification of FP-Labeled Enzyme Activities.
Isolation of FP-labeled proteins was achieved by using biotinylated FPs and an avidin-based affinity purification procedure (13). Avidin-enriched FP-labeled proteins were separated by SDS/PAGE, and the protein bands were excised and digested with trypsin. The resulting peptides were analyzed by a combination of matrix assisted laser desorption mass spectrometry (MS) (Voyager-Elite time-of-flight MS instrument, PerSeptive Biosystems, Framingham, MA) and microcapillary liquid chromatography-electrospray tandem MS [1100 HPLC (Agilent, Palo Alto, CA) combined with a Finnigan LCQ Deca MS (Thermo Finnigan, San Jose, CA)]. The MS data were used to search public databases to identify the FP-labeled proteins as described (13).
Fatty Acid Amide Hydrolase (FAAH) Enzyme Actvity Assays.
FAAH enzyme activity assays were conducted by using 14C-oleamide as a substrate as described (16), with the exception that the reactions were conducted at pH 8.0.
Cluster Analysis of Proteomic Profiles.
Averaged cell line values for each serine hydrolase activity were compared, with the line that expressed the highest level of this activity being defined as 100%. The rest of the cell lines were expressed as a percentage of this highest activity to normalize the data sets. We then applied a hierarchical clustering algorithm to the cell lines by average linkage clustering using the Pearson correlation coefficient as the measure of similarity (Gene Cluster computer package; ref. 17). Additional cluster analyses were performed on enzyme activity profiles of the secreted, membrane, and soluble proteomes separately and in all of their respective pair-wise combinations. Of these six additional cluster analyses, only the “membrane + secreted” analysis produced a dendrogram with increased distances among the three major clusters observed in the “total” serine hydrolase activity analysis.
Invasion Assays.
Cell invasiveness was assessed by using BIOCOAT matrigel invasion chambers (Becton–Dickinson) according to the protocol provided by the manufacturer. Briefly, 1.5 × 105 cells were seeded into each chamber in serum-free conditions, and incubated for 16 h at 37°C, 5% CO2/95% air. Invading cells on the bottom surface of the membrane insert were fixed, stained with crystal violet, and counted. Results expressed as number of invading cells refers to average number of invading cells per 8 fields counted (n = 3–4 for each cell line).
Results
Activity-Based Profiling of Human Cancer Cell Proteomes.
We have previously described the generation of affinity tagged FPs as prototype ABPP probes that target the serine hydrolase superfamily of enzymes (12, 13). Considering that serine hydrolases represent one of the largest and most diverse classes of enzymes in the human proteome, composing approximately 1% of all predicted gene products (18, 19), we hypothesized that a comprehensive examination of their catalytic activities would yield proteomic information of sufficient quantity and quality to portray higher-order cellular properties. To test this hypothesis, we selected a panel of human cancer cell lines for comparative analysis by ABPP based on the following criteria: (i) they exhibit a diverse range of well characterized cellular properties, including differences in hormone responsiveness, invasiveness, and metastatic potential; (ii) they represent multiple lines derived from at least two distinct types of cancer, and therefore permit the comparison of proteomic expression patterns both within and between cancer classes; and (iii) they have previously been analyzed with gene expression microarrays, and therefore allow for a comparison between proteomic data and trancriptional profiles (2, 3, 5). To profile serine hydrolase activities in the context of their subcellular localization, proteomes from each cell line were separated into three fractions (secreted, membrane, and soluble) before treatment with a rhodamine-tagged FP probe (15). Fluorescently labeled proteins were then separated by SDS/PAGE and visualized in-gel by using a flatbed laser-induced fluorescence scanner. Integrated band intensities for each identified enzyme activity were averaged from 4–6 proteomic samples to provide the results presented in Figs. 1–4 (complete results are provided in bar graphs, which are published as supporting information on the PNAS web site, www.pnas.org). In parallel experiments, biotinylated FP probes were used to affinity isolate the active enzymes, which allowed for their molecular identification by mass spectrometry methods.
