Abstract
The heterodimeric Elongin BC complex has been shown to interact in vitro and in cells with a conserved BC-box motif found in an increasing number of proteins including RNA polymerase II elongation factor Elongin A, suppressor of cytokine signaling (SOCS)-box proteins, and the von Hippel–Lindau tumor suppressor protein. Recently, the Elongin BC complex was found to function as an adaptor that links these BC-box proteins to a module composed of Cullin family members Cul2 or Cul5 and RING-H2 finger protein Rbx1 to reconstitute a family of E3 ubiquitin ligases that activate ubiquitylation by the E2 ubiquitin-conjugating enzyme Ubc5. As part of our effort to understand the functions of Elongin BC-based ubiquitin ligases, we exploited a modified yeast two-hybrid screen to identify a mammalian BC-box protein similar in sequence to Saccharomyces cerevisiae Mediator subunit Med8p. In this report we demonstrate (i) that mammalian MED8 is a subunit of the mammalian Mediator complex and (ii) that MED8 can assemble with Elongins B and C, Cul2, and Rbx1 to reconstitute a ubiquitin ligase. Taken together, our findings are consistent with the model that MED8 could function to recruit ubiquitin ligase activity directly to the RNA polymerase II transcriptional machinery.
Eukaryotic messenger RNA synthesis is regulated by DNA-binding transcriptional activators and repressors that communicate with RNA polymerase II and the general initiation factors through the action of a multiprotein coactivator referred to as Mediator. Mediator was first identified in Saccharomyces cerevisiae and was shown to be required for transcriptional activation in vitro and in cells. Biochemical studies revealed that the yeast Mediator is a multiprotein complex composed of more than 20 polypeptides, including the products of the Srb2, Srb4, Srb5, Srb6, Srb7, Srb8, Srb9, Srb10/Ssn3, Srb11/Ssn8, Med1, Med2, Med4, Med6, Med7, Med8, Med11, Rox3, Cse2, Nut1, Nut2/MED10, Pgd1/Hrs1, Gal11, Rgr1, and Sin4 genes (1). Subsequently, multiprotein coactivator complexes with structural and functional similarities to the yeast Mediator have been identified in mammalian cells. To date, characterization of these mammalian Mediator complexes has led to the identification of homologs of yeast Mediator subunits Rgr1p, Med6p, Med7p, Srb4p, Srb7p, Srb10p, Srb11p, and Nut2, as well as additional subunits that seem to be unique to mammalian complexes (1–3).
The Elongin BC complex is a heterodimer composed of the 112-aa Elongin C protein and the 118-aa, ubiquitin-like Elongin B protein. The Elongin BC complex was initially identified as a positive regulator of RNA polymerase II elongation factor Elongin A, which is one of several transcription factors capable of stimulating the rate of elongation by RNA polymerase II in vitro (4, 5). The Elongin BC complex was subsequently found to be a component of the multiprotein von Hippel–Lindau (VHL) tumor suppressor and suppressor of cytokine signaling (SOCS)-box protein complexes (5–8). Interaction of Elongin BC with the Elongin A, SOCS-box, and VHL proteins depends on binding of Elongin C to an ≈10-aa degenerate sequence motif, referred to as the BC-box, which has the consensus sequence [(A,P,S,T)LxxxCxxx(A,I,L,V)]. Analysis of the crystal structure of the VHL–Elongin BC complex (9) revealed that interaction of Elongin BC with the BC-box is governed by interaction of the highly conserved leucine at position 2 in the N terminus of the BC-box motif with a hydrophobic pocket created by residues in the C-terminal half of Elongin C. Elongin B binds to a short N-terminal Elongin C region and does not appear to interact directly with the BC-box.
A role for the Elongin BC complex in ubiquitylation was brought to light by the discovery that the VHL tumor suppressor complex is an E3 ubiquitin ligase that targets the α subunits of the hypoxia-inducible transcription factors HIF1 and HIF2 for ubiquitylation and degradation by the proteosome (10–14). As a component of the VHL complex, the VHL protein functions as the substrate recruitment subunit, while Elongin BC functions as an adaptor that links the VHL protein to a Cul2/Rbx1 module that functions as a potent activator of HIF1α ubiquitylation by the E2 ubiquitin conjugating enzyme Ubc5. Recently, the additional BC-box proteins Elongin A, SOCS1, and WSB1 were found to assemble with Elongin BC and a Cul5/Rbx1 module to reconstitute multisubunit complexes with ubiquitin ligase activity, raising the possibility that the Elongin BC complex could function as an integral component of a larger family of ubiquitin ligases by linking other BC-box substrate recruitment subunits to Cullin/Rbx1 modules (15).
As part of our effort to address this possibility, we have been attempting to identify additional BC-box proteins and to determine whether they might also function as components of ubiquitin ligases. In this report, we have exploited a modified yeast two-hybrid screen (16) to identify a mammalian BC-box protein similar in sequence to S. cerevisiae Mediator subunit Med8p. In this report, we demonstrate (i) that mMED8 is a component of mammalian Mediator and (ii) that it can assemble into a complex that is composed of Elongins B and C, Cul2, and Rbx1 and is associated with ubiquitin ligase activity. In addition, we present evidence that Elongins B and C, Cul2, and Rbx1 can be purified from rat liver nuclear extracts as components of a high molecular mass complex or complexes that cofractionate with mMED8 and other Mediator subunits.
