Abstract
Studies of the mutant gene in Huntington's disease, and for eight related neurodegenerative disorders, have identified polyglutamine (polyQ) expansions as a basis for cellular toxicity. This finding has led to a disease hypothesis that protein aggregation and cellular dysfunction can occur at a threshold of approximately 40 glutamine residues. Here, we test this hypothesis by expression of fluorescently tagged polyQ proteins (Q29, Q33, Q35, Q40, and Q44) in the body wall muscle cells of Caenorhabditis elegans and show that young adults exhibit a sharp boundary at 35–40 glutamines associated with the appearance of protein aggregates and loss of motility. Surprisingly, genetically identical animals expressing near-threshold polyQ repeats exhibited a high degree of variation in the appearance of protein aggregates and cellular toxicity that was dependent on repeat length and exacerbated during aging. The role of genetically determined aging pathways in the progression of age-dependent polyQ-mediated aggregation and cellular toxicity was tested by expressing Q82 in the background of age-1 mutant animals that exhibit an extended lifespan. We observed a dramatic delay of polyQ toxicity and appearance of protein aggregates. These data provide experimental support for the threshold hypothesis of polyQ-mediated toxicity in an experimental organism and emphasize the importance of the threshold as a point at which genetic modifiers and aging influence biochemical environment and protein homeostasis in the cell.
The folding and maintenance of proteins in their native conformation is essential to cellular function. Disruption of protein-folding homeostasis, leading to the appearance of protein aggregates, is associated with an increasing number of human diseases (1, 2). A prototypical class of these disorders, composed of at least eight progressive neurodegenerative diseases including Huntington's disease, is associated with genes containing (CAG)n trinucleotide repeats encoding polyglutamine (polyQ) tracts in otherwise unrelated proteins (3, 4). Expression of expanded polyQ, with or without flanking sequences, is sufficient to recapitulate the pathological features of the diseases in multiple model systems, supporting a central role for the expansion in the etiology of these disorders (5–7).
Molecular genetic studies have established that huntingtin alleles from normal chromosomes contain fewer than 30–34 CAG repeats, whereas those from affected chromosomes contain more than 35–40 repeats (8, 9). These observations have led to the suggestion of a 35–40-residue threshold at which the disease gene products are converted to a proteotoxic state. Analysis of patient databases has established a strong inverse correlation between repeat length and age of onset (9–11). However, both this correlation and disease penetrance are much weaker for repeats of 42 or fewer residues (12), suggesting that substantial variation in the behavior of polyQ-containing proteins can exist at near-threshold repeat lengths, which influences the course of pathology.
Despite intense study, the nature of the transition of polyQ proteins to a toxic form is not well understood. Protein aggregates are a pathological hallmark of polyQ-mediated diseases (13), and aggregate formation has been observed in numerous in vitro and in vivo systems with polyQ proteins, leading to the hypothesis that the molecular events that lead to polyQ aggregation mark the transition to the toxic form (14). However, several studies have described an imperfect or inverse correlation between aggregate formation and toxicity (15–18). Thus, whether aggregate formation is necessary in the transition leading to diminished cellular function remains a central, but unresolved, question.
To address the underlying principles of polyQ-mediated aggregation and cellular toxicity, we have used Caenorhabditis elegans expressing chimeric fusions of polyQ and the yellow fluorescent protein (polyQ-YFP). Whereas previous studies on polyQ-mediated toxicity in animal models have compared the effects of short (<30Q) or long (>60Q) repeats, in this study we have analyzed transgenic lines expressing nine repeat lengths ranging from Q0 to Q82 with an emphasis on polyQ lengths in the range of 30 to 40 glutamine residues (Q29, Q33, Q35, Q40, Q44). We reasoned that such an analysis would allow us to test directly the polyQ threshold hypothesis and would yield insights into the nature of the transition governing conversion of polyQ-containing proteins to toxic forms.
Methods
DNA Cloning.
Plasmids for expression of Q19-YFP and Q82-YFP in C. elegans body wall muscle were described (19). Constructs for expression of the repeat lengths were generated by PCR amplification of the appropriate polyQ-YFP cassette in pEYFP-N1 (CLONTECH) by using oligonucleotides containing restriction sites for NheI and KpnI. PCR amplicons were digested and ligated into the NheI and KpnI sites of pP30.38 containing the promoter and enhancer elements from the unc-54 myosin heavy-chain locus (20). RNA interference (RNAi) constructs were created by reverse transcription–PCR amplification of cDNA corresponding to age-1 (21) or daf-16 (22), digestion with KpnI/XbaI or SacI/SalI, respectively, and ligation into appropriately digested L4440 (ref. 23; gift of A. Fire). Successful construction of plasmids was confirmed by DNA sequencing.