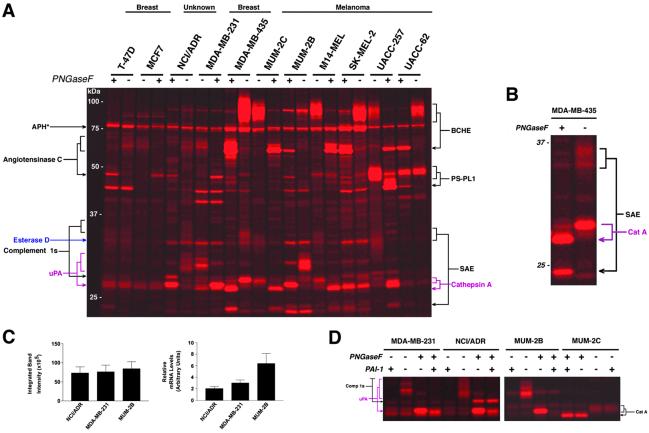
Fig 1.
Serine hydrolase activity profiles of the secreted proteomes of human cancer cell lines. (A) Representative in-gel fluorescence analysis of secreted serine hydrolase activity profiles obtained from reactions between cancer cell line conditioned media and a rhodamine-tagged FP. Enzyme activities are identified on either side of the gel (arrowheads point to the deglycosylated form of each enzyme; see Fig. 3A for complete names of proteins). Deglycosylation was accomplished by treatment of a portion of the FP-labeled proteomes with PNGaseF before analysis. APH* refers to acyl peptide hydrolase, a cytosolic protein detected in the conditioned media. (B) Expanded view of FP-labeled secreted MDA-MB-435 proteome highlights the increased resolution that is achieved for highly glycosylated enzyme activities (e.g., SAE) after treatment with PNGaseF. (C) Levels of active urokinase secreted by cancer cell lines as measured by ABPP (left panel) and urokinase mRNA levels as measured by Northern analysis (n = 3 or 4). mRNA levels are expressed in arbitrary units relative to an internal control. (D) Inhibition of urokinase (uPA) activity by PA-I. Pretreatment of each proteome with PA-I (20 μg/ml) blocked the labeling of uPA by FP-rhodamine, but did not affect the labeling of other serine proteases (e.g., Comp 1s and cat. A).
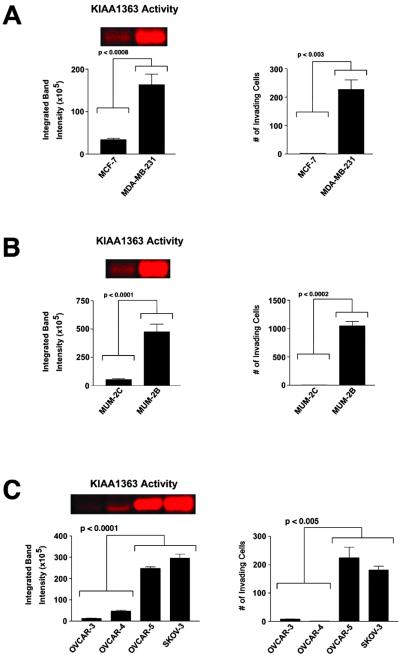
Fig 4.
Correlation between the activity of the membrane-associated hydrolase KIAA1363 and the invasiveness of human cancer cell lines. (A–C) Levels of active KIAA1363 present in cancer cell membrane proteomes as measured by ABPP (Left), and cancer cell invasiveness as measured by matrigel invasion assays (Right). Results expressed as number of invading cells refers to average number of invading cells per 8 fields counted (n = 3–4 for each cell line). (A) Breast carcinoma lines. (B) Melanoma lines. (C) Ovarian carcinoma lines.
Serine Hydrolase Activity Profiles of the Secreted Proteomes of Human Breast and Melanoma Cancer Cells.