Materials and Methods
Yeast Two-Hybrid Screening.
pAS2-Elongin C, pAS2-Elongin B, and yeast strains GC1945, GC1945(EloC), and GC1945(EloB) have been described (16). For two-hybrid screening, yeast strain GC1945(EloC) was transformed with a human B cell cDNA pACT library from S. J. Elledge (Baylor University, Waco, TX). The transformation mix was distributed on SC plates containing 2.5 mM 3-aminotriazole (Sigma), but lacking Trp, Leu, and His. A total of 1 × 106 transformants was assayed for Trp+, Leu+, His+ growth, and β-galactosidase activity. Positive colonies were reassayed for growth on His− media and for β-galactosidase activity and then grown in Leu–Trp+ media to lose pAS2-EloB plasmids. pACT plasmids were recovered, and inserts were sequenced.
Expression of Recombinant Proteins in Escherichia coli.
Human Ubc5a with an N-terminal 6-histidine tag and a C-terminal FLAG tag, S. cerevisiae ubiquitin-activating enzyme Uba1 with an N-terminal Myc tag and a C-terminal 6-histidine tag, and mammalian glutathione S-transferase (GST)-ubiquitin were expressed in E. coli strain BL21(DE3) and purified by Ni2+-agarose or glutathione-Sepharose chromatography (12, 17). After dialysis against 40 mM Hepes⋅NaOH, pH 7.9/60 mM potassium acetate/2 mM DTT/1 mM MgCl2/0.5 mM EDTA/10% (vol/vol) glycerol, proteins were stored at −80°C.
Expression of Recombinant Proteins in Sf21 Insect Cells.
Recombinant baculoviruses encoding C-terminally HPC4-tagged Elongin B (Elongin B-HPC4), C-terminally herpes simplex virus (HSV)-tagged Elongin C (Elongin C-HSV), N-terminally Myc-tagged Rbx1 (Myc-Rbx1), and N-terminally hemagglutinin (HA)-tagged Cul2 (HA-Cul2) were described (7, 12, 17). N-terminally T7-tagged human MED8 (T7-MED8) was subcloned into pBacPAK9 (CLONTECH), and recombinant baculoviruses were generated with the BacPAK baculovirus expression system (CLONTECH). Sf21 cells were cultured at 27°C in Sf-900 II SFM (GIBCO, Invitrogen) with 5% FCS, penicillin (100 units/ml), and streptomycin (100 μg/ml), and infected with the recombinant baculoviruses indicated in the figures. Seventy-two hours after infection, cells were collected and lysed in ice-cold buffer containing 40 mM Hepes⋅NaOH (pH 7.9), 500 mM NaCl, 1 mM DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin and centrifuged at 10,000 × g for 20 min at 4°C.
Expression of Recombinant Proteins in Mammalian Cells.
pcDNA3.1 vector encoding N-terminally Myc-tagged Elongin B (Myc-Elongin B) and pCI-neo vector encoding N-terminally HSV-tagged Elongin C (HSV-Elongin C) have been described (18). N-terminally T7-tagged mMED8 (T7-MED8) was amplified from human expressed sequence tag R60110 (Research Genetics, Huntsville, AL) by PCR with primers containing NheI and BamHI restriction sites and subcloned into pcDNA3.1 vector (Invitrogen). N-terminally T7-tagged mMED8[L143P,C147F] (T7-MED8[L143P,C147F]) was obtained by PCR site-directed mutagenesis and subcloned into pcDNA3.1 vector. 293T mammalian cells were maintained in DMEM (GIBCO/BRL) containing 10% FCS. Cells were transfected with the plasmids indicated in the figures by using Lipofectamine (GIBCO/BRL). After 48 h, cells were collected and lysed in ice-cold buffer containing 40 mM Hepes⋅NaOH (pH 7.9), 500 mM NaCl, 1 mM DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin and centrifuged at 10,000 × g for 20 min at 4°C.
Immunoprecipitation and Immunoblotting.
Anti-Myc (9E10) and anti-HA (12CA5) monoclonal antibodies (Roche Molecular Biochemicals), anti-T7 monoclonal antibody (Novagen), anti-HSV monoclonal antibody (Novagen), anti-GST (4C10) monoclonal antibody (Covance, Princeton, NJ), anti-HPC4 monoclonal antibody (19), anti-Cul2 antibody (Zymed), anti-Elongin B antibody (20), and affinity-purified anti-mMED8 antibody (Cocalico Biologicals, Reamstown, PA) were used in immunoprecipitations and immunoblotting. Immunoprecipitations from lysates of transfected 293T cells or baculovirus-infected Sf21 cells were performed for 1 h at 4°C with the antibodies indicated in the figures. Protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) were then added to immunoprecipitations and incubated for an additional hour at 4°C. Beads were washed twice in buffer containing 40 mM Hepes⋅NaOH (pH 7.9), 500 mM NaCl, 1 mM DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, and once in buffer containing 40 mM Hepes⋅NaOH (pH 7.9), 150 mM NaCl, 1 mM DTT, and 10% (vol/vol) glycerol. Immunoprecipitated proteins were fractionated by SDS-13.5% PAGE, transferred to Hybond membranes (Amersham Pharmacia), and analyzed by immunoblotting with the antibodies indicated in the figures. Immunoblots were visualized with either SuperSignal West Pico (Pierce) or SuperSignal West Dura chemiluminescent reagent (Pierce) according to the manufacturer's instructions.
