Abstract
Background
Recent studies suggest that intratumoral microbiome and altered metabolic networks play crucial roles in pancreatic cancer (PC) progression. However, the precise interplay between microbial communities and tumor metabolism in PC remains poorly understood. This study aims to investigate the impact of the intratumoral microbiome, the metabolic landscape, and their interactions on PC development.
Methods
16S rDNA sequencing and Untargeted metabolomic profiling were performed on 47 paired pancreatic cancer and adjacent normal tissues to analyze their intratumoral microbiome and metabolic landscapes. Bioinformatics tools were used to conduct differential microbiome abundance analysis and pathway enrichment. A correlation analysis was performed to identify key microbiota-metabolite interactions.
Results
16S rDNA sequencing revealed significant differences in the abundance and diversity (α-diversity and β-diversity) of the intratumoral microbiome in PC. The predominant species in pancreatic cancer were Pseudomonas. Enrichment analysis showed that amino acid metabolic pathways, including Arginine and Proline Metabolism, Arginine Biosynthesis, were significantly enriched in PC. Untargeted metabolomics identified 298 metabolites that were significantly altered in PC (fold change > 1.5, P-value < 0.05). These included amino acid metabolites such as Lys-Leu, Pro-Leu, Arg-Leu, Lys-Val, His-Lys, and others. Functional enrichment analysis highlighted several metabolic pathways that play important roles in pancreatic cancer, including Glycine, Serine, and Threonine Metabolism, Amino Acid Biosynthesis, Metabolic Pathways and Cysteine and Methionine Metabolism. Correlation analysis between microbiome and metabolic data revealed significant associations between Pseudomonas and several metabolites, including Alpha-ketoisovaleric acid, 16-hydroxyhexadecanoic acid, Myristic acid, Nonanoic acid (the Spearman correlation coefficient r, 0.5 ≤|r|≤ 1 and P-value < 0.05).
Conclusion
This study suggests a relationship between the microbiome and metabolism in pancreatic cancer. We observed that Pseudomonas contributes to altered amino acid metabolism, but whether this interaction is causal and the mechanisms underlying it remain unclear. Further experimental validation is required before considering microbiome-targeted metabolic interventions as viable therapeutic strategies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-025-02458-9.
Keywords: Pancreatic cancer, Intratumoral microbiome, Metabolic landscape, Microbiota-metabolite interactions
Introduction
Pancreatic cancer (PC) has a rising incidence and is one of the most lethal human malignancies [1]. PC is projected to become the second-leading cause of cancer-related deaths by 2040 in the United States [2], with its 5-year relative survival rate at only 13% [1], which is the lowest among all types of cancer.
The current standard of care for pancreatic cancer is a systemic treatment paradigm that comprising surgery, chemotherapy, immunotherapy and targeted therapy. Early symptoms of pancreatic cancer are often subtle, and the majority of patients are diagnosed at a locally advanced or metastatic stage. Surgical resection is the sole effective method for achieving cure and long-term survival in pancreatic cancer patients; however, less than 20% of patients have the opportunity for curative surgical resection [3], approximately 80% of who undergo curative surgery will experience recurrence within 2 years postoperatively [4]. Chemotherapy is the standard of care for pancreatic cancer patients, but the response rate to chemotherapy is only 30% and is prone to drug resistance [5]. Only a very small number of pancreatic cancer patients benefit from immunotherapies and targeted therapy approaches [4, 6]. Pancreatic cancer has a very poor prognosis due to the difficulty of early diagnosis, low resection rate, and poor therapeutic efficacy. Therefore, there is an urgent need to find novel molecular mechanisms and potential targets.
A defining feature of the tumor microenvironment (TME) in pancreatic cancer is the intricate interplay between the internal and external mechanisms of cancer cells, which gives rise to a TME characterized by a dense extracellular matrix, metabolic reprogramming, and pronounced immunosuppression [7, 8]. To adapt to the severe hypoxia environment, cells have changed their metabolic phenotypes to maintain their survival and proliferation, including enhanced glycolysis metabolism, glutamine metabolism and fatty acids metabolism [9]. Adaptive metabolic changes of cancer cells promote their survival and affect tumor-infiltrating immune cells in the TME, which contributes to the immunosuppressive microenvironment and induces tumor immunotherapy resistance [9, 10].
There is a close link between the intratumoral microbiota and host metabolic state, and this interplay may influence tumor metabolic reprogramming. The intratumoral microbiome exerts tumor-promoting or tumor-suppressive effects by engaging in metabolic reactions within the body, regulating cancer-related signaling pathways, and impacting both host cell function and immune system [11]. Microbiota-derived metabolites represent important mediators of host-microbe interactions and play a profound role in physiological functions and the immune environment. These microbes are involved in regulating host mechanisms such as detoxification and nutritional pathways, as well as influencing tumor metastasis and the efficacy of chemotherapy [12].
Accumulating evidence suggests that microbiota (specifically specific well-studied oral, gastrointestinal, and intrapancreatic microbes, as well as certain hepatotropic viruses and bactibilia) and their metabolites may have potential etiological roles in altering the microenvironment of pancreatic tumors, impacting the functionality of the immune system, and regulating the biological behavior of pancreatic cancer cells, those actions can promote or inhibit the progression of pancreatic tumors [13, 14]. Concrete mechanisms include perpetuating inflammation, regulating the immune system-microbe-tumor axis, affecting metabolism, and altering the tumor microenvironment [14]. Intratumoral microbiota and their products can regulate the microenvironment of pancreatic tumors, the biological behavior of pancreatic cancer cells, and the functionality of the immune system [15]. Three potential mechanisms of intratumoral microbiota promoting pancreatic tumorigenesis: secretion of metabolites to cause DNA damage, activation of oncogenic pathways and alteration the tumor immune microenvironment [15].
Metabolomics reveals the metabolic characteristics of PC. Despite existing in a relatively hypoxic and nutrient-poor environment, PC achieves its metabolic advantage through three mechanisms: (i) reprogramming intracellular energy metabolism, (ii) enhancing nutrient acquisition via scavenging and recycling (autophagy), and (iii) facilitating metabolic interactions with other components in its microenvironment [16]. PC can be stratified into glycolytic and lipogenic subtypes [17]. These subtypes can interconvert (e.g., transitioning from glycolysis to oxidative phosphorylation) under varying metabolic demands. Such plasticity may be closely linked to tumor aggressiveness, therapy resistance, and microenvironmental adaptation. Distinct metabolic profiles may influence the development of PC through metabolites and metabolic pathways [18], underscoring the pivotal role of amino acid metabolism in the progression of PC [19].
Investigating the role of the intratumoral microbiome in the diagnosis and treatment of pancreatic cancer using 16S sequencing has emerged as an active area of research. The tumor microbiome composition correlates with distinct metabolic pathways in PC patients with short-term survival (STS) versus long-term survival (LTS) [20], highlighting that intraltumor microbiome influences the prognosis of PC patients through lipid metabolism and protein anabolic pathways. Studies have revealed that the microbiota of pancreatic cancer can reshape the immune microenvironment through their metabolites; however, the roles of specific metabolites (such as amino acids) and their metabolic pathways remain insufficiently characterized [21, 22].
As seen from the above discussion, current research predominantly focuses on the association of individual microbiota or metabolites with PC; however, the causal mechanisms governing their dynamic interplay (e.g., how specific microbial communities mediate oncogenic metabolite synthesis) remain systematically underexplored. Traditional single-omics approaches fail to capture complex interactions within multifaceted biomolecular networks, resulting in fragmented insights into host-microbiota metabolic crosstalk. Therefore, systematically mapping the interactions between intratumoral microbiota and metabolism in PC is critical for advancing early diagnosis and targeted therapy of the disease.
Microbiome-induced alterations in human metabolism may contribute to diverse metabolic diseases and cancers [23]. The intratumoural microbiota modulate cancer initiation and progression through metabolic regulation [24], including in hepatocellular carcinoma [25], breast cancer [26], gastric cancer [27], and cervical cancer [28]. Most current studies have focused on the effects of gut microbes on PC [13]. Changes in gut microbiota may affect host metabolic status, tumor metabolic reprogramming and influence pancreatic cancer progression by modulating immune responses [13, 29]. Although studies have performed microbiome and metabolome analyses to investigate how smoking influences PC progression via alterations in microbial communities and metabolic pathways, the relationship between specific microbes and amino acid metabolism remains unexplored [30]. Therefore, studies on the integration of microbes and metabolites in intratumoral tissues are still insufficient. The relevance of specific intratumoral microbiota to metabolites and metabolic pathways in PC tissues and their potential mechanisms of action remain unknown.
The integration of multi-omics approaches—leveraging data-level cross-validation and interaction model construction—can elucidate key regulatory hubs in tumor progression and holds promise for advancing precision oncology [31]. Multi-omics approaches, which are novel frameworks, integrate multiple omics datasets generated from the same patients to better understand the molecular and clinical features of cancer [32]. Zhu et al.’s study, employing microbiome and spatially resolved metabolomics analyses, revealed how Akkermansia muciniphila exerts anticancer effects in a murine model through interactions with intratumoral microbiota and metabolic reprogramming [33], providing a new potential target for the prevention and treatment of gastric cancer. Another study integrated multi-omics data (microbiome, transcriptome, and metabolome) to reveal that Streptococcus anginosus modulates the arginine metabolic pathway, elevating ornithine levels and thereby remodeling the tumor immune microenvironment (TIME) to promote gastric cancer progression [27]. Multi-omics approach elucidated the impact of intratumoral microbes on neuron-related gene expression in glioma, mediated by microbial metabolites, suggesting that Fusobacterium nucleatum may serve as a potential diagnostic and therapeutic target for glioma [34]. Furthermore, flora analysis, mass spectrometry (MS), and integrated metabolomics demonstrate that coordinated microbiota modulation alters the abundance and diversity of intratumoral microbiota and disrupts metabolic pathways within both the microbiota and tumor microenvironment [35]. Through these approaches, distinct signaling pathways and biosynthetic demands or metabolites influencing tumor progression have been identified. The clear underlying principles of microbiome-regulated tumorigenesis and the established microbiome metabolism regulation methods provide distinctive insights into tumor therapy [35].