Fig. 1A shows a representative in-gel fluorescence analysis of the secreted serine hydrolase activity profiles of human cancer cell lines. Initial profiles revealed that several enzyme activities migrated as faint, diffuse bands, suggesting that they existed in a highly glycosylated state. Therefore, a portion of each FP-labeled proteome was deglycosylated before separation by SDS/PAGE, resulting in a striking increase in the resolution of these proteins [for example, see sialic acid 9-O-acetylesterase (SAE); Fig. 1B]. Most of the secreted serine hydrolase activities exhibited a restricted pattern of distribution among the human cancer lines. For example, three secreted enzyme activities, SAE, butyrylcholinesterase, and cathepsin A, were up-regulated in most melanoma lines relative to breast carcinomas. Notably, however, the estrogen receptor negative [ER(−)] breast line MDA-MB-435 secreted high levels of these activities (Fig. 1A). Interestingly, a recent cDNA microarray analysis revealed that the transcriptional profile of the MDA-MB-435 line more closely resembled those of melanoma cells than breast carcinoma cells, suggesting that this line may be misclassified (2). Thus, several secreted serine hydrolase activities were identified that appeared to represent markers for cells of melanoma origin. It was therefore surprising to observe that all of these proteins were dramatically down-regulated in the highly invasive melanoma line, MUM-2B. Instead, MUM-2B cells secreted high levels of active urokinase and esterase D, two serine hydrolases that were also up-regulated in other aggressive lines examined, including the ER(−) breast carcinoma MDA-MB-231 and the multidrug-resistant NCI/ADR line.
Identification of Posttranslationally Regulated Serine Proteases.
The identification of active urokinase in several invasive cancer lines was intriguing considering that this serine protease is an established marker of human cancer progression in vivo (20). However, a major open question regarding urokinase is the degree to which its mRNA and/or protein levels in tumor samples reflect the state of activity of the protein. Urokinase activity is regulated by a host of posttranslational mechanisms including zymogen processing and interactions with multiple endogenous inhibitory proteins (PAI-1, PAI-2, maspin, myoepithelium-derived serine proteinase inhibitor), that also have perceived roles in tumorigenesis (21–25). With these factors in mind, it is noteworthy that urokinase mRNA levels failed to directly correlate with urokinase activity in the cancer lines examined. Whereas approximately equal levels of active urokinase were observed in the NCI/ADR, MDA-MB-231, and MUM-2B lines (Fig. 1C Left), 1.5- and 3-fold more urokinase transcript were observed in the latter two lines (Fig. 1C Right), respectively, suggesting that posttranscriptional events regulated urokinase activity in these cells. To confirm that ABPP probes could detect shifts in the balance of proteases and their endogenous regulatory proteins, excess PAI-1 was applied to each secreted proteome before treatment with FP-rhodamine. The addition of PAI-1 blocked more than 85% of the observed urokinase activity in each of the cancer lines without affecting the activity of other proteases (Fig. 1D; PAI-1 inhibition of urokinase activity: MDA-MB-231, 95% ± 3%; NCI/ADR, 88% ± 4%, MUM-2B, 98% ± 1%; n = 3 or 4 per cell line). Collectively, these findings underscore the value of activity-based proteomics methodologies that can measure the functional outcome of posttranslational events that regulate enzyme activity in vivo.
Serine Hydrolase Activity Profiles of the Membrane and Soluble Proteomes of Human Cancer Cells.