C. elegans Methods.
Nematodes were handled by using standard methods (24). For generation of transgenic animals, plasmid DNAs encoding polyQ-YFP in pPD30.38 were linearized with PvuII and mixed (at 1 μg/ml) with PvuII-digested C. elegans genomic DNA (100 μg/ml). Mixtures were microinjected into the gonads of adult hermaphrodite N2 or age-1(hx546) animals. Transgenic F1 progeny were selected on the basis of fluorescence in muscle cells. Individual fluorescent F2 animals were picked to establish transgenic lines. At least three independent lines for each transgene were isolated and analyzed with similar results. Synchronized populations were isolated by collecting embryos from gravid adults after treatment with alkaline hypochlorite (2:5, vol/vol, bleach/1 M NaOH) for 10 min (25) or by collecting embryos laid by adult animals in a 6-h period. RNAi experiments were performed by growing animals on Escherichia coli strain HT115(DE3) transformed with the indicated plasmid or empty vector L4440 essentially as described (23).
Fluorescence Recovery After Photobleaching (FRAP).
Animals were mounted on a 2% agar pad on a glass slide, immobilized in 1 mM levamisole, and subjected to FRAP analysis using a Zeiss LSM 510 confocal microscope imaged through a 40× 1.0 numerical aperture objective with the 488-nm line for excitation. Areas indicated by boxes were bleached for 10 s at 100% power, and recovery images were acquired at the indicated times by using 7% power. Scanning time was 3 s.
Motility Assays.
Individual animals were picked to fresh spread plates and their tracks were recorded at different intervals by using a charge-coupled device camera and Leica dissection stereomicroscope (magnification, ×8). Digital images of the tracks were analyzed to determine the average velocity of the animals. Pixels in the images were converted to distances by using a ruler calibration macro in the OPENLAB (Improvision, Lexington, MA) software program. The distance traveled by each animal was determined by tracing its tracks in the image. Each data point was the average of two independent tracings of the same tracks. Dividing this distance by the time interval gave the motility index for each animal. Statistical significance of the results was determined by a χ2 test. For blinded Q40 motility assays (see Fig. 3F), adult animals were randomly picked from populations and measured for motility without knowledge of the aggregation phenotype. Once all motility assays were completed, the same animals were viewed by using fluorescence microscopy, and the number of aggregates was counted (see below). Because the polyQ transgenes were carried on extrachromosomal arrays, some animals in the population were nontransgenic and consequently provided internal controls. Motility values for wild-type (N2) and nontransgenic control groups were indistinguishable from one another.
Fig 3.
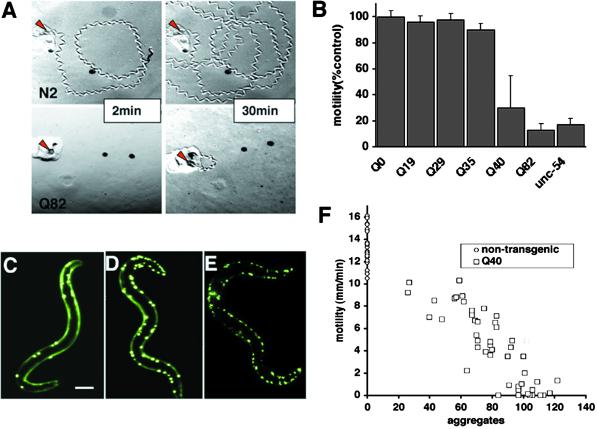
Expression of polyQ expansions in C. elegans muscle results in a motility defect that directly corresponds to aggregate formation. (A) Time-lapse micrographs illustrating tracks left by 5-day-old wild-type (N2) and Q82 animals 2 and 30 min after being placed at the position marked by the red arrow. (B) Quantitation of motility index for 4- to 5-day-old Q0, Q19, Q29, Q35, Q40, Q82, and unc-54(r293) animals. Data are mean ± SD for at least 50 animals of each type as a percentage of N2 motility. (C–E) Epifluoresence micrographs of late larval/young adult Q40 animals illustrating various numbers of aggregates in different animals. (Bar = 0.1 mm.) (F) Comparison of motility and aggregate number in adult Q40 animals (squares) and nontransgenic siblings (circles, no aggregates). Aggregate number is representative of number of muscle cells affected as the body-wall muscle cells had, on average, 1.3 ± 0.6 aggregates per cell (n = 212).