Therefore, we proposed the scientific hypothesis of this project: specific intratumoral microbiota within pancreatic cancer tissues are involved in pancreatic cancer progression by regulating metabolism-related products (e.g., amino acids). The aim of this article is to combine 16S sequencing (microbiome) and untargeted metabolomics data to systematically reveal the microbial-metabolite interaction network in pancreatic cancer tissues and to elucidate its role in tumor progression.
Methods
Pancreatic samples sources
All human samples (including 47 cases of pancreatic cancer and adjacent tissues), as well as the clinical data, were obtained from our hospital. This study was conducted in accordance with the regulations of the tissue biobank and approval by the Ethics Committee of our hospital. All patients provided written informed consent.
Experimental techniques and procedures
Untargeted metabolomic analysis(LC–MS/MS)
PC samples were thawed slowly at 4℃ and metabolites were extracted according to the experimental procedure. LC–MS/MS analysis was performed using an UHPLC (1290 Infinity LC, Agilent Technologies) coupled to a quadrupole time-of-flight (AB Sciex TripleTOF 6600), in both positive and negative ion modes. The raw MS data were converted to MzXML files using ProteoWizard MSConvert before importing into freely available XCMS software. After sum-normalization, the processed data were analyzed by R package (ropls). Data analysis includes univariate statistical analysis, multidimensional statistical analysis, differential metabolite screening, differential metabolite metabolic pathway enrichment, and other bioinformatics analyses. Differential analysis between two sample groups often utilizes univariate statistical methods, including Fold Change Analysis (FC Analysis) and non-parametric tests (e.g., t-test). Based on the results of univariate analysis, differential analysis is performed on all detected metabolites, including those that are unidentified. Metabolites were classified as differential if they meet the criteria of FC > 1.5 or FC < 0.67 and have a P value < 0.05 (t-test). These differential metabolites were then visualized using a volcano plot. Multivariate data analysis was performed with orthogonal partial least-squares discriminant analysis (OPLS-DA). The variable weight values obtained from the OPLS-DA model, known as Variable Importance for the Projection (VIP), can be used to measure the strength of influence and explanatory power of the expression patterns of metabolites on the classification and discrimination of samples between groups. Metabolites with a VIP value > 1 were generally considered to have a significant contribution to the model interpretation. In this experiment, metabolites with an OPLS-DA VIP value > 1 and a T-test P value < 0.05 were selected as significantly different metabolites for subsequent bioinformatics analysis. Functional analysis of differential metabolites encompasses KEGG pathway annotation, MetPA enrichment, and metabolite set enrichment analysis (MSEA).
Microbiome analysis (16S rDNA sequencing)
DNA extraction, detection, PCR amplification, purification of amplification products, fluorescence quantification, library preparation, and V3-V4 variable region amplification and sequencing were performed using the Illumina NovaSeq 6000 sequencing platform. Raw data obtained from sequencing were processed using the default parameters of QIIME2 for quality control analysis to obtain Amplicon Sequence Variants (ASVs) for ASV/Operational Taxonomic Units (OUT) clustering and annotation. Subsequently, the data analysis was carried out, including community structure analysis, alpha (α) diversity analysis, beta (β) diversity analysis, species difference analysis and function prediction. Specifically, α and β diversity indices were calculated using QIIME2 software (with default parameters). α diversity (including Chao 1, Shannon and Simpson indices) was used to analyze the diversity of microbial communities within the sample. β Diversity compared the microbial community composition of different samples. Anosim analysis was used to examines whether the differences between groups were significantly greater than the differences within groups. The Wilcoxon rank-sum test was used to determine whether differences in species diversity between groups were statistically significant (P < 0.05 indicates a significant difference). Linear discriminant analysis effect size analysis (LEfSe, LDA analysis) was used to identify differentially abundant species. With only those having an LDA Score > 2 (default set at 2) displayed in the figure. The length of the bar represents the magnitude of the LDA value. Microbial function and pathway enrichment were analyzed based on KEGG database by the PICRUSt software (with the default parameters) to infer the Functional gene composition of samples by comparing the species composition information obtained from 16S sequencing data, this allowed for the analysis of Functional differences between different samples or groups. Due to the wide variation in the expression of individual abundance of intratumoral microbiota in pancreatic adenomas, only 16 patients in the microbiome were included in the subsequent data analysis based on PCoA component analysis.
Multi-omics correlation analysis
Microbiota with significant differences identified in the 16S microbiome were integrated with the metabolites that showed significant differences in the metabolome. Spearman correlation analysis was used to identify associations between significantly different flora (LEfSe LDA > 2 and P value < 0.05) and significantly different metabolites (VIP > 1 and P value < 0.05). A microbial-metabolite interaction network was then constructed, including Spearman rank correlation analysis and Mantel test network graphs), focusing on analyzing the functional roles of core microbiota and metabolic nodes.
Correlation Analysis of Clinical Data
①Correlation analysis of clinical staging: The correlation analysis between alpha diversity indices (ACE, Chao1, Shannon, and Simpson indices) and clinical staging was conducted. Additionally, the relative abundance of Pseudomonas was analyzed in relation to clinical staging. The analysis was performed using R software, employing t-tests, with a P-value less than 0.05 indicating a statistically significant difference. ②Survival analysis of Pseudomonas: Survival analysis was first performed using public data from the TCMbio database (https://microbiomex.sdu.edu.cn/). To conduct the analysis, navigate to "Survival Analysis—Microbe," select "PAAD" as the Cancer Type, and enter "Pseudomonas" in the "Enter microbe name" field. For overall survival (OS) analysis, set the Group Cutoff value to 70%, and for disease-free interval (DFI), use the default Median (50%). Keep all other settings as default and click "Plot" to obtain the survival curves. In our cohort, Kaplan–Meier survival analysis was performed using the median abundance of Pseudomonas as the grouping criterion.
Statistical analysis
Data were analyzed by R (version 3.4.2). Differences between two groups were considered statistically significant at P < 0.05.
Results
Significant differences between the microbiomes of pancreatic cancer and adjacent tissues
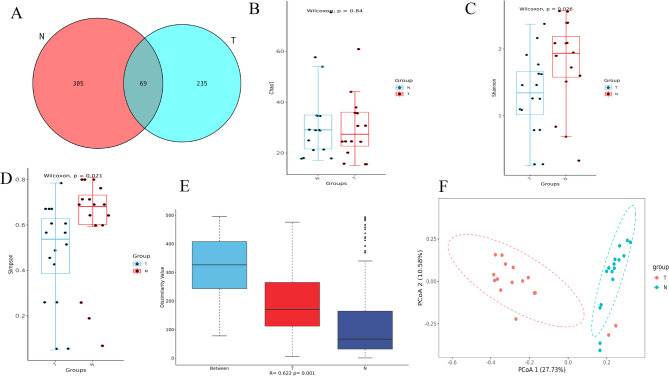
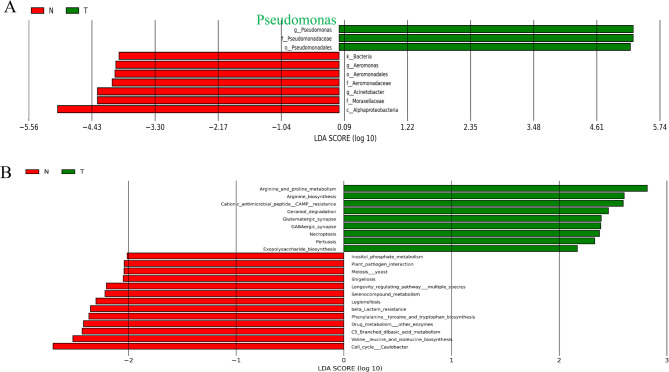
Differences in the number of species detected were observed between the pancreatic cancer tissues (T group) and the adjacent tissues (N group) (Fig. 1A). Analysis of α-diversity using the Shannon and Simpson indices (Fig. 1B-D) and β-diversity using ANOSIM analysis (Fig. 1E), and the PCOA analysis (Fig. 1F) based on the Braycurtis distance revealed that significant differences in the α-diversity and β-diversity between cancerous tissues (T) and adjacent (N). These results revealed the significant differences in species richness and community composition between cancerous tissues (T) and adjacent tissues (N), suggesting the distinct intratumoral microbiota compositions between the two groups (Fig. 1). LEfSe analysis was used to determine the dominant species between two groups. Using an LDA score > 2 as the screening criterion, the dominant species in the intratumoral tissues of pancreatic cancer were Pseudomonadales, Pseudomonadacea, and Pseudomonas (Fig. 2A), while those in the paracancerous tissues were Alphaproteobacteria, Moraxellaceae, Acinetobacter, Aeromonadaceae, Aeromonadales,Aeromonas. KEGG analysis (based on computer calculations) highlighted several key metabolic pathways in pancreatic cancer: arginine and proline metabolism, arginine biosynthesis, cationic antimicrobial peptide (CAMP) resistance, geraniol degradation, glutamatergic synapse, GABAergic_synapse, necroptosis and (Fig. 2B).
Fig. 1.