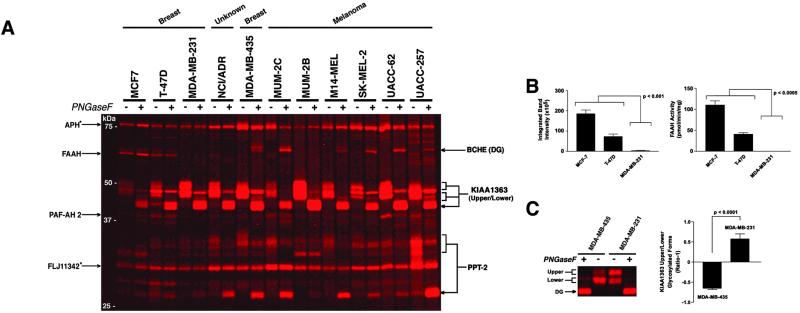
Several membrane-associated serine hydrolase activities also exhibited restricted patterns of distribution across the cancer lines (Fig. 2A). Notably, the integral membrane enzyme, FAAH, was detected exclusively in the poorly invasive breast cancer lines MCF7 and T-47D. The graded distribution of FAAH among breast cancer lines was used as a model to test the accuracy and sensitivity with which ABPP could measure moderate as well as extreme differences in enzyme activity. FAAH activity was estimated by ABBP to be 2.5-fold higher in MCF7 cells relative to T-47D cells (Fig. 2B Left), and a nearly identical ratio was calculated with assays using the radiolabeled FAAH subtrate 14C-oleamide (FAAHMCF7/FAAHT-47D = 2.6; Fig. 2B Right). Taking into account the kcat of FAAH for oleamide (approximately 2 s−1 at pH 8.0; ref. 26) and the amount of total membrane protein loaded in each gel lane (15 μg), we estimate that the fluorescent signal observed for FAAH in the T-47D membrane protein sample corresponded to approximately 600 pg of active protein, or less than 0.0005% of the total T-47D cell proteome. This measure of FAAH protein matched closely the value calculated by comparing the T-47D FAAH signal to signals of a serial dilution of purified FAAH protein labeled to completion with FP-rhodamine (800 pg; data not shown). These findings highlight that ABPP methods can detect changes in enzyme activity with a level of accuracy and sensitivity compatible with profiling low abundance proteins in complex proteomes.
Fig 2.
Serine hydrolase activity profiles of the membrane proteomes of human cancer cell lines. (A) Representative in-gel fluorescence analyses of the serine hydrolase activity profiles of cancer cell membrane proteomes. Enzyme activities are identified on either side of the gels (arrowheads point to the deglycosylated form of each enzyme; see Fig. 3A for full names of proteins). Proteins marked with an asterisk represent soluble hydrolases also detected in the membrane proteome. DG, deglycosylated. (B) The activity of FAAH in breast cancer membranes as measured by ABPP (Left) and FAAH substrate (Right) assays. (C) Relative activity levels for upper and lower glycosylated forms of the membrane hydrolase KIAA1363 in MDA-MB-231 and MDA-MB-435 lines. Shown are a representative in-gel fluorescence analysis (Left) and the ratio of upper to lower glycosylated forms, expressed as ratio-1 (Right).
A second membrane-associated serine hydrolase activity, protein KIAA1363, displayed a cellular expression profile similar to that of uPA, being strongly up-regulated in both invasive melanoma (MUM-2B) and breast carcinoma (MDA-MB-231) lines. Interestingly, this enzyme was found to exist in two discrete glycosylation states that were themselves differentially expressed among the cancer lines. For example, the ratios of the upper to lower glycosylated forms of KIAA1363 were inversely related in the MDA-MB-231 and MDA-MB-435 lines (Fig. 2C).
In contrast to the diverse patterns of enzyme activity observed in the secreted and membrane proteomes of the cancer cell lines, the activity profiles of the soluble proteomes appeared quite similar, with few enzymes exhibiting restricted patterns of distribution (see Fig. 5 and bar graphs).