Ubiquitylation Assays.
Reaction mixtures (10 μl) contained 40 mM Hepes⋅NaOH (pH 7.9), 60 mM potassium acetate, 2 mM DTT, 1 mM MgCl2, 0.5 mM EDTA (pH 7.9), 10% (vol/vol) glycerol, 1.5 mM ATP, ≈50 ng of Uba1, ≈100 ng of Ubc5a, ≈150 ng of GST-ubiquitin, and the protein fractions indicated in the figures. Reaction mixtures were incubated for 40 min at 30°C. Reaction products were fractionated by SDS/7.5% PAGE and analyzed by immunoblotting with anti-GST antibodies.
Purification of a mMED8-Containing Complex from Rat Liver.
Nuclear extract prepared from the livers of ≈100 male Sprague–Dawley rats (200–300 g) was fractionated by 0–38% NH2(SO4)2 precipitation followed by chromatography on consecutive DEAE-cellulose, phosphocellulose, and TSK phenyl 5-PW columns as described (21). mMED8-containing fractions were identified by immunoblotting by using affinity-purified antibodies against a synthetic peptide corresponding to mMED8 amino acids 247–268. The peak mMED8-containing fractions from TSK phenyl 5-PW (Tosoh, Tokyo) were dialyzed against buffer C with 100 mM KCl. After centrifugation for 20 min at 60,000 × g, the resulting supernatant was applied to a 7.5 × 75 mm TSK DEAE 5-PW column (Tosoh) equilibrated in buffer C [40 mM Tris⋅HCl (pH 7.9)/0.5 mM EDTA/1 mM DTT/10% (vol/vol) glycerol] containing 50 mM KCl. The column was eluted at 1 ml/min with a 30-ml linear gradient from 50 to 500 mM KCl in buffer C. One-milliliter fractions were collected, and those containing mMED8 were pooled, concentrated by precipitation with (NH4)2SO4 (0.436 g/ml), and resuspended in 200 μl of buffer G [40 mM Hepes⋅NaOH (pH 7.0)/0.5 mM EDTA/1 mM DTT/10% (vol/vol) glycerol], and applied at 0.5 ml/min to a 7.5 mm × 60 cm TSK-G4000SW gel filtration column (Tosoh) equilibrated in buffer G containing 400 mM KCl. Fractions (0.5-ml) were collected. Fractions containing mMED8 were pooled and dialyzed against buffer C with 100 mM KCl, and applied to a Mono Q PC 1.6/5 column (Amersham Pharmacia) equilibrated in buffer C containing 100 mM KCl. The column was eluted at 50 μl/min with a 1-ml linear gradient from 100 to 500 mM KCl in buffer C. Fifty-microliter fractions were collected.
Results
Identification of a Mammalian MED8 Homolog As an Elongin BC-Interacting Protein.
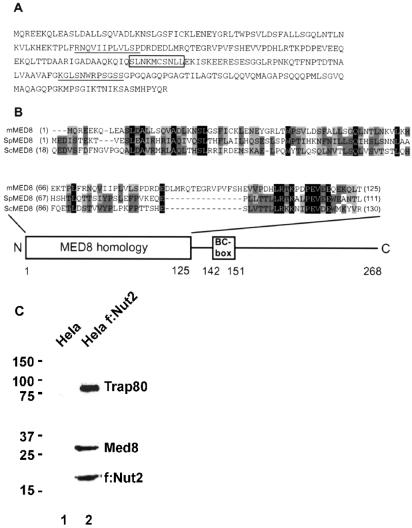
In a previous study, interaction of the Elongin BC-box-containing VHL tumor suppressor protein with the Elongin BC complex was investigated by using a modified yeast two-hybrid system (16). In this system, interaction of the VHL protein with Elongin B was detected only in a yeast strain constitutively expressing Elongin C, and interaction of the VHL protein with Elongin C was detected only in a yeast strain constitutively expressing Elongin B. As part of our effort to identify Elongin BC-box proteins, we exploited this modified yeast two-hybrid system to screen a GAL4-activation domain-B cell cDNA library with a GAL4-DNA-binding domain-Elongin B fusion protein in a yeast strain constitutively expressing mammalian Elongin C from a chromosomally integrated copy of the gene. One cDNA identified in this two-hybrid screen was found to encode a 268-aa protein (Fig. 1A) that shares significant sequence similarity throughout its N-terminal ≈120 aa with the N termini of Schizosaccharomyces pombe (22) and S. cerevisiae (23) Mediator subunits Med8p (Fig. 1B). Suggesting that mMED8 is a subunit of the mammalian Mediator complex, inspection of the amino acid sequence of mMED8 revealed that it corresponds to the p32 subunit of the human ARC Mediator complex isolated by Tjian and coworkers (2). In addition, Gustafsson and coworkers independently identified mMED8 through database searches as a homolog of Sch. pombe MED8 present in their purified Sch. pombe Mediator preparations (24). To confirm the association of mMED8 with Mediator, we took advantage of a HeLa cell line that stably expresses FLAG-tagged Mediator subunit NUT2 and that has been used for immunoaffinity purification of the transcriptionally active mammalian Mediator complex, thyroid hormone receptor-associated protein/SRB-Med-containing cofactor (TRAP/SMCC) (25). Western blotting of immunoaffinity-purified HeLa cell TRAP/SMCC with affinity-purified anti-mMED8 antibodies raised against a peptide corresponding to mMED8 C-terminal residues 247–268 revealed that mMED8 is specifically coprecipitated with the FLAG-tagged Mediator subunit Nut2, Trap80, and other Mediator subunits (Fig. 1C and data not shown), indicating that mammalian MED8, like S. cerevisiae Med8p, is a Mediator subunit.