Aggregate Quantitation.
Animals were viewed at ×100 magnification with a stereomicroscope equipped for epifluorescence, and the number of polyQ aggregates was counted. Aggregates were defined as discrete structures with boundaries distinguishable from surrounding fluorescence on all sides. Aggregate size, measured by using confocal microscopy, typically ranged from 1 to 5 μm. At ×100 magnification, we were able to detect >80% of aggregates observable at higher magnifications. Repeated aggregate counts by the same observer and independent observers varied by less than 10%.
Results
Length-Dependent Threshold for Aggregation and Toxicity of polyQ Proteins.
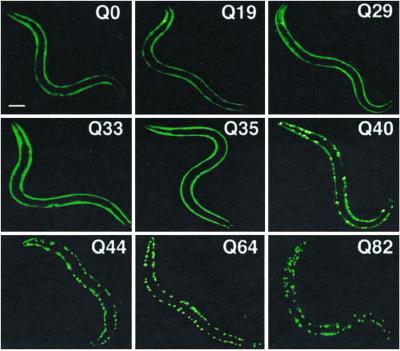
We previously described the formation of discrete cytoplasmic aggregates in body-wall muscle cells of C. elegans expressing Q82-YFP under the control of the unc-54 myosin heavy-chain promoter (19). In this study, we examined animals expressing Q0, Q19, Q29, Q33, Q35, Q40, Q44, Q64, and Q82 as chimeric fusions to YFP. In young adult animals (days 3–4) expressing repeats of Q35 or fewer, we observed diffuse fluorescence distribution in all expressing cells (Fig. 1). In contrast, animals expressing Q44, Q64, or Q82 exhibited focal fluorescence distribution corresponding to protein aggregates. Q40 animals displayed a striking polymorphic distribution with diffuse fluorescence in some cells and foci in others (Fig. 1). These results demonstrate a shift in the cellular distribution of the protein in young adult animals between Q35 and Q40.
Fig 1.
Length-dependent aggregation of polyQ-YFP fusion proteins in C. elegans. Epifluoresence micrographs of 3- to 4-day-old C. elegans expressing different lengths of polyQ-YFP (Q0, Q19, Q29, Q33, Q35, Q40, Q44, Q64, Q82). (Bar = 0.1 mm.)
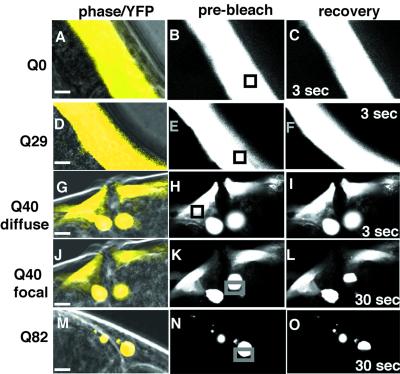
The change from diffuse to focal fluorescence in animals expressing Q19 or Q82, respectively, corresponds to a conversion of the biochemical state of the polyQ proteins from soluble to aggregate as detected in whole animal extracts (19). However, to investigate whether the cell-to-cell variation observed in Q40 animals reflected different in vivo states of polyQ proteins, we used a noninvasive method, FRAP. We reasoned that soluble YFP-tagged proteins in the cytoplasm would diffuse freely and recover rapidly after photobleaching as has been demonstrated for green fluorescent protein in solution and in tissue culture cells (26). After photobleaching, the fluorescence of Q0-YFP and Q29-YFP recovered completely within 3 s (Fig. 2 A–F), suggesting that both YFP alone (Q0) and Q29-YFP exhibited biochemical properties as soluble proteins in vivo. In contrast, fluorescence of Q82-YFP did not recover within 30 s after photobleaching (Fig. 2 M–O) and remained bleached after 5 min (data not shown), consistent with its properties as an aggregate. These data indicated that FRAP could be used as a tool to distinguish between different states of Q40-YFP expressed in adjacent cells of individual animals. Whereas diffuse Q40-YFP exhibited rapid recovery after photobleaching (Fig. 2 G–I), similar to that observed for Q0 and Q29, focal Q40-YFP exhibited slow recovery indicative of protein aggregates (Fig. 2 J–L). Although FRAP does not directly assess biochemical properties, the different recovery rates observed for diffuse and focal Q40-YFP are consistent with different biochemical states of Q40 within adjacent cells of the same animal.