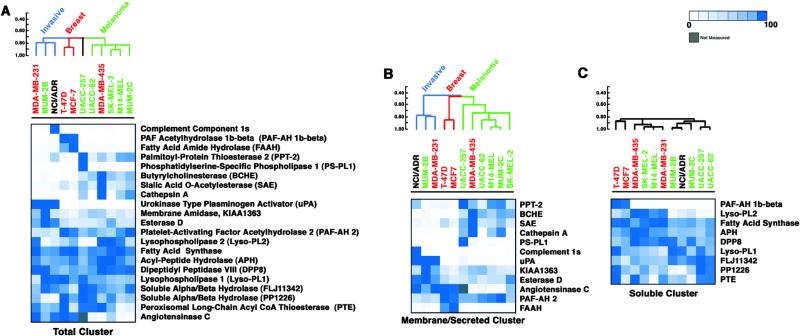
Classification of Human Cancer Cells Based on Serine Hydrolase Activity Profiles.
Serine hydrolase activity profiles of the secreted, membrane, and soluble proteomes for each cancer cell line were merged and the resulting data sets analyzed with a hierachical clustering algorithm and a psuedo-color visualization matrix (Fig. 3A) (17). Cancer cell lines were found to segregate into three major clusters that could be generally described as follows: a melanoma cluster (UACC-62, MDA-MB-435, SK-MEL-2, M14-MEL, MUM-2C), a breast carcinoma cluster (T-47D, MCF7), and an invasive cancer cluster (MDA-MB-231, MUM-2B, NCI/ADR). Notably, the presence of the ER(−) breast cancer line MDA-MB-435 in the melanoma cluster provides the first proteomic support for the recent transcriptome-based hypothesis that this cell line may represent a misclassified melanoma line (2).
Fig 3.
Cluster analysis of the serine hydrolase activity profiles of human cancer cell lines. (A–C) A hierarchical clustering algorithm was applied to the cell lines by average linkage clustering using the Pearson correlation coefficient as the measure of similarity (CLUSTER computer package). Bars to the left of the dendrograms represent similarity scores. Shown are the results of cluster analyses conducted on total (A), membrane/secreted (B), and soluble (C) serine hydrolase activity profiles. The intensity of blue color scales directly with the relative activity of each hydrolase among the cell lines (0–100%, where for each enzyme, 100% represents the cell line with the highest activity, and the rest of the cell lines are expressed as a percentage of this highest activity to normalize the data sets). Gray, not measured. Red, breast cancer lines. Green, melanoma cancer lines. Black, NCI/ADR is of unknown origin.
To understand the features of the enzyme activity profiles responsible for both origin-driven and phenotype-driven classifications, we performed additional cluster analyses in which enzyme activities from different subcellular fractions were separately examined. Strikingly, nearly all of the serine hydrolase activities that contributed to the observed classifications were found to reside in the secreted and membrane proteomes (Fig. 3B). In contrast, serine hydrolase activities from the soluble proteome mostly antagonized the observed classifications, possibly reflecting the presence of many broadly expressed “housekeeping” enzymes in this proteomic fraction (Fig. 3C). Several secreted and membrane-associated enzyme activities were expressed selectively by either breast carcinomas (e.g., FAAH, angiotensinase C) or melanomas (e.g., butyrylcholinesterase, cathepsin A, PPT2, SAE), providing a driving force for the origin-based clusters (Fig. 3B). However, the majority of these enzymes activities were strongly down-regulated in the invasive melanoma and breast carcinoma lines, MUM-2B and MDA-MB-231, respectively, which instead up-regulated a distinct set of secreted and membrane-associated enzyme activities (Fig. 3B) that included urokinase, a serine protease with a well-characterized association with tumorigenesis (20, 21), and the novel membrane-associated enzyme KIAA1363.
Characterization of a Membrane-Associated Serine Hydrolase KIAA1363 as a Marker for Cancer Cell Invasiveness.