Analysis of intra-tissue bacterial diversity between PC (T) and adjacent (N) group. A Venn diagram of detected between PC and adjacent group; B Chao 1 index; C Simpson index; D Shannon index; E anosim index; F PCoA showing significant differences between two groups (the horizontal axis shows the first principal component (PCoA1), and the vertical axis shows the second principal component (PCoA2). The percentages show how much each component contributes to the differences between the samples). Note: group: T; adjacent group: N. A-E The horizontal axis represents different groups; the vertical axis represents the alpha and beta diversity index (Chao 1, P = 0.84; Simpson, P = 0.021; Shannon, P = 0.026; and anosim index, P = 0.001). P < 0.05 indicates statistical significance
Fig. 2.
Functional analysis of dominant species and differential intratumoral microbiota between PC (T) and adjacent (N) groups. A Significantly different intratumoral microbiota between PC and adjacent (N) group; B KEGG pathway enrichment bar graphs of different species between two groups. Note: The LDA value distribution bar chart features a red area for the adjacent tissue group (N) and a green area for the tumor group (T), indicating distinct groups. In the tree branches, red nodes represent important microbial taxa in the adjacent tissue group (N), while green nodes represent those in the tumor group (T). The figure shows species with an LDA Score > 2 (The default cutoff value for LDA is set at 2), with the length of each bar reflecting the magnitude of the LDA value
Metabolomics results showed significantly different metabolites and their pathways in pancreatic cancer
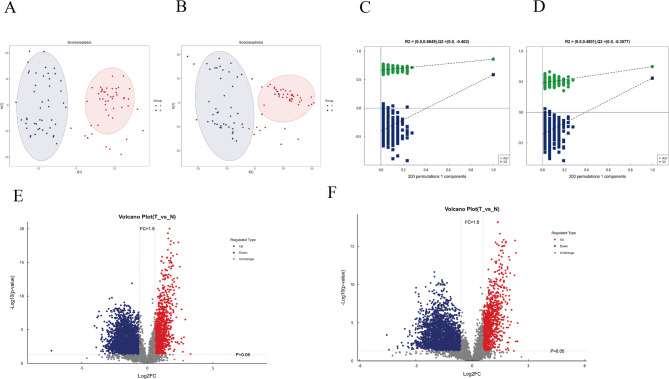
Untargeted metabonomics was used to compare metabolites difference between pancreatic cancer group (T) and adjacent group (N). The model demonstrated good stability and effectiveness (Fig. 3A-D). A volcano map identified 298 significantly different metabolites between two groups, including both up-regulated and downregulated metabolites (Fig. 3E-F).
Fig. 3.
Analysis of model stability and metabolite changes in PC (T) and adjacent group (N). A-B metabolite OPLS-DA score chart (positive and negative ions). The figure illustrates the first principal component on the horizontal axis and the second principal component on the vertical axis, with ellipses indicating the 95% confidence intervals. Points of the same color represent biological replicates within a group, and their distribution demonstrates the differences both between and within groups; C-D Histogram of permutation of metabolites (positive and negative ions). In the figure, the horizontal axis represents the retention rate of permutations, which is the proportion consistent with the original order of the Y variable in the model. The vertical axis indicates the values of R2 and Q2. The green points denote R2, while the blue points denote Q2. The two dashed lines represent the regression lines for R2 and Q2, respectively. The R2 and Q2 values in the upper right corner correspond to a retention rate of 1, which means they are the R2 and Q2 values of the original model; E–F. Volcanic diagram of differential metabolites (positive and negative ions). The horizontal axis shows the log2 value of the fold change (log2 Fold Change), while the vertical axis shows the negative log10 value of the significance p-value (-log10 p-value). Metabolites with a fold change > 1.5 and P < 0.05 are highlighted in red, while those with a fold change < 0.67 and P < 0.05 are highlighted in blue. Non-significantly different metabolites are indicated in grey
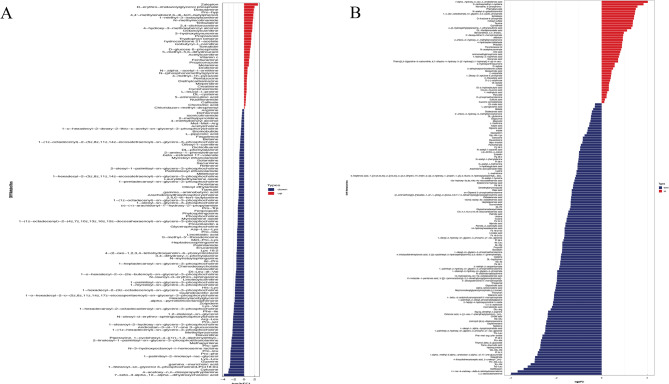
Analysis of metabolite differences revealed that the following metabolites in positive ion mode were significantly upregulated in cancer tissues: Zaleplon, Derythromimidazolyl Glycol Phosphate, Dobutamine, Pro − hyp, 1 − methyl − 3 − isobutylxanthine, N − methylnicotinamide, Terbutaline, Octanoylcarnitine and Tryptophan betaine. Conversely, the following metabolites were downregulated: Lys − Leu, Pro − phe, Pro − leu, Pro − gln, methyltyrosinate, Pro − ser, Arg − Leu, Phe − Ile, Lys − Val, His − Lys, Dl − Leu − dl − Val, Met − Pro − Lys and Asp − Leu − Lys (Fig. 4A). In negative ion mode, the following metabolites were significantly upregulated: α- Hydroxy3oxo4cholestronic acid, S − carboxymethyl − l − cysteine, N − acetyl − d − galactosaminitol and D − fructose 6 − phosphate. Meanwhile, the following metabolites were downregulated: Trp − Leu, Trp − Val, Thr − Val − Leu, Val − Gly − Va, Pro − Ala, Thr − Leu, Val − Glu, Glu − Arg, Val − Ile and Ile − Thr (Fig. 4B). These results demonstrate significant differences in amino acid metabolism in pancreatic cancer tissues (T) compared to adjacent tissues (N).
Fig. 4.
Screening of metabolites with significant differences between two group (T vs. N). A Metabolites (positive ions) with significant differences between PC and N group; B Metabolites (negative ions) with significant differences between PC and N group. Note: Significant differential metabolites are intuitively displayed using a butterfly plot. The horizontal axis shows the log2 fold change (FC) values of differential metabolites, derived from base 2 logarithms of their fold changes. The vertical axis displays the significantly different metabolites, with red indicating upregulated and blue indicating downregulated metabolites
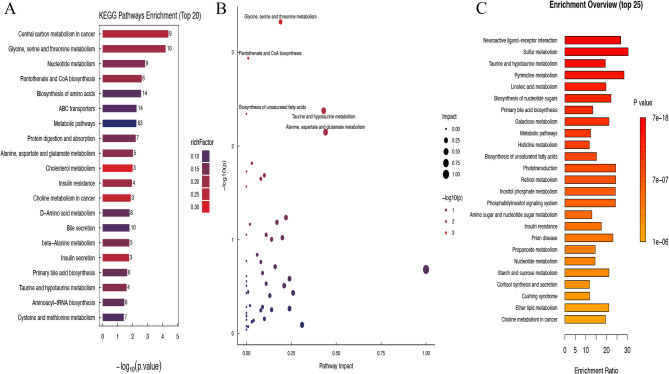
KEGG analysis identified several metabolic pathways that play important roles in pancreatic cancer: Glycine, serine and threonine metabolism, Nucleotide metabolism, Pantothenate and CoA biosynthesis、Biosynthesis of amino acids, ABC transporters, Metabolic pathways, Protein digestion and absorption、Alanine, aspartate and glutamate metabolism, Cholesterol metabolism, Insulin resistance, Choline metabolism in cancer, D − Amino acid metabolism, Bile secretion, beta − Alanine metabolism, Insulin secretion, Primary bile acid biosynthesis, Taurine and hypotaurine metabolism, Aminoacyl − tRNA biosynthesis、Cysteine and methionine metabolism (Fig. 5A). Met PA analysis highlighted the most relevant metabolic pathways, including alanine, aspartate and glutamate metabolism, taurine and hypo taurine metabolism, glycine, serine and threonine metabolism, pantothenate and CoA biosynthesis and biosynthesis of unsecured fatty acids (Fig. 5B). MSEA analysis, a supplementary mode of KEGG analysis, further identified highly correlated pathways such as: Neuroactive Ligand Receiver Interaction, Sulfur metabolism, Taurine and Hypo Taurine Metabolism, Pyrimidine metabolism, Linoleic acid metabolism、Biosynthesis of nucleotide sugars、Primary bile acid biosynthesis、Galactose metabolism、Metabolic pathways、 Histidine metabolism and biosynthesis of unstructured fatty acids (Fig. 5C). Therefore, we will focus on amino acid metabolism, particularly glutamate metabolism (involved in glutamine metabolism pathway) and its intermediates, in pancreatic cancer.
Fig. 5.