Fig 2.
Determination of polyQ-YFP solubility in living animals by using FRAP. (Left: A, D, G, J, M) Merged phase-contrast and fluorescence images (pseudocolored yellow). (Center: B, E, H, K, N) Fluorescence images of the same region before photobleaching (prebleach). Boxes indicate the area that was subjected to photobleaching. (Right: C, F, I, L, O) Fluorescence images of recovery at the indicated times after photobleaching. The earliest time point possible to assess recovery of the chimeric YFP signal was at 3 s. (Bars = 3 μm.)
To determine whether the appearance of polyQ aggregates was associated with cellular dysfunction, we examined the motility of 4-day-old adult animals expressing polyQ tracts of 0, 19, 29, 35, 40, or 82 residues. C. elegans are maintained on agar plates with a lawn of E. coli. Consequently, as the animals move, their tracks can be visualized and quantified to establish a motility index. After 2 min, wild-type (N2) animals had moved 10 to 20 body lengths from the point of origin, whereas Q82 animals remained at or near the point of origin (Fig. 3A). Quantitation of these results revealed a ≈10-fold reduction in motility of young adult Q82 animals, corresponding to a defect similar to animals expressing mutant unc-54 myosin heavy chain (Fig. 3B). In contrast, Q19-, Q29-, or Q35-expressing animals that did not have polyQ aggregates exhibited motility similar to wild type (Fig. 3B). Q40 animals, which had aggregates in some cells but not in others, exhibited an intermediate motility defect with a high degree of variation in the intensity of loss of motility across a population (Fig. 3B).
Variation in Aggregate Formation Underlies Polymorphism in Q40-Mediated Motility Defect.
In addition to cell-to-cell differences in aggregate formation in any given animal, we had observed striking variation in Q40 populations regarding the number of aggregates observed in individuals (Fig. 3 C–E). Within a population of Q40 young adults, we observed animals with as few as 5 and as many as 140 aggregates despite similar expression levels of Q40-YFP protein, as determined by Western blotting of whole-animal extracts with antibody to green fluorescent protein followed by scanning densitometry (data not shown). To address whether variation in Q40 aggregation phenotypes was due to a heritable factor, we examined whether the number of aggregates in the parent influenced the number of aggregates in the progeny. Because C. elegans can reproduce as a hermaphrodite, three individuals with fewer than 20 polyQ aggregates (Q40 “low”, Fig. 3C) and four animals with more than 80 aggregates (Q40 “high”, Fig. 3E) were allowed to lay eggs for 6 h, and aggregates were counted in the progeny. The average number of aggregates per animal after 3 days was similar whether the parent had few or many aggregates (Q40 low progeny = 54 ± 21, n = 80; Q40 high progeny = 54 ± 21, n = 100). These results suggest that substantial variation can exist at intermediate polyQ lengths even in a uniform genetic background. Other explanations for this polymorphism include polyQ repeat expansion or contraction; however, if the repeats were dramatically unstable we might have expected strains of Q19 or Q29 that exhibited aggregates or strains of Q40 that were no longer polymorphic. We have not observed any drift in these transgenic strains, which have been maintained in continuous culture for more than 2 years and retained their original phenotypes.
The polymorphism of aggregation phenotypes (Fig. 3 C–E) and motility defects (Fig. 3B) in Q40 animals provided an opportunity to test whether the formation of aggregates was directly linked to cellular toxicity, which was accomplished by measuring motility and subsequently assessing, by fluorescence microscopy, the number of polyQ-YFP aggregates in the same animal. Q40 animals with the fewest aggregates exhibited a motility index that overlaps with that observed for the nontransgenic animals in the same population and separately with wild-type N2 animals, whereas Q40 animals with the largest number of aggregates exhibited a reduced motility similar to Q82 animals (Fig. 3F). Linear regression analysis resulted in an R2 value of −0.93, which reveals that greater than 90% of the variation in toxicity can be explained by the extent to which the protein has formed aggregates. Although these results provide evidence that formation of aggregates is correlated directly with toxicity, we cannot distinguish between aggregates themselves causing toxicity or a common mechanism leading to both aggregate formation and cellular dysfunction.
Aging-Dependent Shift in the Threshold for polyQ Aggregation and Toxicity.