The up-regulation of KIAA1363 in invasive cancer lines suggested that this enzyme may represent a new marker of tumor progression. Consistent with this notion, database searches revealed that the gene encoding KIAA1363 localizes to 3q26, a chromosomal region highly amplified in a variety of malignant cancers (27), including nearly 50% of advanced stage ovarian tumors (28). To further explore the relationship between KIAA1363 activity and cancer cell invasiveness, we determined the levels of activity of this enzyme across a panel of human ovarian cancer lines and correlated these values with measurements of invasiveness. We selected for analysis a group of four ovarian carcinoma lines that, despite forming a discrete cluster based on global gene expression profiles (2), were otherwise relatively uncharacterized in terms of their cell biological properties, including invasiveness. The strong positive correlation that we observed between the levels of active KIAA1363 and cell invasiveness in breast carcinoma (Fig. 4A) and melanoma (Fig. 4B) lines directly extended to the ovarian carcinoma lines (Fig. 4C). Specifically, the two ovarian carcinoma lines that displayed high invasiveness (OVCAR-5, SKOV-3) were found to exhibit 5- to 25-fold higher levels of active KIAA1363 than the two noninvasive ovarian carcinoma lines (OVCAR-3, OVCAR-4). Thus, activity levels of the novel membrane-associated enzyme KIAA1363 correlated with pronounced differences in the invasiveness of cell lines derived from three distinct types of cancer, even in a case where this cellular phenotype was not reflected at the level of global gene expression profiles.
Discussion
Here, we have shown that a proteome-wide analysis of variations in serine hydrolase activity permits the classification of human cancer lines into functional subtypes based on tissue of origin and state of invasiveness. Considering that most of the enzyme activities that contributed to the observed classifications resided in the secreted and membrane proteomes, we speculate that these cellular fractions may contain molecular markers especially representative of differences in cancer cell behavior. Furthermore, many of the secreted and membrane-associated enzymes were posttranslationally modified not only by glycosylation, but also by processing, as at least four enzymes, complement 1s protease, cathepsin A, urokinase, and SAE, were detected as single molecular species with masses 10–20 kDa lower than that predicted from their ORFs. The identification of SAE as an FP-reactive protein was particularly noteworthy given that this enzyme has not been classified in public databases as a serine hydrolase and shares no discernible sequence homology to any other functionally characterized protein. The reactivity of SAE with FP-rhodamine, coupled with the enzyme's ability to hydrolyze esterified sialic acid groups (29), suggests that the protein may be a member of the serine hydrolase superfamily. If confirmed, this finding would indicate that even for intensively studied enzyme classes like the serine hydrolases, unrecognized members may exist in the human proteome that resist classification by primary sequence alignment.
The surprising finding that highly invasive cancer cells displayed secreted/membrane serine hydrolase activity profiles nearly orthogonal to those displayed by their less aggressive counterparts suggests that invasive cancers may share proteomic signatures that are more reflective of their cellular phenotype than tissue of origin. These results lend support to the hypothesis that the advancement of cancers from a variety of origins may be accompanied by reversion to a common pluripotent embryonic-like state (30). Accordingly, enzyme activities, like KIAA1363, that are consistently up-regulated in invasive cancer lines derived from several different tumor types may represent new biomarkers and/or targets for the diagnosis and treatment of cancer. In general support of this notion, uPA, the enzyme activity that displayed the most similar profile to KIAA1363, is a well-established marker of tumor metastasis in vivo (20, 21) and a current target for multiple cancer drug development programs (31). Additionally, the localization of the KIAA1363 gene to 3q26, a chromosomal region highly amplified in a variety of cancers (27), including many advanced-staged ovarian tumors (28), supports a potential relationship between the overexpression of this enzyme and tumorigenesis. Nonetheless, specific antibodies and/or inhibitors of KIAA1363 will likely be required to validate whether this enzyme is associated with, or plays a role in, supporting cancer invasiveness in vivo.