Fig 1.
Sequence of mammalian MED8 and homology to Sch. pombe and S. cerevisiae MED8 proteins. (A) Amino acid sequence of human mMED8. The Elongin BC-box is boxed. Peptide sequences derived from ARCp32 by Tjian and coworkers (2) are underlined. (B) Alignment of the N termini of human, Sch. pombe, and S. cerevisiae MED8 proteins, generated by using the ALIGNX program from Vector NTI. (C) Anti-FLAG immunoprecipitations were performed essentially as described (31) with nuclear extracts prepared according to Dignam et al. (36) from ≈2.5 × 109 HeLa cells (lane 1) or HeLa M10 cells (25) stably expressing FLAG-tagged NUT2 (lane 2). Nuclear extracts were adjusted to 300 mM KCl and 0.5% Triton X-100 and incubated with M2-agarose beads (Sigma). Beads were washed with buffer containing 50 mM Hepes⋅NaOH (pH 7.9), 5 mM MgCl2, 20% glycerol, 300 mM KCl, and 0.1% Triton X-100, then eluted with the same buffer containing 100 mM KCl, 0.05% Triton X-100, and 0.2 mg FLAG peptide (Sigma) per milliliter. Precipitated proteins were analyzed by blotting with anti-FLAG, anti-mMed8, or anti-Trap80 antibodies. The positions and relative molecular masses in kDa of protein size markers are indicated on the left (Prestained Precision Protein Standards, Bio-Rad).
mMED8 Binds to the Elongin BC Complex Through a Canonical BC-Box.
Inspection of the mMED8 amino acid sequence revealed that it contains a potential Elongin BC-box motif, SLNKMCSNLL, located between amino acids 142 and 151 (Fig. 1). This potential BC-box motif was present in each of three different mMED8 clones identified in the yeast two-hybrid screen (data not shown). In addition, characterization of the interaction of mMED8 with Elongins B and C in the modified yeast two-hybrid system revealed that, like interaction of the BC-box-containing VHL protein with Elongins B and C (16), detectable interaction of mMED8 with Elongin B occurs only in a yeast strain expressing Elongin C, and detectable interaction of mMED8 with Elongin C occurs only in a yeast strain expressing Elongin B (Table 1).
Table 1.
Characterization of the interactions of mMED8 and Elongins B and C in the modified yeast two-hybrid system
| Strain | DBD | AD | −HIS | βGal |
|---|---|---|---|---|
| GC1945(wt) | MED8 | Empty | − | − |
| GC1945(wt) | MED8 | EloC | − | − |
| GC1945(wt) | MED8 | EloB | − | − |
| GC1945(EloB) | MED8 | Empty | − | − |
| GC1945(EloB) | MED8 | EloC | + | + |
| GC1945(EloB) | MED8 | EloB | − | − |
| GC1945(EloC) | EloB | MED8 | + | + |
| GC1945(EloC) | MED8 | Empty | − | − |
| GC1945(EloC) | MED8 | EloC | − | − |
| GC1945(EloC) | MED8 | EloB | + | + |
Yeast strains GC1945, GC1945(EloC), which constitutively expresses Elongin C, and GC1945(EloB), which constitutively expresses Elongin B, were transformed with the indicated plasmids (DBD, DNA-binding domain plasmid pAS2-1, and AD, activation domain plasmid pACT2). β-Galactosidase activity (βGal) and growth of yeast in the absence of Trp, Leu, and His (−HIS) were measured as described under Materials and Methods. MED8, mMED8; EloB, Elongin B; EloC, Elongin C.
To confirm that the mMED8 protein is capable of interacting with the Elongin BC complex in cells and to determine whether mMED8 interacts with Elongin BC through the potential BC-box motif located between amino acids 142 and 151, human 293T cells were cotransfected with vectors encoding epitope-tagged Elongins B and C and either T7-MED8 or a T7-MED8 double-point mutant T7-MED8[L143P;C147F], which contains BC-box mutations corresponding to those shown to disrupt binding of Elongin A, VHL, and SOCS1 to the Elongin BC complex (7, 26, 27). Consistent with results of yeast two-hybrid experiments, the Elongin BC complex could be specifically coimmunoprecipitated with wild-type mMED8 from 293T cell lysates (Fig. 2). In addition, interaction of mMED8 with Elongins B and C strongly depended on the presence of an intact mMED8 BC-box, because Elongins B and C were not efficiently coimmunoprecipitated with the MED8[L143P;C147F] mutant from 293T cell lysates (Fig. 2).