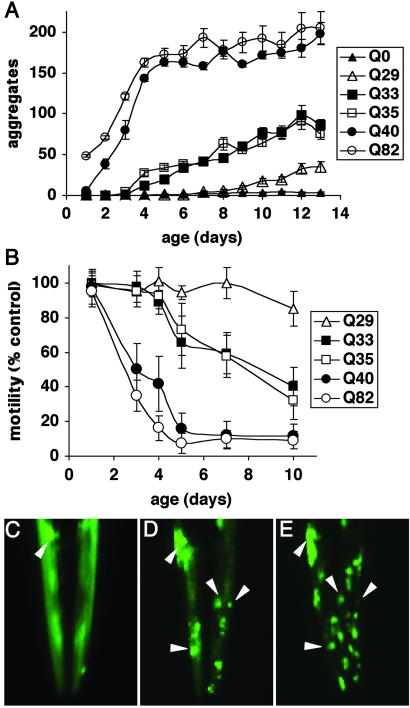
In further support for the existence of polymorphism at the threshold, we observed the appearance of protein aggregates as Q33 and Q35 animals aged (>4–5 days), which led us to perform an experiment in which individual Q0, Q29, Q33, Q35, Q40, and Q82 animals were examined daily for the appearance of protein aggregates and motility (Fig. 4 A and B). Relative to Q40 and Q82 animals that quickly accumulated aggregates and exhibited a rapid decline in motility (Fig. 4 A and B), Q33 and Q35 animals exhibited an initial lag before the gradual accumulation of aggregates to levels much lower, however, than for Q40 or Q82 (Fig. 4A). For example, aging-dependent aggregate accumulation can be seen by comparison of the same Q35 animal at 4, 7, and 10 days (Fig. 4 C–E). Q33 and Q35 animals also exhibited an age-dependent decline in motility (Fig. 4B). Q35-YFP fluorescence in young adults recovered rapidly after photobleaching, similar to that observed for Q0 or Q29 animals, whereas the Q35-YFP in older animals did not recover, consistent with conversion to the aggregated state (data not shown). These results reveal that the threshold for polyQ aggregation and toxicity is not static. At 3 days of age or less, only animals expressing Q40 or greater exhibit aggregates (Figs. 1 and 4A). However, at 4–5 days of age the threshold shifts as aggregates appear in Q33 and Q35 animals (Fig. 4A). The threshold again shifts to Q29 in aged animals (>9–10 days) (Fig. 4A). Thus, the threshold for polyQ aggregation is dynamic and likely reflects a balance of different factors including repeat length and changes in the cellular protein-folding environment over time.
Fig 4.
Influence of aging on polyQ aggregation and toxicity. (A) Accumulation of aggregates in Q82 (○), Q40 (•), Q35 (□), Q33 (▪), Q29 (▵), and Q0 (▴) during aging. Data are mean ± SEM. Twenty-four animals of each type are represented at day 1. Cohort sizes decreased as animals died during the experiment, but each data point represents at least five animals. (B) Motility index as a function of age for the same cohorts of animals described in A. Data are mean ± SD as a percentage of age-matched Q0 animals. (C–E) Epifluorescence micrographs of the head of an individual Q35 animal at 4 (C), 7 (D), and 10 (E) days of age, illustrating age-dependent accumulation of aggregates. Arrowheads indicate positions of the same aggregates on different days. In E, the animal is rotated slightly relative to its position in D.
Lifespan-Extending Mutation Delays the Onset of polyQ-Mediated Aggregation and Toxicity.
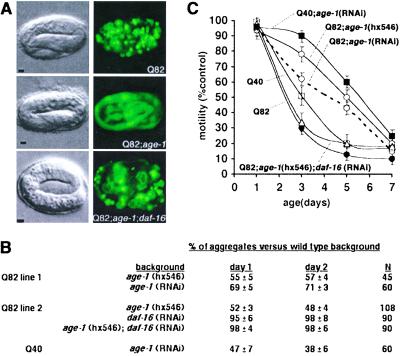
Our results reveal that the threshold for polyQ aggregation and cytotoxicity in vivo is dynamic throughout the lifetime of an animal. Does this dynamic behavior result from the intrinsic properties of a protein motif, or do changes over time reflect the influence of aging-related alterations in the cell? The availability of C. elegans mutants with extended lifespans allowed us to test these ideas directly. To accomplish this testing, we generated transgenic animals expressing Q82-YFP in the background of the age-1(hx546) mutation or age-1 RNAi. age-1 encodes a phosphoinositide 3-kinase that functions in an insulin-like signaling pathway, and mutations in this gene can extend the lifespan (21, 27, 28). Q82-YFP in the age-1(hx546) background (Q82; age-1) exhibited reduced aggregate formation in embryos relative to Q82-YFP in the wild-type background (Fig. 5A). Q82 aggregate formation was also reduced 30–50% during larval stages (1–2 days old) in age-1 animals compared with wild-type animals and was significantly lower until 4–5 days of age (data not shown). Parallel motility assays also demonstrated a striking delay in onset of the motility defect, consistent with slower aggregate accumulation in Q82;age-1 animals (Fig. 5C).