In summary, these studies highlight that proteomic approaches, like ABPP, that can analyze technically challenging fractions of the proteome (e.g., membrane, glycosylated, and low abundance proteins) are capable of generating molecular profiles that accurately depict higher-order cellular properties. Moreover, because ABPP measures changes in enzyme activity rather than enzyme abundance, this method can detect in complex proteomes the functional outcome of a network of potential protease-protease inhibitor interactions, like the uPA-PAI-1 system, that may be critically important for cancer biology and other complex cellular processes. Finally, ABPP is a rapid and sensitive method for the comparative characterization of large numbers of proteomic samples, meaning that numerous cell types under a variety of experimental conditions can be analyzed in parallel, thereby accelerating the discovery of novel enzymes like KIAA1363, whose activities correlate with higher-order cellular properties. On this note, by using conventional two-dimensional gel electrophoresis proteomics technologies, a comparable analysis to the one described here would have required over 400 two-dimensional gels. Collectively, these findings suggest that activity-based chemical proteomics methods that target serine hydrolases, as well as other classes of enzymes (14, 32, 33), should prove of general value for the identification of active proteins associated with many cellular traits, including for example, proliferation, migration, differentiation, and drug resistance.
Supplementary Material
Acknowledgments
We thank A. Sharabi for technical assistance during the early stages of this project; K. Demarest for assistance with the statistical analysis of the data; M. Hendrix for MUM-2B and MUM-2C cells; Gary Siuzdak and Jane Wu for assistance with mass spectrometry analysis; Activx Biosciences for rhodamine-FPs; and T. Bartfai, P. Schimmel, V. Quaranta, and the entire Cravatt group for helpful discussions and critical reading of the manuscript. Supported by the National Cancer Institute of the National Institutes of Health (CA87660), the California Breast Cancer Research Program, Activx Biosciences, and the Skaggs Institute for Chemical Biology.
Abbreviations
ABPP, activity-based protein profiling
FP, fluorophosphonate
PAI, plasmid activator inhibitor
FAAH, fatty acid amide hydrolase
SAE, sialic acid 9-O-acetylesterase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Golub T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- 2.Ross D. T., Scherf, U., Eisen, M. B., Perou, C. M, Rees, C., Spellman, P., Iyer, V., Jeffrey, S. S., Van de Rijn, M., Waltham, M., et al. (2000) Nat. Genet. 24, 227-235. [DOI] [PubMed] [Google Scholar]
- 3.Bittner M., Meltzer, P., Chen, Y., Jiang, Y., Seftor, E., Hendrix, M., Radmacher, M., Simon, R., Yakhini, Z., Ben-Dor, A., et al. (2000) Nature (London) 406, 536-540. [DOI] [PubMed] [Google Scholar]
- 4.Perou C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature (London) 406, 747-752. [DOI] [PubMed] [Google Scholar]
- 5.Scherf U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236-244. [DOI] [PubMed] [Google Scholar]
- 6.Cravatt B. F. & Sorensen, E. J. (2000) Curr. Opin. Chem. Biol. 4, 663-668. [DOI] [PubMed] [Google Scholar]
- 7.Griffin T. J. & Aebersold, R. (2001) J. Biol. Chem. 276, 45497-45500. [DOI] [PubMed] [Google Scholar]
- 8.Corthals G. L., Wasinger, V. C., Hochstrasser, D. F. & Sanchez, J. C. (2000) Electrophoresis 21, 1104-1115. [DOI] [PubMed] [Google Scholar]
- 9.Santoni V., Molloy, M. & Rabilloud, T. (2000) Electrophoresis 21, 1054-1070. [DOI] [PubMed] [Google Scholar]