Enrichment pathway analysis of significantly different metabolites between PC group (T) and adjacent group (N). A Bar graph of significantly different metabolite KEGG enrichment analysis pathway between the two groups (The bar chart illustrates KEGG metabolic pathways on the vertical axis and the p-value of the enrichment analysis on the horizontal axis, expressed as the negative common logarithm (-log10(p-value)). Each bar's height indicates the number of differentially abundant metabolites enriched in the corresponding pathway. Additionally, the color of the bars represents the rich factor for each pathway, with darker shades of red indicating higher values. The rich factor shows the proportion of differentially abundant metabolites relative to the total annotated metabolites in the pathway); B Bubble graph of significantly different metabolite Met PA analysis pathway between the two groups (Each circle represents a distinct metabolic pathway. The horizontal axis indicates how much the pathway is impacted, referred to as Pathway Impact. The size of each circle correlates with Pathway Impact; larger impact values produce larger circles. The vertical axis shows the negative logarithm of the P-value from the pathway enrichment analysis. The color gradient, ranging from blue to red, indicates a positive correlation with the negative logarithm of the P-value for pathway changes. The redder the color, the more significantly enriched the pathway is.); C Bar chart of significantly different metabolite MSEA analysis pathway between two groups (The vertical axis shows the names of the metabolite sets and their annotated p-values. In contrast, the horizontal axis illustrates the degree of enrichment, where the color corresponds to the p-value from the enrichment analysis. As the color darkens, the p-value decreases, indicating a more significant enrichment analysis)
Interaction between significantly different intratumoral microbiota and significantly different metabolites
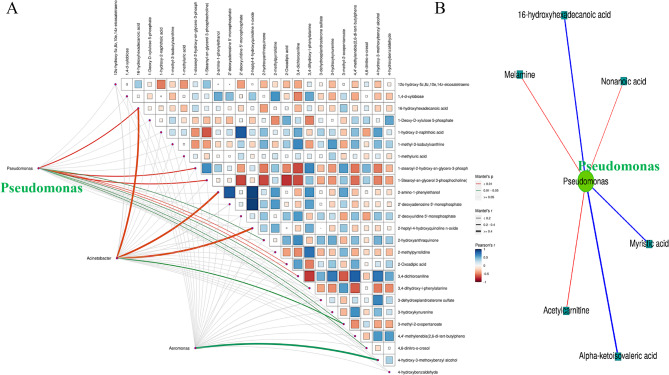
The association analysis between 16S sequencing data and metabolomics primarily utilized statistical algorithms to identify significant correlations between differentially abundant intratumoral microbiota and metabolites. Spearman correlation analysis revealed significant associations between differentially abundant microbiota and metabolites, as well as among the metabolites themselves and within the intratumoral microbiota. These correlations were visualized as a heatmap of the correlation coefficient matrix (Supplementary Fig. 1). Spearman and Mantel test correlation network plots showed that Pseudomonas may negatively regulate tumor progression by downregulating metabolites such as Alpha-ketoisovaleric acid (a metabolic intermediate of valine, a branched-chain amino acid [BCAA]), 16-hydroxyhexadecanoic acid, and Myristic acid. Conversely, Pseudomonas may promote tumor progression by upregulating metabolites like Acetylcarnitine, Nonanoic acid, and Melamine (Fig. 6). Combined analysis of metabolomes and microbiomes highlighted the prominent role of amino acid metabolism in PC. Therefore, the interaction among Pseudomonas, amino acids, and their intermediate metabolites may regulate the initiation and progression of pancreatic cancer.
Fig. 6.
Network diagram of correlation between significantly differential intratumoral microbiota and significant differential metabolites in PC and adjacent group A Mantel Test analysis of correlation for the top 25 differential metabolites and differential flora with the smallest significant p-value. (Note: The thickness of the connecting lines indicates the correlation between the two matrices, while the color of the lines represents the significance of the correlation's p-value; the matrix plot illustrates the correlation among differentially abundant metabolites) B Spearman correlation analysis network plot of significant differential intratumoral microbiota with significant differential metabolites (Note: In the figure, circles represent significantly different microbial genera, and rectangles represent significantly different metabolites. The color of the lines indicates the sign of the correlation coefficient between them: blue for negative correlation, red for positive correlation. While the thickness of the lines is proportional to the absolute value of the correlation coefficient. Node size is positively correlated with its degree; higher-degree nodes are larger
The clinical significance of differential abundance of Pseudomonas
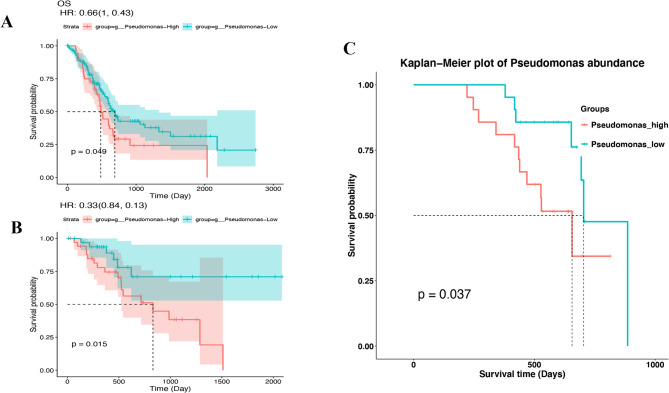
No significant correlation between alpha diversity (ACE, Chao 1, Shannon, and Simpson indices) and the stages (stages I, II, and III) was found in pancreatic cancer (Supplementary Fig. 2). Similarly, there was no significant correlation between the relative abundance of Pseudomonas and the T stage, N stage, or clinical stages (Supplementary Fig. 3). Overall survival (OS) analysis by the Cancer Microbiome Database (TCMbio) showed that higher levels of Pseudomonas are linked to shorter overall survival (OS, P = 0.049) and disease-free intervals (DFI, P = 0.015) in patients with pancreatic cancer (Fig. 7 A-B). Kaplan–Meier curves revealed that high abundance of Pseudomonas in pancreatic cancer is associated with reduced overall survival in our cohort (log-rank P = 0.037, Fig. 7C).
Fig. 7.
Kaplan–Meier survival analysis stratified by Pseudomonas abundance. A Overall survival analysis of Pseudomonas based on TCMbio database, B Disease-Free Interval analysis of Pseudomonas based on TCMbio database, C Overall survival analysis of Pseudomonas from our cohort (n = 42)
Discussion
Microbiome analysis revealed that the predominant intratumoral microbiota in pancreatic cancer tissues is Pseudomonas, while metabolomic profiling identified significant alterations in amino acid metabolism particularly in glutamate metabolism, which is involved in glutamine metabolism pathway. Multi-omics correlation analysis further demonstrated a strong association between Pseudomonas and amino acid metabolic pathways, including alpha − ketoisovaleric acid (a metabolite derived from the branched -chain amino acid valine [36]), suggesting that the interaction may serve as a key regulatory mechanism within the pancreatic cancer microenvironment. The results of this study not only uncover a specific regulatory mechanism in the pancreatic cancer microenvironment (i.e., the enrichment of Pseudomonas and its significant correlation with amino acid metabolic pathways), but also expand the frontier of research on metabolism-microbe interactions. Specifically, differences in amino acid metabolism are a critical feature of metabolic reprogramming in pancreatic cancer cells. Pseudomonas may influence tumor cell growth and invasive potential by modulating specific amino acid metabolic pathways.
Our findings indicated that Pseudomonas was significantly enriched in pancreatic cancer tissues, where it serves as the predominant intratumoral microbiota. The severe hypoxia, immunosuppressive microenvironment, and disorganized vascular system within tumors collectively create favorable conditions that facilitate rapid bacterial colonization, proliferation, and replication within tumor tissues [11, 37]. Furthermore, the distribution of microbiota within a tumor is not random; rather, it is highly organized into microniches that promote cancer progression through interactions with immune and epithelial cells [38]. The intratumor bacteria are mostly intracellular and are present in both cancer and immune cells. Therefore, bacteria are more preferentially present in tumor tissues rather than in the surrounding normal tissues [39]. More importantly, studies found that the cancerous pancreas harbors a markedly more abundant microbiome compared with normal pancreas in both mice and humans [21, 22]. Multi-omics analysis further suggested that Pseudomonas plays a pivotal role in shaping the pancreatic cancer microenvironment, particularly through the regulation of amino acid metabolism. Studies have shown that the intratumoral microbiota in pancreatic cancer is closely linked to tumorigenesis, progression, immune response, drug resistance, and prognosis [14, 15, 22, 40, 41]. While Pseudomonas has been implicated in inflammation and metabolic disorders in some tumors [42, 43], and Pseudomonas has been identified as the most abundant genus of bacteria transported from the gut to the pancreas in patients with PC [21] and is significantly associated with prognosis [20]. However, its specific functions and underlying mechanisms in pancreatic cancer remain poorly understood. In the present study, we demonstrated that Pseudomonas acts as a dominant bacterial species within the pancreatic cancer microenvironment, regulating key metabolic pathways. Our findings underscore the critical role of microbe-metabolite interactions in the pathogenesis of pancreatic cancer by integrating microbiomic and metabolomic analyses.
Untargeted metabolomic analysis revealed significant abnormalities in amino acid metabolism in pancreatic cancer tissues. Pathway analysis identified several amino acid metabolic pathways (such as those involving glycine, serine, and aspartic acid) as being prominently altered in pancreatic cancer, with a particular emphasis on glutamate metabolism, which is involved in glutamine metabolism pathway. Metabolic reprogramming in pancreatic cancer cells, which enables adaptation to increased energy demands, includes enhanced glucose metabolism, glutamine metabolism, and fatty acid oxidation. These changes have been extensively documented [44, 45]. Dysregulation of amino acid metabolism in pancreatic cancer significantly influences cancer cell behavior, systemic metabolism, and various biological processes, including stemness, proliferation, invasion, migration, autophagy, and apoptosis. These metabolic alterations also modulate interactions with the tumor microenvironment, highlighting their potential as diagnostic, prognostic, and therapeutic targets [19]. Previous studies have primarily focused on the metabolic reprogramming occurring within tumor cells. In contrast, the present study integrated metabolic alterations with the microbiome, emphasizing the potential role of microbes in mediating changes to the tumor microenvironment through metabolic regulation. This approach offers a novel perspective on the underlying pathological mechanisms.