Fig 2.
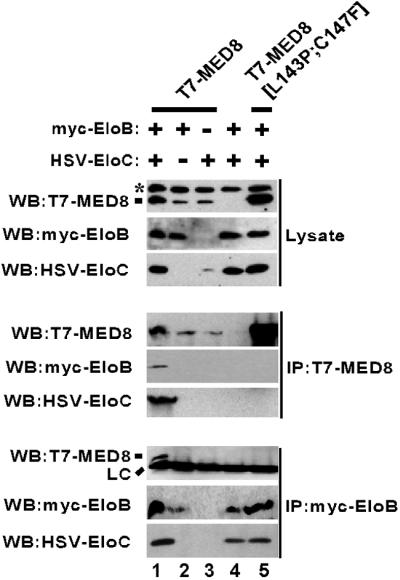
mMED8 interacts with the Elongin BC complex through a canonical BC-box. 293T cells were transfected with the indicated expression vectors, and immunoprecipitations were performed with the indicated antibodies. Total cell lysates were fractionated on SDS/polyacrylamide gels and analyzed by immunoblotting. WB, immunoblot; IP, immunoprecipitate; LC, light chain. The asterisk denotes a nonspecific background band observed in anti-mMED8 immunoblots of 293T cell lysates.
The mMED8–Elongin BC Complex Interacts with Cul2 and Rbx1.
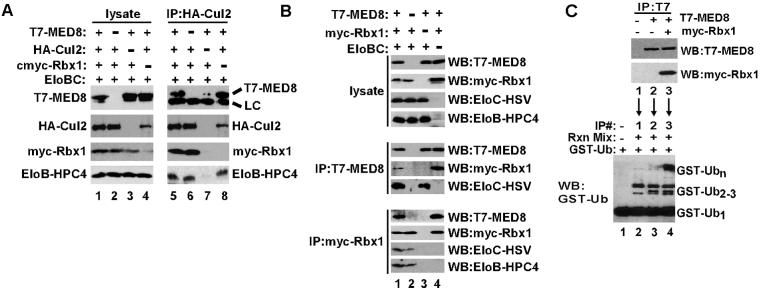
VHL and other Elongin BC-box proteins can assemble with the Elongin BC complex and a heterodimeric module composed of a member of the Cullin protein family and RING-H2 finger protein Rbx1 to form multiprotein ubiquitin ligase complexes, as shown (12, 15, 17, 28, 29). To determine whether the mMED8–Elongin BC complex can assemble into similar complexes, Sf21 insect cells were infected with combinations of baculoviruses encoding T7-MED8, Elongin B-HPC4, Elongin C-HSV, Myc-Rbx1, and HA-tagged members of the Cullin family. When immunoprecipitations were performed with anti-HA antibodies, mMED8, Elongins B and C, and Rbx1 could be specifically coimmunoprecipitated with HA-Cul2 (Fig. 3A), but not with HA-Cul1, HA-Cul3, HA-Cul4, or HA-Cul5 (data not shown). In addition, like the VHL–Elongin BC complex, the mMED8–Elongin BC complex interacts with Rbx1, because Rbx1 could be specifically coimmunoprecipitated with mMED8 and Elongins B and C (Fig. 3B). Thus, like the VHL–Elongin BC complex, the mMED8–Elongin BC complex can assemble into complexes containing Cul2 and Rbx1.
Fig 3.
The mMED8–Elongin BC complex assembles with a Cul2/Rbx1 module. (A and B) Sf21 insect cells were coinfected with the indicated baculoviruses. Immunoprecipitations were performed with the indicated antibodies. Total cell lysates were fractionated by SDS/PAGE and analyzed by immunoblotting. (C) Sf21 cells were coinfected with the indicated baculoviruses. Anti-T7 immunoprecipitations and ubiquitylation assays were performed as described under Materials and Methods. Rxn Mix, reaction mix with Uba1, Ubc5a, GST-ubiquitin, and ATP; GST-Ub, GST-ubiquitin; LC, light chain; WB, immunoblot; IP, immunoprecipitate.
To determine whether mMED8 can assemble into a complex possessing ubiquitin ligase activity, Sf21 insect cells were infected with baculoviruses encoding either T7-MED8 or T7-MED8 and Myc-Rbx1. Anti-T7 immunoprecipitates from Sf21 cell lysates were then assayed for their ability to activate formation of polyubiquitin conjugates by the E2 ubiquitin-conjugating enzyme Ubc5 in the presence of the E1 ubiquitin-activating enzyme Uba1, GST-ubiquitin, and ATP. As shown in Fig. 3C, anti-T7 immunoprecipitates from lysates of insect cells expressing both mMED8 and Rbx1 strongly stimulated formation of polyubiquitin conjugates by Ubc5; in contrast, anti-T7 immunoprecipitates from lysates of insect cells not expressing mMED8 or expressing mMED8 alone supported only formation of mono- and diubiquitin conjugates. In light of observations that the ubiquitin ligase activity of the VHL complex depends strongly on the presence of Cul2 and Elongins B and C (12), it is noteworthy that mMED8 complexes from cells that did not overexpress these proteins could still activate Ubc5a. It is likely that Rbx1 is linked to mMED8 through endogenous insect cell Cul2 and Elongins B and C; however, we cannot rule out the possibility that Rbx1 is bound directly to mMED8 in these assays. In addition, these data show only that mMED8 can assemble into complexes that activate Ubc5-dependent ubiquitin conjugation and not that it is essential for this function. Indeed, in assays similar to these, Rbx1 and the RING domains of other ubiquitin ligases have been shown to be able to activate E2s to form nonspecific multiubiquitin chains in the absence of substrate recognition subunits or domains.