- 10.Gygi S. P., Corthals, G. L., Zhang, Y., Rochon, Y. & Aebersold, R. (2000) Proc. Natl. Acad. Sci. USA 97, 9390-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson N. L. & Anderson, N. G. (1998) Electrophoresis 19, 1853-1861. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Patricelli, M. P. & Cravatt, B. F. (1999) Proc. Natl. Acad. Sci. USA 96, 14694-14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidd D., Liu, Y. & Cravatt, B. F. (2001) Biochemistry 40, 4005-4015. [DOI] [PubMed] [Google Scholar]
- 14.Adam G. C., Cravatt, B. F. & Sorensen, E. J. (2001) Chem. Biol. 8, 81-95. [DOI] [PubMed] [Google Scholar]
- 15.Patricelli M. P., Giang, D. K., Stamp, L. M. & Burbaum, J. J. (2001) Proteomics 1, 1067-1071. [DOI] [PubMed] [Google Scholar]
- 16.Cravatt B. F., Giang, D. K., Mayfield, S. P., Boger, D. L., Lerner, R. A. & Gilula, N. B. (1996) Nature (London) 384, 83-87. [DOI] [PubMed] [Google Scholar]
- 17.Eisen M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature (London) 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 19.Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 20.Konecny G., Untch, M., Arboleda, J., Wilson, C., Kahlert, S., Boettcher, B., Felber, M., Beryt, M., Lude, S., Hepp, H., et al. (2001) Clin. Cancer Res. 7, 2448-2457. [PubMed] [Google Scholar]
- 21.Bouchet C., Spyratos, F., Martin, P. M., Hacene, K., Gentile, A. & Oglobine, J. (1994) Br. J. Cancer 69, 398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou Z., Anisowicz, A., Hendrix, M. J., Thor, A., Neveu, M., Sheng, S., Rafidi, K., Seftor, E. & Sager, R. (1994) Science 263, 526-529. [DOI] [PubMed] [Google Scholar]
- 23.Xiao G., Liu, Y. E., Gentz, R., Sang, Q. A., Ni, J., Goldberg, I. D. & Shi, Y. E. (1999) Proc. Natl. Acad. Sci. USA 96, 3700-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeClerck Y. A., Imren, S., Montgomery, A. M., Mueller, B. M., Reisfel, R. A. & Laug, W. E. (1997) Adv. Exp. Med. Biol. 425, 89-97. [DOI] [PubMed] [Google Scholar]
- 25.Silverman G. A., Bird, P. I., Carrell, R. W., Church, F. C., Coughlin, P. B., Gettins, P. G., Irving, J. A., Lomas, D. A., Luke, C. J., Moyer, R. W., et al. (1999) J. Biol. Chem. 276, 33293-33296. [DOI] [PubMed] [Google Scholar]
- 26.Patricelli M. P., Lashuel, H. A., Giang, D. K., Kelly, J. W. & Cravatt, B. F. (1998) Biochemistry 37, 15177-15187. [DOI] [PubMed] [Google Scholar]
- 27.Rooney P. H., Murray, G. I., Stevenson, D. A., Haites, N. E., Cassidy, J. & McLeod, H. L. (1999) Br. J. Cancer 80, 862-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonoda G., Palazzo, J., du Manoir, S., Godwin, A. K., Feder, M., Yakushiji, M. & Testa, J. R. (1997) Genes Chromosomes Cancer 20, 320-328. [PubMed] [Google Scholar]
- 29.Guimaraes M. J., Bazan, J. F., Castagnola, J., Diaz, S., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Varki, A. & Zlotnik, A. (1996) J. Biol. Chem. 271, 13697-13705. [DOI] [PubMed] [Google Scholar]
- 30.Maniotis A. J., Folberg, R., Hess, A., Seftor, E. A., Gardner, L. M., Pe'er, J., Trent, J. M., Meltzer, P. S. & Hendrix, M. J. (1999) Am. J. Pathol. 155, 739-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muehlenweg B., Sperl, S., Magdolen, V., Schmitt, M. & Harbeck, N. (2001) Expert Opin. Biol. Ther. 1, 683-691. [DOI] [PubMed] [Google Scholar]
- 32.Greenbaum D., Medzihradszky, K. F., Burlingame, A. & Bogyo, M. (2000) Chem. Biol. 7, 569-581. [DOI] [PubMed] [Google Scholar]
- 33.Adam, G. C., Sorensen, E. J. & Cravatt, B. F. (2002) Nat. Biotechnol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.