Fig 5.
An extended lifespan mutation delays polyQ aggregate accumulation and onset of toxicity. (A) Differential interference contrast (Left) and epifluorescence (Right) micrographs showing embryos expressing Q82 in wild-type (Top), age-1(hx546) (Middle), or age-1(hx546);daf-16(RNAi) (Bottom) genetic backgrounds. (Bars = 5 μm.) (B) Aggregate accumulation in larval animals expressing Q40 or Q82 in the indicated genetic backgrounds relative to aggregate accumulation in wild-type background. Mean ± SEM. (C) Motility index for animals expressing Q40 or Q82 in the indicated genetic backgrounds. Data are mean ± SEM for 30 animals of each type. Motility of nontransgenic wild-type and age-1 animals was similar to that of wild-type (N2).
To test whether loss of age-1 function would also influence aggregation and toxicity of other polyQ lengths, we subjected Q40 animals to age-1 RNAi. Both aggregation and onset of motility defects were delayed in Q40;age-1(RNAi) animals (Fig. 5 B and C). In wild-type animals, the kinase activity of AGE-1 is required in a signaling cascade that results in constitutive repression of the forkhead transcription factor DAF-16, leading to normal lifespan (22, 28, 29). Derepression of DAF-16 in age-1 animals results in an extended lifespan, and daf-16 mutations suppress the longevity phenotype (22, 29). To examine whether age-1 effects on longevity and polyQ aggregation and toxicity are mediated through similar regulatory pathways, we tested whether age-1 suppression of Q82 phenotypes was affected by inactivation of daf-16 by using RNAi. Q82;age-1;daf-16 animals exhibited aggregation and motility phenotypes similar to Q82 expressed in wild-type background, suggesting that lifespan extension and polyQ toxicity suppression mediated by age-1 share a common genetic pathway (Fig. 5 A–C).
Discussion
polyQ expansions are typically associated with neurodegenerative diseases in humans, yet the underlying principles of protein homeostasis and protein misfolding are universal properties of proteins in all cell types. Consistent with this premise is the appearance of polyQ-expansion protein aggregates in the yeast Saccharomyces cerevisiae and the expression of polyQ and α-synuclein protein aggregates in Drosophila (7, 30–32). Previous studies describing expression of polyQ-containing proteins in sensory neurons of C. elegans have demonstrated that numerous pathological features can be recapitulated (33, 34). The polyQ-length dependence of toxicity and variability among animals expressing near-threshold repeat lengths observed here suggests that, despite the obvious differences between C. elegans muscle cells and human neurons, the biochemical fates of polyQ proteins are indistinguishable. The demonstration that protein aggregation and toxicity are intensified during aging and the role of the age-1 mutation in suppressing these phenotypes highlight the utility of C. elegans as an animal model system to address these complex biochemical and behavioral phenotypes.
How might polyQ-initiated aggregates mediate the development of cellular toxicity in C. elegans? One explanation for aggregate-mediated toxicity results from observations that expanded polyQ tracts can sequester cellular proteins containing shorter polyQ domains, including transcription factors or coactivators such as CREB-binding protein (CBP) (35, 36). Recruitment of CBP into aggregates was shown to be associated with neuronal toxicity; moreover, reduced transcription from CBP-dependent genes was rescued by overexpression of CBP (36). Likewise, in C. elegans, we have shown that Q82 aggregates cause the relocalization of normally soluble Q19-YFP and a nuclear glutamine-rich splicing factor (HRP-1) into cytoplasmic aggregates (19). The predicted C. elegans proteome contains ≈200 proteins with polyQ or polar amino-acid-rich motifs, including the worm ortholog of CBP (37). It is not unreasonable, therefore, to suggest that some of these proteins are sequestered over time by the aggregates and have a role in polyQ-mediated toxicity in C. elegans. Another potential explanation for myocyte dysfunction could be disruption of the actin and myosin myofibrillar networks by polyQ aggregates (data not shown). It is not clear whether the size or location of the aggregates are important in myocyte dysfunction. polyQ aggregate-mediated disruption of neurofilament networks has been observed in cultured neuroblasts and has been suggested to contribute to polyQ-mediated cellular toxicity (38).