Purification of a High Molecular Mass mMED8-Containing Complex from Rat Liver.
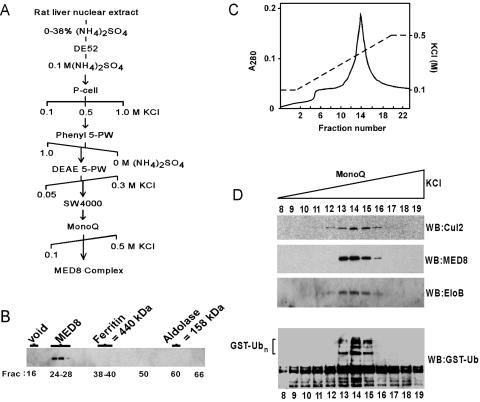
To address the question whether endogenous mMED8 is associated with Elongins B and C, Cul2, and Rbx1, we purified mMED8 from rat liver. Purification of mMED8 was monitored by immunoblotting of aliquots of column fractions by using affinity-purified anti-mMED8 antibodies raised against a peptide corresponding to mMED8 C-terminal residues 247–268. As summarized in Fig. 4A, all of the detectable mMED8 in rat liver homogenates was present in the nuclear extract and could be purified by 0–38% (NH4)2SO4 fractionation followed by chromatography on consecutive DEAE-cellulose, phosphocellulose, TSK phenyl 5-PW, TSK DEAE 5-PW, TSK SW4000 gel filtration, and Mono Q columns. The mMED8-containing species exhibited an apparent native molecular mass greater than 1,000 kDa by TSK SW4000 gel filtration (Fig. 4B). Analysis of aliquots of fractions from the final Mono Q column revealed that mMED8 eluted in a discrete protein peak (Fig. 4C), copurifying closely with Elongin B and Cul2 (Fig. 4D) and with Rbx1 (data not shown). Assay of aliquots of fractions from Mono Q for their abilities to activate formation of polyubiquitin conjugates by the E2 ubiquitin-conjugating enzyme Ubc5 in the presence of the E1 ubiquitin-activating enzyme Uba1, GST-ubiquitin, and ATP revealed that mMED8 copurified closely with ubiquitin ligase activity. To confirm association of the endogenous rat liver mMED8 with Mediator subunits, aliquots of peak fractions of the Mono Q column were fractionated by SDS/4–15% polyacrylamide gradient gel electrophoresis, and proteins in gel slices were subjected to in-gel reduction, S-carboxyamidomethylation, and tryptic digestion. Peptide sequences were determined in a single run by microcapillary reversed-phase chromatography coupled to the electrospray ionization source of a quadrupole ion trap mass spectrometer (Finnigan LCQ, San Jose, CA). Identification of proteins present in mMED8-containing fractions was facilitated by the algorithm SEQUEST and by programs developed in the Harvard Microchemistry and Proteomics Analysis Facility (30). Supporting the identification of mMED8 as an integral component of mammalian Mediator, mass spectroscopy fingerprinting identified the mammalian Mediator subunits (1, 3) TRAP230, TRAP220, SRB4/TRAP80, SRB10/CDK8, SRB11/cyclin C, MED6, MED7, TRAP37, TRFP, p28b, NUT2/MED10, SOH1, the TRAP25 (31), and mammalian homologs of Drosophila MED21, MED23, and MED24 (32) as components of the endogenous mMED8-containing fraction from rat liver.
Fig 4.
Purification of a mMED8-containing complex from rat liver. (A) Outline of purification of the mMED8-containing complex. DE52, DEAE-cellulose; P-cell, phosphocellulose. (B) TSK SW4000 gel filtration of the mMED8-containing complex. Aliquots of column fractions were analyzed by SDS/PAGE, and the mMED8 protein was visualized by immunoblotting. (C) A280 profile of the Mono Q column. (D) Cochromatography of mMED8 with components of the mMED8 ubiquitin ligase and with ubiquitin ligase activity. Aliquots of column fractions from Mono Q were analyzed by SDS/PAGE; mMED8, Cul2, and Elongin B proteins were visualized by immunoblotting.
Discussion
Med8p was originally identified in S. cerevisiae as a subunit of the transcriptional coactivator complex called Mediator (reviewed in ref. 1). Little is known about the role of Med8p within the Mediator complex; however, genetic analyses indicate that the MED8 gene is essential for viability of both S. cerevisiae and Sch. pombe (22, 23). In this report, we identify a mammalian homolog of S. cerevisiae Mediator subunit MED8 and demonstrate (i) that it is a subunit of the mammalian Mediator complex, (ii) that it is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute an E3 ubiquitin ligase, and (iii) that endogenous mMED8 can be purified from rat liver nuclear extracts as a component of a high molecular mass complex or complexes that cofractionate with other Mediator subunits and with ubiquitin ligase activity and ubiquitin ligase subunits.