Microbiomic and metabolomic association analyses revealed significant correlations between Pseudomonas and the metabolism of specific amino acids. Our functional analysis results highlighted the glutamate metabolism pathway in PC, where glutamate participates in the synthesis and catabolism of glutamine. And the correlation analysis also underscored the significant association between Pseudomonas and alpha − ketoisovaleric acid (a metabolite derived from the BCAA-valine). PC cells utilize glutamine to support proliferation and redox balance [46]. Glutamine and BCAA are crucial in regulating immune cell functions and tumor growth in PC [44]. Amino acid metabolism, including glutamine, leucine (a branched-chain amino acid), glutamate, and asparagine, regulates the mammalian target of rapamycin (mTOR) signaling pathway in pancreatic cancer cells [19, 44]. The phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) signaling pathway is critical in the pathogenesis of PC, affecting both its initiation and progression [47]. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway [48]. Moreover, amino acid metabolism modulates autophagy in pancreatic cancer by acting both upstream and downstream [19]. Kras-mutant PC cells require autophagy for their growth [49, 50]. mTORC1 blocks autophagy, and glutamine metabolism is important for regulating this process [19]. Therefore, we proposed the hypothesis between Pseudomonas and amino acid metabolism: our analysis suggests that Pseudomonas species within pancreatic tumors are associated with BCAA metabolism, potentially linked to mTOR signaling pathway activity. However, this association requires functional validation to establish causality. On the other hand, study have revealed that administration of Pseudomonas aeruginosa to the cancer patients induces autophagy [51], and autophagy activation is associated with tumor cells becoming resistant to chemotherapy and radiation therapy [52]. More importantly, Pseudomonas aeruginosa induces autophagy in human breast cancer cells through endoplasmic reticulum stress-mediated activation of the IRE1 signaling pathway [53]. We therefore propose the following hypothesis: our analysis indicates that Pseudomonas species within pancreatic tumors show potential association with IRE1 pathway-induced autophagy. However, functional validation (e.g., bacterial colonization/eradication experiments) is required to establish causality. However, the aforementioned mechanistic hypothesis requires further experimental validation. Collectively, these two hypotheses are hypothesized pathways based on literature and remain experimentally unverified. However, this microbial-metabolic interaction may represent a critical mechanism driving the progression of pancreatic cancer. In recent years, multi-omics analyses have become increasingly prominent in pancreatic cancer research, offering valuable insights into the systems biology mechanisms that drive tumor progression [54]. Despite existing studies on the characterization and role of the microbiome and metabolome in diseases such as inflammatory bowel disease (IBD) and uterine cancer [55, 56], research on microbe-metabolism interactions in pancreatic cancer remains limited. This study was the first to establish a clear association between Pseudomonas and amino acid metabolism in pancreatic cancer, suggesting its potential role as a key regulator. These findings offer new insights into the systemic mechanisms underlying the pancreatic cancer microenvironment. This study aligns with previous research in emphasizing the central role of microenvironmental regulatory mechanisms and metabolic reprogramming in the progression of pancreatic cancer. However, its uniqueness lies in identifying Pseudomonas as a dominant intratumoral microbiota and elucidating the mechanism by which it influences pancreatic cancer through amino acid metabolic pathways. The study presentes a novel hypothesis and provides preliminary evidence, supported by integrated multi-omics analysis, to substantiate this hypothesis.
From a clinical perspective, the results of our study offer several potential applications for the diagnosis and treatment of pancreatic cancer: (i) Discovery of potential diagnostic and therapeutic targets: The abundance of Pseudomonas and its regulation of amino acid metabolites (e.g., glutamine, and branched-chain amino acid metabolites) may serve as potential diagnostic and prognostic biomarkers of pancreatic cancer: ①Intratumoral microbial biomarkers: investigating the correlation of Pseudomonas abundance with early diagnosis and prognosis of PC. A significant difference in the abundance of microbiota was observed between adjacent tissues and pancreatic tumors and the intratumoral microbiota may serve as a biomarker for both early detection and prognosis prediction in PC [15]. Early diagnosis of pancreatic cancer is crucial for improving patient survival rates. Intra-tumoral microbes can significantly increase the risk of early-onset cancers, including early-onset pancreatic cancer [57]. Our study found that Pseudomonas was the dominant bacterial group in PC, and high abundance of Pseudomonas was associated with poor prognosis. Therefore, Pseudomonas may be a potential marker of diagnosis and prognosis for patients with PC. ②Biomarkers of amino acid metabolite: Investigating the spatial distribution and enrichment levels of glutamate, glutamine, branched-chain amino acid metabolism, and intermediate products (e.g., α-ketoisovaleric acid) in pancreatic cancer tissues using spatial metabolomics imaging, and their association with early diagnosis and prognosis. The alterations of amino acids and their metabolism have great potential in PC early detection [19, 58, 59]. Our study found that glutamate metabolism, glutamine metabolism and BCAA metabolism were significantly enriched in pancreatic cancer, and thus may serve as potential diagnostic and prognostic biomarkers. ③Microbiota-metabolite combined biomarker panel: The ratio of Pseudomonas abundance to branched-chain amino acid metabolites (e.g., alpha-ketoisovaleric acid) could also be a promising direction for diagnosis and prognosis. More importantly, the abundance of Pseudomonas and its regulation of amino acid metabolites (e.g., glutamine, and branched-chain amino acid metabolites) can also be promising therapeutic targets: ① Direct targeting of Pseudomonas (e.g., using sensitive antibiotics such as amikacin): Intra-tumoral microbes can impact carcinogenesis and therapy responses via several mechanisms. Intra-tumoral bacteria may negatively impact anti-tumor immunity by promoting a tolerogenic immune microenvironment, characterized by the immunosuppressive activity of tumor-associated macrophages [21]. They may also induce chemoresistance by breaking down chemotherapeutic agents into inactive forms. For example, Gammaproteobacteria can metabolize the chemotherapeutic drug gemcitabine (2',2'-difluorodeoxycytidine) into its inactive form, 2',2'-difluorodeoxyuridine, thereby contributing to gemcitabine resistance in colon cancer and pancreatic cancer [40]. Moreover, the combined use of antibiotics has been shown to enhance the antitumor response to gemcitabine. Antibiotics can suppress pancreatic cancer progression by eradicating pro-tumorigenic intratumoral microbiota, thereby improving survival rates in patients with advanced disease [60]. Therefore, targeting the microbiota in pancreatic tumors increases the effectiveness of immune therapy and chemotherapy [15, 61]. Therefore, Pseudomonas may serve as a potential therapeutic target, with the potential to improve treatment outcomes by modulating its role within the tumor microenvironment.②Blocking the microbiota-metabolite-host axis (glutaminase inhibitors): Devimistat (CPI-613®), a novel mitochondrial metabolism inhibitor, disrupts the tricarboxylic acid (TCA) cycle by targeting two critical carbon entry points. Specifically, it inhibits the pyruvate dehydrogenase and α-ketoglutarate dehydrogenase complexes, thereby blocking the incorporation of glucose- and glutamine-derived carbon units into mitochondrial energy production. Furthermore, amino acid metabolism (e.g., glutamine, branched-chain amino acid-leucine and glutamate), regulates the mammalian target of rapamycin (mTOR) signaling pathway in pancreatic cancer cells [19, 44]. Thus, inhibiting Pseudomonas-induced glutamate (glutamine) catabolism may synergistically enhance the efficacy of mTOR inhibitors (e.g., everolimus) and chemotherapy regimens (e.g., gemcitabine or FOLFIRINOX). Therefore, intervention strategies targeting the Pseudomonas-amino acid metabolism axis could offer novel therapeutic approaches for the treatment of pancreatic cancer. In summary, Pseudomonas, glutamate, glutamine, and branched-chain amino acid metabolites (e.g., alpha-ketoisovaleric acid) may emerge as novel biomarkers and therapeutic targets for pancreatic cancer. (ii)Postoperative recurrence monitoring and prognostic assessment: Pseudomonas and its associated metabolites could serve as dynamic biomarkers for monitoring postoperative recurrence in pancreatic cancer. By tracking changes in microbial abundance and metabolite levels, the risk of recurrence may be detected earlier. This approach could complement traditional imaging-based monitoring, offering a more sensitive and timely follow-up strategy. (iii) Promoting the development of precision medicine: The heterogeneity in microbiota and metabolite profiles among pancreatic cancer patients provides a foundation for personalized treatment strategies. By analyzing the microbiome and metabolic profiles of individual patients, treatment responsiveness can be predicted, enabling the development of personalized interventions, such as microbial therapies or metabolism-modifying drugs. These findings not only enhance our understanding of the mechanisms underlying pancreatic cancer but also provide a foundation for the development of innovative diagnostic and therapeutic strategies targeting microbial and metabolic pathways. Such approaches hold the potential to significantly improve the prognosis and treatment outcomes for pancreatic cancer.