The threshold hypothesis of polyQ-mediated cytotoxicity suggests that expansion of a glutamine homopolymer beyond a critical length results in a transition in the disposition or activities of the disease gene products. Consistent with this idea, in vitro studies on polyQ peptides of various lengths have demonstrated nucleation-dependent aggregation kinetics with a lag phase and rate of accumulation that depends on repeat length (39, 40). The lag period and rate of aggregate accumulation, however, are not linear. For example, synthetic peptides of Q44 exhibit a lag period of several hours followed by very rapid aggregate accumulation, peptides of Q37 and Q41 aggregate less rapidly after a lag period of approximately 20 h, and peptides of Q25–Q32 aggregate very slowly with lag times of up to 100 h (40). These results establish that the intrinsic properties of polyQ proteins are consistent with the inverse correlation between repeat length and age-of-onset observed in human polyQ diseases. However, what had not been addressed were the properties of polyQ proteins at threshold in the crowded macromolecular environment of the cell.
Our ability to monitor the biochemical properties of polyQ proteins in transparent C. elegans provided an opportunity to examine aggregation kinetics and its effects on cellular function throughout the lifespan of a living organism. The kinetics and length dependence of aggregation observed in living C. elegans exhibited striking similarity to those observed in vitro (39, 40). Animals expressing Q82 rapidly accumulated aggregates starting in embryos. Aggregates developed in Q40 animals at a similar rate, but with a delay of 1–2 days. Q33 and Q35 animals accumulated aggregates at a much slower rate and to smaller numbers only after a lag period of 4–5 days, and aggregates in Q29 animals appeared only after a week. For each polyQ length tested, the development of a motility defect paralleled the rate of aggregate accumulation. Taken together, these data suggest that the intrinsic parameters governing self-association of polyQ motifs derived from studies with synthetic peptides are manifest in living animals and may underlie the relationship between repeat length and age-of-onset in human polyQ diseases. However, patients with the same repeat length, especially near the threshold, can exhibit markedly different ages-of-onset and disease courses (9–12), suggesting that these intrinsic parameters interact with modifiers of pathology.
The identification of age-1 as a genetic modifier of protein aggregation is intriguing, and one interpretation of these results is that age-1 and daf-16 define a genetic pathway that governs aging and may do so by influencing the biochemical events that that have an impact on protein homeostasis as monitored by the appearance of protein aggregates. For example, daf-16 could be a regulator of chaperone or proteasome activity or indirectly have effects by changes in metabolism influencing the overall synthesis or degradation rate of proteins. Insulin-like signaling in C. elegans regulates not only longevity but also entry into the alternative developmental state of dauer diapause (21, 22, 27–29). Entry into the dauer state results in elevated levels of molecular chaperones, such as Hsp70 and Hsp90 (41, 42). However, sequence analysis of the promoter regions from Hsp16, Hsp70, and Hsp90 homologs in the C. elegans genome did not reveal the presence of the previously characterized daf-16 family binding element TTGTTTAC (43), arguing against direct transcriptional regulation of chaperone genes by DAF-16.
Loss of age-1 function is pleiotropic, and animals bearing this mutation are resistant to a variety of environmental stresses, including heat shock and reactive oxygen species (44–46). No precedent exists for direct effects of insulin-like signaling on stress-response regulators, such as the heat-shock transcription factor. However, previous studies have shown that the heat-shock response is induced poorly during aging as a result of reduced heat-shock transcription factor activity (47, 48). Consequently, the ability of chaperone networks to respond to the appearance of misfolded and aggregation-prone proteins during aging would be compromised, which is consistent with forward and reverse genetic approaches that have identified molecular chaperones as suppressors in models of protein aggregation-related diseases (49–52).
The delay in onset of polyQ-associated phenotypes observed in age-1 animals implies that the rate of progression for proteinopathies is linked with the genetic regulation of aging. This finding, although unexpected, is supported by observations that the time until polyQ-mediated pathology develops (days in C. elegans, weeks in Drosophila, months in mice, and years in humans) correlates approximately with the lifespan of the organism. This link suggests that strategies to extend lifespan could also delay the onset of aging-related diseases characterized by the appearance of misfolded and aggregation-prone proteins.