The functional significance of the mMED8-associated ubiquitin ligase activity in the context of Mediator remains to be established. Database searching reveals that the BC-box is conserved throughout the chordata; however, no easily recognizable binding site for Elongin C exists in the MED8 orthologs from S. cerevisiae, Caenorhabditis elegans, or insects (data not shown), suggesting that MED8 may have functions in Mediator that are independent of its ability to recruit ubiquitin ligase subunits. Indeed, Cul2, Rbx1, and Elongins B and C do not seem to be associated with all Mediator complexes, because results of Western blot analyses of the most highly purified mMED8 fractions from rat liver are consistent with the notion that Cul2, Elongins B and C, and Rbx1 are present in these fractions in substoichiometric amounts relative to mMED8 (data not shown). In addition, we have been unable to detect the ubiquitin ligase subunits in the TRAP/SMCC Mediator complexes purified through FLAG-tagged Nut2 subunit, suggesting either (i) that the FLAG-tag on Nut2 is inaccessible when the ubiquitin ligase subunits are bound or blocks association of ubiquitin ligase subunits with Mediator, (ii) that the mMED8-containing Mediator and E3 ubiquitin ligase complexes are distinct, >1 MDa complexes that copurify on Mono Q, or (iii) that the interaction of Mediator with ubiquitin ligase subunits is negatively regulated in HeLa cells under the growth conditions used.
Although we cannot rule out the possibility that mMED8-containing Mediator and E3 ubiquitin ligase are present in separate complexes, it would not be surprising if the ubiqutin ligase subunits were present in only a subfraction of MED8-containing complexes. First, evidence suggests that recruitment of Cullin/Rbx1 modules to the related SCF E3 ubiquitin ligase complexes results in the rapid ubiquitylation and turnover of their substrate-recognition subunits (33–35). Hence, many fully assembled SCF ubiquitin ligase complexes are highly unstable in cells. Second, although targets of mMED8 ubiquitin ligase have not yet been identified, it is possible that binding of ubiquitin ligase subunits to mMED8 could result in ubiquitylation and degradation of Mediator subunits. Consequently, it is possible that an mMED8 complex containing the ubiquitin ligase subunits would be present only transiently in cells. In any case, our findings raise the possibility that mammalian mMED8 might regulate transcription at least in part by recruiting ubiquitin ligase activity directly to transcription initiation complexes, perhaps in response to specific signals, and that it could alter the function of RNA polymerase II, transcription factors, or the Mediator complex itself through ubiquitylation of specific subunits. Experiments to investigate this possibility and to identify potential targets for ubiquitylation by the mMED8 ubiquitin ligase among the more than 50 proteins present in the RNA polymerase II initiation complex are underway.
Acknowledgments
We thank A. Dvir, D. Haque, and M. Conrad for assistance with parts of this research and S. J. Elledge for the human B cell two-hybrid library. This work was supported in part by National Institutes of Health Grant R37 GM41628 (to R.C.C.).
Abbreviations
GST, glutathione S-transferase
VHL, von Hippel–Lindau
HSV, herpes simplex virus
HA, hemagglutinin
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF521562).
References
- 1.Myers L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729-749. [DOI] [PubMed] [Google Scholar]
- 2.Naar A. M., Beaurang, P. A., Zhou, S., Abraham, S., Solomon, W. & Tjian, R. (1999) Nature (London) 398, 828-832. [DOI] [PubMed] [Google Scholar]
- 3.Malik S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277-283. [DOI] [PubMed] [Google Scholar]
- 4.Reines D., Conaway, R. C. & Conaway, J. W. (1999) Curr. Opin. Cell Biol. 11, 342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conaway J. W. & Conaway, R. C. (1999) Annu. Rev. Biochem. 68, 301-319. [DOI] [PubMed] [Google Scholar]
- 6.Gnarra J. R., Duan, D. R., Weng, Y., Humphrey, J. S., Chen, D. Y., Lee, S., Pause, A., Dudley, C. F., Latif, F., Kuzmin, I., et al. (1996) Biochim. Biophys. Acta 1242, 201-210. [DOI] [PubMed] [Google Scholar]
- 7.Kamura T., Sato, S., Haque, D., Liu, L., Kaelin, W. G., Conaway, R. C. & Conaway, J. W. (1998) Genes Dev. 12, 3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J. G., Farley, A., Nicholson, S. E., Willson, T. A., Zugaro, L. M., Simpson, R. J., Moritz, R. L., Cary, D., Richardson, R., Hausmann, G., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stebbins C. E., Kaelin, W. G. & Pavletich, N. P. (1999) Science 284, 455-461. [DOI] [PubMed] [Google Scholar]