Limitations of this study and strategies to address are as follow: (i) Sample size: The sample size in this study is relatively small, especially the reduction of samples for 16S sequencing, which may limit the ability to fully capture the diversity and heterogeneity of the microbiome and metabolome in pancreatic cancer patients. The implications of this reduction on the generalizability of results: ①Insufficient Representativeness: Given the substantial inter-individual variability in microbiomes, analyzing only 16 samples may not adequately reflect the microbiome characteristics of the entire study population. This reduction in sample size may lead to biased research outcomes, failing to accurately capture the comprehensive relationship between the microbiome and the study variables. ②Reduced Statistical Power: A smaller sample size may in result insufficient statistical power, thereby increasing the risk of Type I errors (false positives) and Type II errors (false negatives). This can make it difficult to detect biologically significant differences. Consequently, some potential microbiome features or biomarkers may be overlooked, thereby affecting the reliability of the study conclusions. ③Limited Extrapolation Capability: Conclusions drawn from a smaller sample may be restricted when extrapolated to or larger more complex populations. The composition and function of the microbiome are influenced by a variety of factors, such as diet, environment, and genetic background. For instance, the alpha diversity and abundance of Pseudomonas were likely not significantly influenced by tumor staging, based on our results. (ii) The absence of stage IV (M1) patients: this finding may be limited because all included samples were M0, and no stage IV (M1) cases were included. We acknowledge the limitation of lacking stage IV (M1) samples. Advanced-stage microbiome profiles likely diverge significantly from early/mid-stage patients. This sampling gap constrains comprehensive understanding of microbiome evolution throughout pancreatic cancer progression, thereby constraining clinical generalizability across disease stages. To address this issue, it has to explicitly states: ①conclusions are generalizable to resectable pancreatic cancer cohorts, with no direct extrapolation to stage IV or non-surgical candidates. ②Multicenter trials with expanded cohorts (incorporating stage IV patients) will evaluate population-wide generalizability and strengthen conclusion validity. Therefore, it is necessary to validate these findings in a larger cohort. Our explanations and proposed solutions are as follows: ①The heterogeneity observed within the sample cohort is driven by tumor microenvironment heterogeneity in pancreatic cancer [62], variability in intratumoral microbiome distribution [38], and the random selection of the 47 samples included. ②We acknowledge that the reduced sample size for 16S sequencing is relatively small, which may limit the generalizability of the results. However. To ensure the quality of the intergroup difference analysis (tumor vs. adjacent tissue), we employed strict selection criteria to exclude samples with high dispersion in a paired manner. This approach ensured a relatively ideal consistency within the samples of each group, thereby enhancing the reliability of the intergroup comparison results from a statistical and methodological rigor. ③The validity of using small sample cohorts in exploratory research is well-recognized(sample size varying from 12 to 20 case) [20, 21, 63–65]. And small sample cohorts in exploratory research are beneficial as they provide rapid preliminary results that can guide future large-scale studies. ④Validation in a larger, more diverse cohort (To enhance the generalizability of our research findings, we plan to expand the sample size to better capture the diversity of the microbiome in future study. We intend to initiate a multicenter study cohort, incorporating pancreatic cancer patients from five high-volume pancreatic centers across three regions in China in subsequent studies.) Furthermore, integrating and analyzing existing large-scale microbiome datasets (e.g., TCMbio) may help identify more robust microbiome features. These will include patients with different stages (I- IV) and pathological subtypes of PC, as well as healthy controls, would strengthen comparisons and provide more robust validation. In summary, although a reduced sample size may have certain impacts on the generalizability of the research findings, expanding the sample size and conducting validation studies can significantly enhance the reliability and applicability of the research conclusions. (iii) Bias in microbiome data: Microbiome analysis based on 16S rDNA sequencing may introduce biases. This technique does not comprehensively characterize all bacterial species, and non-bacterial components of the microbiome, such as Fungi and viruses, are often overlooked. Additionally, 16S rDNA sequencing has Limitations in identifying species at the strain level. However, the primary objective was to identify bacterial community structure shifts associated with host metabolic phenotypes. 16S sequencing remains the gold standard for such species and strain-level microbiome analysis, with proven reproducibility across large cohorts [66]. Given the need for paired multi-omics data (metabolomics) from clinical samples, 16S sequencing provided cost-effective bacterial profiling while preserving budget for functional assays. This study also employed metabolomic sequencing and analysis. We correlated PICRUSt2-predicted metabolic functions (based on computer calculations) from 16S sequencing data with metabolomic data, including pathways such as arginine and proline metabolism, arginine biosynthesis, and glutamate metabolism. This approach validated the reliability of our conclusions. Therefore, although PICRUSt2 functional prediction has inherent biases, our study can enhance the reliability and scientific rigor of the research results to some extent by cautiously interpreting the results, combining with other methods (such as metabolomics), and keeping track of database updates. While less precise than metagenomics, this approach predicts the metabolic functions of the microbial community and correlates them with relevant modules in metabolomics. However, complementary approaches, such as metagenomics or metatranscriptomics, and the inclusion of other omics technologies (e.g., proteomics), can provide comprehensive genomic information about microbial communities, enabling strain-level identification and functional gene analysis and reveal dynamic expression profiles of these communities [67–69]. These new technologies could explore the structure and function of intratumoral microbiota more thoroughly and comprehensively in pancreatic cancer [69]. Therefore, we will validate our findings by metagenomics or metatranscriptomics in a larger cohort in subsequent research. (iv) Deficiencies in causality: Current data indicate potential associations between intra-tumoral Pseudomonas species and amino acid (BCAA) metabolic reprogramming. However, mechanistic inferences tracing microbiota-metabolism-signaling cascades (specifically Pseudomonas → IRE1 → autophagy and Pseudomonas → BCAA → mTOR pathways) remain hypothetical statements, requiring causal validation through future experiments investigating these pathways' regulatory roles in pancreatic cancer development. Such causal validation will employ in vivo models (animal models) or pancreatic cancer organoid/Pseudomonas co-cultures with independent BCAA metabolism and IRE1 signaling interventions. Although a significant correlation between Pseudomonas and amino acid metabolism was identified, the causal relationship remains unclear. Further in vitro and in vivo experiments, including animal models, are necessary to validate whether Pseudomonas influences pancreatic cancer progression through amino acid metabolism. The proposed study will utilize germ-free KPC mice to investigate the changes in metabolic profiles following colonization with Pseudomonas (n = 10 per group). The experimental design was divided into four groups, namely Experimental Group 1 (Colonization with Pseudomonas, clinically isolated), Control Groups 1 (Colonization with non-pathogenic Escherichia coli, negative control), Control Group 2(Germ-free control group) and Control Group 3(Colonization with Pseudomonas + treatment with a sensitive antibiotic: amikacin). Then metabolite detection at multiple time points within 2 weeks (e.g., 1 day, 3 days, 7 days) to elucidate the distribution and changes of amino acid metabolites by spatial metabolite imaging. By comparing the metabolomic data between the colonization group and the control groups, the causal effects of Pseudomonas on host metabolism can be directly assessed. Additionally, advanced tools such as CRISPR could be used to investigate the functional role of these metabolic pathways within the intratumoral microbiota. CRISPR knockout strains of key amino acid metabolism genes in Pseudomonas (e.g., genes encoding key enzymes in branched-chain amino acid metabolism or glutamine metabolism) will be constructed and compared with wild-type (WT) strains. These strains will be inoculated into germ-free C57BL/6 mice (n = 10 per group) to assess changes in amino acid metabolism levels by spatial metabolite imaging between the knockout and control groups. (v) Hypothesis-Verification Gap. This study proposes two Pseudomonas-focused research hypotheses with experimental verification gaps: (1) BCAA-mTOR Axis Hypothesis: Pseudomonas species may activate mTOR signaling by regulating BCAA metabolites within the tumor microenvironment. Supported by: ① observed correlations between Pseudomonas and BCAA metabolites; Verification gaps: ① Pseudomonas colonization/eradication functional validation; ② BCAA metabolic flux tracing; ③ mTOR pathway activity modulation assays. (2) IRE1-Autophagy Axis Hypothesis: Pseudomonas species may trigger endoplasmic reticulum stress → IRE1 activation → autophagy. Supported by: ①literature-documented interactions between microbiota (P. aeruginosa), endoplasmic reticulum stress, IRE1, and autophagy. Verification gaps: ① Pseudomonas colonization/eradication studies; ② IRE1 inhibitor interventions; ③ functional assays (e.g., TEM autophagy flux quantification). This work advances novel hypotheses regarding Pseudomonas-mediated regulatory mechanisms in pancreatic cancer progression, necessitating validation through follow-up studies such as (but not limited to) pancreatic cancer organoid/Pseudomonas co-cultures and germ-free murine models. (vi) Biases introduced by sample handling or experimental variability and corresponding processing strategies: ①Sample handling biases include sample collection and preservation (the timing, location, and methods of sample collection, as well as the conditions of sample preservation, can influence the composition of microbial communities and metabolites), DNA or metabolite extraction (different methods for DNA or metabolite extraction can lead to variations in extraction efficiency and quality, thereby affecting the results of 16S sequencing and metabolomics analysis) and sample contamination (contamination during sample collection, storage, and extraction of DNA can interfere with the analysis of target intratumoral microbiota).②Experimental variation biases include PCR amplification (differences in PCR primers, reaction conditions, or operators can lead to variations in amplification efficiency, affecting the accuracy of 16S sequencing), sequencing platforms (different sequencing platforms or batches can result in variations in data quality, impacting the analysis of microbial communities), metabolomics analysis (different instruments, analytical methods, or operators can cause systematic biases in metabolite detection, affecting the results of metabolomics analysis) and data processing (different data processing methods or analytical software can lead to differences in results, affecting the final biological interpretation). ③Specific strategies to reduce biases about sample handling include standardized operating procedures (develop and strictly adhere to standardized protocols for sample collection and preservation), standardized sample reservation conditions (samples are processed or stored under appropriate conditions to prevent changes in the composition of microbial communities and metabolites), and validated DNA and metabolite extraction methods (select validated methods and use the same kits and operators in experiments to reduce differences in extraction efficiency). ④ Addressing experimental variation, we can minimize the possibility of batch effects in the experimental design phase, regularly calibrate instruments to ensure consistent performance, and standardize and homogenize training for operators to ensure consistent experimental operations. For data processing and analysis, we can implement rigorous quality control during data processing and analysis to exclude low-quality data, use appropriate statistical methods (e.g., the ComBat algorithm) to detect and correct batch effects before differential analysis, and apply bioinformatics tools (e.g., Decontam) to filter out contaminants [70]. In summary, these limitations highlight areas for improvement in future studies. Expanding the sample size (including all clinical stage), integrating multi-omics data, conducting mechanistic validation experiments, and addressing sample handling and experimental bias will enhance both the scientific understanding and clinical applicability of these findings.
The following directions deserve further exploration to deepen the understanding of the pancreatic cancer microenvironment and to facilitate the advancement of basic research to clinical translation: (i) Validation of the causal relationship between Pseudomonas and amino acid metabolism. To verify whether Pseudomonas directly regulates pancreatic cancer development and progression through amino acid metabolism, animal models or organoid models should be utilized. This would clarify the causal relationship between Pseudomonas and key metabolites (e.g., α-ketoisovaleric acid), providing a foundation for the subsequent development of therapeutic targets. (ii) Association between Pseudomonas and the efficacy of anticancer drugs. It is crucial to explore whether Pseudomonas and its metabolic activities influence the sensitivity of pancreatic cancer patients to conventional chemotherapies (e.g., gemcitabine or FOLFIRINOX) or targeted therapies. Understanding this relationship could inform the development of personalized treatment regimens and potentially reduce therapeutic resistance. (iii) Metabolite target development and validation. Developing inhibitors or mimetics targeting key metabolites (e.g., α-ketoisovaleric acid) and investigating their therapeutic potential would offer valuable insights for drug development based on metabolic targets. This could open new avenues for the treatment of pancreatic cancer by modulating specific metabolic pathways. (iv) Mechanisms of the bacterial-metabolite-immunity axis. Future studies should focus on understanding how Pseudomonas and its associated metabolites contribute to pancreatic cancer progression by regulating the tumor immune microenvironment, including immunosuppressive cells and inflammatory factors. Identifying key nodes in the microbial-metabolic-immune network could lead to novel strategies for immunotherapy. (v) Promoting multidisciplinary integration in pancreatic cancer research. Integrating microbiomics, metabolomics, and cancer biology provides a comprehensive approach to studying the complex microenvironment of pancreatic cancer. This multidimensional analysis enhances our understanding of tumor biology and facilitate the development of more effective diagnostic and therapeutic strategies. (vi) Validation with a larger cohort is needed. We plan to initiate multicenter cooperation to build a multicenter cohort of pancreatic cancer patients, incorporating more samples to validate our findings, and enhance the generalizability of our research results. The multicenter cohort design is as follows: ①Cohort 1 (Retrospective Validation): Integrate microbiome, metabolome, and clinical data related to pancreatic cancer from three high-volume pancreatic centers of three large tertiary-A level hospitals affiliated with our university (n = 300) to verify the consistency of the research findings.② Cohort 2 (Prospective Multicenter Cohort): Collaborate with five large tertiary-A level hospitals in three regions in China to establish a multicenter pancreatic cancer cohort (n = 500) to assess the generalizability of the study's conclusions. Furthermore, we plan to collaborate with two existing partner-pancreatic cancer research institutions to share data and conduct integrated analyses to further validate our findings.
Conclusions
Collectively, this study provides preliminary evidence of an interaction between Pseudomonas and amino acids in the pancreatic cancer microenvironment, suggesting a potential association with its regulation. It proposes a novel theoretical framework for the "microbe-metabolism-cancer" mechanism, which could enhance our understanding of pancreatic cancer pathogenesis. However, the causal relationship and underlying mechanisms remain unexplored experimentally and require further validation. These findings highlight future research possibilities for exploring diagnostic and therapeutic implications in pancreatic cancer, but they do not establish direct applications for early diagnosis, personalized treatment, or prognosis improvement.
Supplementary Information
Supplementary Material 1. Supplementary Figure 1 Spearman correlation coefficient matrix heatmap of significantly different microbial communities and significantly different metabolites. Note: This matrix heatmap displayed the correlation between significantly different microbial communities and significantly different metabolites, as well as the correlation among significantly different metabolites and among significantly different microbial communities. The heatmap is divided into four quadrants by the blue dashed line (┄┄┄). The upper-left quadrant displays the correlation among significantly different microbial communities, while the lower-right quadrant shows the correlation among significantly different metabolites. The upper-right and lower-left quadrants display the correlation between significantly different microbial communities and significantly different metabolites, which are mirror-symmetrical. The Spearman correlation coefficient value (r) ranges from -1 to +1. The correlation coefficient (r) is represented by color, with an r value greater than 0 indicating a positive correlation (represented by red). The deeper the color, the stronger the correlation.
Supplementary Material 2. Supplementary Figure 2 Correlation analysis between alpha diversity and stages (I-III). (A) Raincloud plots of the correlation analysis between ACE index and clinical staging, (B) Raincloud plots of the correlation analysis between Chao1 index and clinical staging, (C) Raincloud plots of the correlation analysis between Shannon index and clinical staging, (D) Raincloud plots of the correlation analysis between Simpson index and clinical staging.
Supplementary Material 3. Supplementary Figure 3 Correlation analysis between relative abundance of Pseudomonas and clinical stages.(A) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and T stage (T1-3), (B) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and N stage (N0-2), (C) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and clinical stage(I-III).
Acknowledgements
Dr. Dong Luo appreciates the valuable support from Tao Lab at Cancer Research Institute, Central South University, Chang Lab (led by Prof. Shi Chang) in the Department of General Surgery, Xiangya Hospital, Central South University, and colleagues including Prof. Huihuan Tang, Prof. Jun Zhou, Prof. Shuai Liang et al. in the Department of Pancreatic Surgery, Xiangya Hospital, Central South University. Dr. Dong Luo also appreciate the nice support from Xiaolin Dou from Department of General Surgery, Xiangya Hospital, Central South University during the revisions.
Authors’ contributions
Dong Luo: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing–original draft, Writing–review and editing Yixiong Li: Writing – review & editing, Project administration, Funding acquisition Qizhen Chen: Writing –review & editing, Methodology. Jianbo Yang: Formal analysis, Investigation, Methodology. Yongguang Tao: Conceptualization, Writing–review and editing. Liandong Ji: Writing – review & editing, Project administration, Investigation, Conceptualization, Supervision. Xuejun Gong: Writing – review & editing, Methodology, Funding acquisition, Conceptualization, Supervision.
Funding
This research was supported by The Science and Technology Innovation Program (The Key Research and Development Program) of Hunan Province of China 2024 (2024JK2111).
Data availability
The data that support the findings of this study are provided within the manuscript and reviewers have access to the supporting data by the link: https://pan.baidu.com/s/1UQMUwhwnhmVhhP48Fpu_Kg (Click the above link, and then enter the extraction code: 6666).
Declarations
Ethics approval and consent to participate
All the studies were conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. CommonRule) and tissue and clinical information were used after approval from the Ethics Committee of Xiangya hospital of Central South University (2025081285), which approved the patient consent forms or waiver of consent. Pancreatic cancer samples were provided by the Biobank of Xiangya Hospital, Central South University, which has been accredited by China National Accreditation Service for Conformity Assessment (CNAS). The collection, storage, and use of samples were in accordance with ISO 20387: 2018 Biotechnology-Biobanking-General Requirements for Biobanking (CNAS-CL10 Accreditation Criteria for the Quality and Competence of Biobank).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liandong Ji, Email: ja19841013@csu.edu.cn.
Xuejun Gong, Email: peigong158@csu.edu.cn.
References
- 1.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4:e214708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16:11–26. [DOI] [PubMed] [Google Scholar]
- 4.Xu J. China Anti-Cancer Association guidelines for the holistic integrative management of pancreatic cancer (abridged version). Chin J Clin Oncol 2023; 50:487-496.