- 10.Lisztwan J., Imbert, G., Wirbelauer, C., Gstaiger, M. & Krek, W. (1999) Genes Dev. 13, 1822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwai K., Yamanaka, K., Kamura, T., Minato, N., Conaway, R. C., Conaway, J. W., Klausner, R. D. & Pause, A. (1999) Proc. Natl. Acad. Sci. USA 96, 12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamura T., Sato, S., Iwai, K., Czyzyk-Krezeska, M. F., Conaway, R. C. & Conaway, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, 10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohh M., Park, C. W., Ivan, M., Hoffman, M. A., Kim, T. Y., Huang, L. E., Pavletich, N., Chau, V. & Kaelin, W. G. (2000) Nat. Cell Biol. 2, 423-427. [DOI] [PubMed] [Google Scholar]
- 14.Cockman M. E., Masson, N., Mole, D. R., Jaakola, P., Chang, G. W., Clifford, S. C., Maher, E. R., Pugh, C. W., Ratcliffe, P. J. & Maxwell, P. H. (2000) J. Biol. Chem. 275, 25733-25741. [DOI] [PubMed] [Google Scholar]
- 15.Kamura T., Burian, D., Yan, Q., Schmidt, S. L., Lane, W. S., Querido, E., Branton, P. E., Shilatifard, A., Conaway, R. C. & Conaway, J. W. (2001) J. Biol. Chem. 276, 29748-29753. [DOI] [PubMed] [Google Scholar]
- 16.Pause A., Peterson, B., Schaffar, G., Stearman, R. & Klausner, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 9533-9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamura T., Koepp, D. M., Conrad, M. N., Skowyra, D., Moreland, R. J., Iliopoulos, O., Lane, W. S., Kaelin, W. G., Elledge, S. J., Conaway, R. C., et al. (1999) Science 284, 657-661. [DOI] [PubMed] [Google Scholar]
- 18.Brower C. S., Shilatifard, A., Mather, T., Kamura, T., Takagi, Y., Haque, D., Treharne, A., Foundling, S. I., Conaway, J. W. & Conaway, R. C. (1999) J. Biol. Chem. 274, 13269-13636. [DOI] [PubMed] [Google Scholar]
- 19.Stearns D. J., Kurosawa, S., Sims, P. J., Esmon, N. L. & Esmon, C. T. (1988) J. Biol. Chem. 263, 826-832. [PubMed] [Google Scholar]
- 20.Garrett K. P., Aso, T., Bradsher, J. N., Foundling, S. I., Lane, W. S., Conaway, R. C. & Conaway, J. W. (1995) Proc. Natl. Acad. Sci. USA 92, 7172-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conaway R. C., Reines, D., Garrett, K. P., Powell, W. & Conaway, J. W. (1996) Methods Enzymol. 273, 194-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilahi E., Miklos, I. & Sipiczki, M. (2000) Curr. Genet. 38, 227-232. [DOI] [PubMed] [Google Scholar]
- 23.Myers L. C., Gustafsson, C. M., Bushnell, D. A., Lui, M., Erdjument-Bromage, H., Tempst, P. & Kornberg, R. D. (1998) Genes Dev. 12, 45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spahr H., Samuelsen, C. O., Baraznenok, V., Ernest, I., Huylebroeck, D., Remacle, J. E., Samuelsson, T., Kieselbach, T., Holmberg, S. & Gustafsson, C. M. (2001) Proc. Natl. Acad. Sci. USA 98, 11985-11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik S., Gu, W., Wu, W., Qin, J. & Roeder, R. G. (2000) Mol. Cell 5, 753-760. [DOI] [PubMed] [Google Scholar]
- 26.Aso T., Haque, D., Barstead, R. J., Conaway, R. C. & Conaway, J. W. (1996) EMBO. J. 15, 5557-5566. [PMC free article] [PubMed] [Google Scholar]
- 27.Ohh M., Takagi, Y., Aso, T., Stebbins, C. E., Pavletich, N. P., Zbar, B., Conaway, R. C., Conaway, J. W. & Kaelin, W. G. (1999) J. Clin. Invest. 104, 1583-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonergan K. M., Iliopoulos, O., Ohh, M., Kamura, T., Conaway, R. C., Conaway, J. W. & Kaelin, W. G. (1998) Mol. Cell. Biol. 18, 732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pause A., Lee, S., Worrell, R. A., Chen, D. Y. T., Burgess, W. H., Linehan, W. M. & Klausner, R. D. (1997) Proc. Natl. Acad. Sci. USA 94, 2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chittum H. S., Lane, W. S., Carson, B. A., Roller, P. P., Lung, F. D., Lee, B. J. & Hatfield, D. L. (1998) Biochemistry 37, 10866-10870. [DOI] [PubMed] [Google Scholar]
- 31.Baek H. J., Malik, S., Qin, J. & Roeder, R. G. (2002) Mol. Cell. Biol. 22, 2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu, J. Y., Park, J. M., Song, E. J., Mizuguchi, G., Yoon, J. H., Jeongsil, K. H., Lee, K. J. & Kim, Y. J. (2002) J. Biol. Chem., 10.1074/jbc.M204144200.
- 33.Zhou P. & Howley, P. M. (1998) Mol. Cell 2, 571-580. [DOI] [PubMed] [Google Scholar]
- 34.Wirbelauer C., Sutterluty, H., Blondel, M., Gstaiger, M., Peter, M., Reymond, F. & Krek, W. (2000) EMBO J. 19, 5362-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galan J.-M. & Peter, M. (1999) Proc. Natl. Acad. Sci. USA 96, 9124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dignam J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]