- 5.Meneses-Medina MI, Gervaso L, Cella CA, Pellicori S, Gandini S, Sousa MJ, et al. Chemotherapy in pancreatic ductal adenocarcinoma: when cytoreduction is the aim. A systematic review and meta-analysis. Cancer Treat Rev. 2022;104:102338. [DOI] [PubMed] [Google Scholar]
- 6.Del Chiaro M, Sugawara T, Karam SD, Messersmith WA. Advances in the management of pancreatic cancer. BMJ. 2023;383:e073995. [DOI] [PubMed] [Google Scholar]
- 7.Sherman MH, Beatty GL. Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance. Annu Rev Pathol. 2023;18:123–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ullman NA, Burchard PR, Dunne RF, Linehan DC. Immunologic strategies in pancreatic cancer: making cold tumors hot. J Clin Oncol. 2022;40:2789–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao X, Ren Y, Feng M, Wang Q, Wang Y. Metabolic reprogramming due to hypoxia in pancreatic cancer: implications for tumor formation, immunity, and more. Biomed Pharmacother. 2021;141:111798. [DOI] [PubMed] [Google Scholar]
- 10.Ren J, Ren B, Liu X, Cui M, Fang Y, Wang X, et al. Crosstalk between metabolic remodeling and epigenetic reprogramming: a new perspective on pancreatic cancer. Cancer Lett. 2024;587:216649. [DOI] [PubMed] [Google Scholar]
- 11.Meng YF, Fan ZY, Zhou B, Zhan HX. Role of the intratumoral microbiome in tumor progression and therapeutics implications. Biochimica et Biophysica Acta (BBA). 2023;1878:189014. [DOI] [PubMed] [Google Scholar]
- 12.Huang JT, Mao YQ. The impact of the microbiome in cancer: targeting metabolism of cancer cells and host. Front Oncol. 2022;12:1029033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Jin M, Liu Y, Jin L. Gut microbiota: its potential roles in pancreatic cancer. Front Cell Infect Microbiol. 2020;10:572492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Z, Zhang W, Zhang Z, Sha G, Wang D, Tang D. Intratumoral microbiota: a new force in diagnosing and treating pancreatic cancer. Cancer Lett. 2023;554:216031. [DOI] [PubMed] [Google Scholar]
- 16.Picozzi VJ. Pancreatic cancer: new approaches to drug therapy. Int J Surg. 2024;110:6070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong Z, Yang J, Liu J, Meng Q, Hua J, Tan Z, et al. Dense stroma activates the TGF-beta1/FBW7 axis to induce metabolic subtype switching in pancreatic cancer. Int J Surg. 2025;111:1891–903. [DOI] [PubMed] [Google Scholar]
- 18.Luo D, Li Y, Ji L, Gong X. 249P the intratumoral metabolic characterization and potential mechanisms in pancreatic ductal adenocarcinoma. Ann Oncol. 2024;35:S1497. [Google Scholar]
- 19.Fu S, Xu S, Zhang S. The role of amino acid metabolism alterations in pancreatic cancer: from mechanism to application. Biochimica et Biophysica Acta (BBA). 2023;1878:188893. [DOI] [PubMed] [Google Scholar]
- 20.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(795–806):e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell. 2022;40:1240-1253.e1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, et al. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Cancer Res. 2017;77:1783–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, Xia H, Tan X, Shi C, Ma Y, Meng D, et al. Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduct Target Ther. 2024;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue C, Jia J, Gu X, Zhou L, Lu J, Zheng Q, et al. Intratumoral bacteria interact with metabolites and genetic alterations in hepatocellular carcinoma. Signal Transduct Target Ther. 2022;7:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan L, Pan L, Wang Y, Zhao J, Fang L, Zhou Y, et al. Characterization of the landscape of the intratumoral microbiota reveals that Streptococcus anginosus increases the risk of gastric cancer initiation and progression. Cell Discov. 2024;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilhan ZE, Łaniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine. 2019;44:675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. 2023;615:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, Liu X, Ren J, Wang X, Zhou F, Huang S, et al. Integrated analysis of microbiome and metabolome reveals signatures in PDAC tumorigenesis and prognosis. Microbiol Spectr. 2024;12:e0096224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40:920–38. [DOI] [PubMed] [Google Scholar]
- 32.Heo YJ, Hwa C, Lee GH, Park JM, An JY. Integrative multi-omics approaches in cancer research: from biological networks to clinical subtypes. Mol Cells. 2021;44:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. 2023;15:2166700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, Zhao Z, Peng M, Zhang L, Wang C, Luo F, et al. Multi-omics analysis reveals the interplay between intratumoral bacteria and glioma. mSystems. 2025;10:e0045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong F, Fang C, Zhang Y, Duan L, Du D, Xu G, et al. Abundance and metabolism disruptions of intratumoral microbiota by chemical and physical actions unfreeze tumor treatment resistance. Adv Sci. 2022;9:e2105523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kand’ár R, Záková P, Jirosová J, Sladká M. Determination of branched chain amino acids, methionine, phenylalanine, tyrosine and alpha-keto acids in plasma and dried blood samples using HPLC with fluorescence detection. Clin Chem Lab Med. 2009;47:565–72. [DOI] [PubMed] [Google Scholar]
- 37.Walker SP, Tangney M, Claesson MJ. Sequence-based characterization of intratumoral bacteria-a guide to best practice. Front Oncol. 2020;10:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galeano Niño JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611:810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178:795-806.e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taglialegna A. Pseudomonas against cancer. Nat Rev Microbiol. 2023;21:131. [DOI] [PubMed] [Google Scholar]
- 43.Choi JK, Naffouje SA, Goto M, Wang J, Christov K, Rademacher DJ, et al. Cross-talk between cancer and Pseudomonas aeruginosa mediates tumor suppression. Commun Biol. 2023;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Santis MC, Bockorny B, Hirsch E, Cappello P, Martini M. Exploiting pancreatic cancer metabolism: challenges and opportunities. Trends Mol Med. 2024;30:592–604. [DOI] [PubMed] [Google Scholar]
- 45.Wu H, Fu M, Wu M, Cao Z, Zhang Q, Liu Z. Emerging mechanisms and promising approaches in pancreatic cancer metabolism. Cell Death Dis. 2024;15:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Encarnación-Rosado J, Sohn ASW, Biancur DE, Lin EY, Osorio-Vasquez V, Rodrick T, et al. Targeting pancreatic cancer metabolic dependencies through glutamine antagonism. Nat Cancer. 2024;5:85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouissam AJ, Hind C, Sami Aziz B, Said A. Inhibition of the PI3K/AKT/mTOR pathway in pancreatic cancer: is it a worthwhile endeavor? Ther Adv Med Oncol. 2024;16:17588359241284912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki T, Kishikawa T, Sato T, Takeda N, Sugiura Y, Seimiya T, et al. Mutant KRAS drives metabolic reprogramming and autophagic flux in premalignant pancreatic cells. Cancer Gene Ther. 2022;29:505–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018;8:276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang Z, Xu Y, Zhu Q. Early growth response 1 suppresses macrophage phagocytosis by inhibiting NRF2 activation through upregulation of autophagy during Pseudomonas aeruginosa infection. Front Cell Infect Microbiol. 2021;11:773665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Miguel-Perez D, Ortega FG, Tejada RG, Martínez-Única A, Peterson CB, Russo A, et al. Baseline extracellular vesicle miRNA-30c and autophagic CTCs predict chemoradiotherapy resistance and outcomes in patients with lung cancer. Biomark Res. 2023;11:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng X, Fang Y, Zou X, Wang X, Li Z. Therapeutic potential of Pseudomonas aeruginosa-mannose sensitive hemagglutinin (PA-MSHA) in cancer treatment. Microb Pathog. 2023;185:106422. [DOI] [PubMed] [Google Scholar]
- 54.Yousuf S, Qiu M, Voith von Voithenberg L, Hulkkonen J, Macinkovic I, Schulz AR, et al. Spatially Resolved Multi-Omics Single-Cell Analyses Inform Mechanisms of Immune Dysfunction in Pancreatic Cancer. Gastroenterology. 2023;165:891-908.e814. [DOI] [PubMed] [Google Scholar]
- 55.Colbert LE, El Alam MB, Wang R, Karpinets T, Lo D, Lynn EJ, et al. Tumor-resident Lactobacillus iners confer chemoradiation resistance through lactate-induced metabolic rewiring. Cancer Cell. 2023;41:1945-1962.e1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ning L, Zhou YL, Sun H, Zhang Y, Shen C, Wang Z, et al. Microbiome and metabolome features in inflammatory bowel disease via multi-omics integration analyses across cohorts. Nat Commun. 2023;14:7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mima K, Hamada T, Inamura K, Baba H, Ugai T, Ogino S. The microbiome and rise of early-onset cancers: knowledge gaps and research opportunities. Gut Microbes. 2023;15:2269623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang S, Li M, Yan L, He M, Lin H, Xu Y, et al. Metabolomics study reveals systematic metabolic dysregulation and early detection markers associated with incident pancreatic cancer. Int J Cancer. 2022;150:1091–100. [DOI] [PubMed] [Google Scholar]
- 59.Zuzčák M, Trnka J. Cellular metabolism in pancreatic cancer as a tool for prognosis and treatment (Review). Int J Oncol. 2022. 10.3892/ijo.2022.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohindroo C, Hasanov M, Rogers JE, Dong W, Prakash LR, Baydogan S, et al. Antibiotic use influences outcomes in advanced pancreatic adenocarcinoma patients. Cancer Med. 2021;10:5041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359:1366–70. [DOI] [PubMed] [Google Scholar]
- 62.Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22:131–42. [DOI] [PubMed] [Google Scholar]
- 63.Jeong JY, Kim TB, Kim J, Choi HW, Kim EJ, Yoo HJ, et al. Diversity in the extracellular vesicle-derived microbiome of tissues according to tumor progression in pancreatic cancer. Cancers (Basel). 2020. 10.3390/cancers12092346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. 2022;14:2073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. 2018;39:1068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlopoulos GA, Baltoumas FA, Liu S, Selvitopi O, Camargo AP, Nayfach S, et al. Unraveling the functional dark matter through global metagenomics. Nature. 2023;622:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aplakidou E, Vergoulidis N, Chasapi M, Venetsianou NK, Kokoli M, Panagiotopoulou E, et al. Visualizing metagenomic and metatranscriptomic data: a comprehensive review. Comput Struct Biotechnol J. 2024;23:2011–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang N, Kandalai S, Zhou X, Hossain F, Zheng Q. Applying multi-omics toward tumor microbiome research. Imeta. 2023;2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. Supplementary Figure 1 Spearman correlation coefficient matrix heatmap of significantly different microbial communities and significantly different metabolites. Note: This matrix heatmap displayed the correlation between significantly different microbial communities and significantly different metabolites, as well as the correlation among significantly different metabolites and among significantly different microbial communities. The heatmap is divided into four quadrants by the blue dashed line (┄┄┄). The upper-left quadrant displays the correlation among significantly different microbial communities, while the lower-right quadrant shows the correlation among significantly different metabolites. The upper-right and lower-left quadrants display the correlation between significantly different microbial communities and significantly different metabolites, which are mirror-symmetrical. The Spearman correlation coefficient value (r) ranges from -1 to +1. The correlation coefficient (r) is represented by color, with an r value greater than 0 indicating a positive correlation (represented by red). The deeper the color, the stronger the correlation.
Supplementary Material 2. Supplementary Figure 2 Correlation analysis between alpha diversity and stages (I-III). (A) Raincloud plots of the correlation analysis between ACE index and clinical staging, (B) Raincloud plots of the correlation analysis between Chao1 index and clinical staging, (C) Raincloud plots of the correlation analysis between Shannon index and clinical staging, (D) Raincloud plots of the correlation analysis between Simpson index and clinical staging.
Supplementary Material 3. Supplementary Figure 3 Correlation analysis between relative abundance of Pseudomonas and clinical stages.(A) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and T stage (T1-3), (B) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and N stage (N0-2), (C) Raincloud plots of the correlation analysis between relative abundance of Pseudomonas and clinical stage(I-III).
Data Availability Statement
The data that support the findings of this study are provided within the manuscript and reviewers have access to the supporting data by the link: https://pan.baidu.com/s/1UQMUwhwnhmVhhP48Fpu_Kg (Click the above link, and then enter the extraction code: 6666).