Acknowledgments
We thank Ellen Nollen, Sally McFall, Li Tai, and other members of the Morimoto Laboratory for discussions and advice. We are grateful to Chris Ross and Greg Beitel for their advice and suggestions on the manuscript. We acknowledge the Caenorhabditis Genetics Center, supported by a grant from the National Institutes of Health, for providing C. elegans strains and the Cell Imaging Facilities both at Northwestern University Medical School and at the Evanston campus for microscope use. J.F.M. is supported by National Institute of General Medical Sciences Medical Scientist Training Program Fellowship GM08152. H.R.B. is supported by National Institute of General Medical Sciences Cellular and Molecular Biology of Disease Training Grant T32 GM08061. R.I.M. is supported by National Institutes of Health Grant GM38109, the Gollob Foundation, the Hereditary Disease Foundation, and a Coalition for the Cure grant from the Huntington's Disease Society of America.
Abbreviations
polyQ, polyglutamine
YFP, yellow fluorescent protein
FRAP, fluorescence recovery after photobleaching
RNAi, RNA interference
CBP, CREB-binding protein
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kopito R. R. & Ron, D. (2000) Nat. Cell Biol. 2, E207-E209. [DOI] [PubMed] [Google Scholar]
- 2.Horwich A. L. & Weissman, J. S. (1997) Cell 89, 499-510. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M. F. (1999) Trends Biochem. Sci. 24, 58-63. [DOI] [PubMed] [Google Scholar]
- 4.Orr H. T. (2001) Genes Dev. 15, 925-932. [DOI] [PubMed] [Google Scholar]
- 5.Mangiarini L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W. & Bates, G. P. (1996) Cell 87, 493-506. [DOI] [PubMed] [Google Scholar]
- 6.Ordway J. M., Tallaksen-Greene, S., Gutekunst, C. A., Bernstein, E. M., Cearley, J. A., Wiener, H. W., Dure, L. S., IV, Lindsey, R., Hersch, S. M., Jope, R. S., et al. (1997) Cell 91, 753-763. [DOI] [PubMed] [Google Scholar]
- 7.Warrick J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N. & Bonini, N. M. (1998) Cell 93, 939-949. [DOI] [PubMed] [Google Scholar]
- 8.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 9.Duyao M., Ambrose, C., Myers, R., Novelletto, A., Persichetti, F., Frontali, M., Folstein, S., Ross, C., Franz, M., Abbott, M., et al. (1993) Nat. Genet. 4, 387-392. [DOI] [PubMed] [Google Scholar]
- 10.Snell R. G., MacMillan, J. C., Cheadle, J. P., Fenton, I., Lazarou, L. P., Davies, P., MacDonald, M. E., Gusella, J. F., Harper, P. S. & Shaw, D. J. (1993) Nat. Genet. 4, 393-397. [DOI] [PubMed] [Google Scholar]
- 11.Andrew S. E., Goldberg, Y. P., Kremer, B., Telenius, H., Theilmann, J., Adam, S., Starr, E., Squitieri, F., Lin, B., Kalchman, M. A., et al. (1993) Nat. Genet. 4, 398-403. [DOI] [PubMed] [Google Scholar]
- 12.Brinkman R. R, Mezei, M. M., Theilmann, J., Almqvist, E. & Hayden, M. R. (1997) Am. J. Hum. Genet. 60, 1202-1210. [PMC free article] [PubMed] [Google Scholar]
- 13.Davies S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 14.Paulson H. L. (1999) Am. J. Hum. Genet. 64, 339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klement I. A., Skinner, P. J., Kaytor, M. D., Yi, H., Hersch, S. M., Clark, H. B., Zoghbi, H. Y. & Orr, H. T. (1998) Cell 95, 41-53. [DOI] [PubMed] [Google Scholar]
- 16.Saudou F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 17.Simeoni S., Mancini, M. A., Stenoien, D. L., Marcelli, M., Weigel, N. L., Zanisi, M., Martini, L. & Poletti, A. (2000) Hum. Mol. Genet. 9, 133-144. [DOI] [PubMed] [Google Scholar]
- 18.Huynh D. P., Figueroa, K., Hoang, N. & Pulst, S. M. (2000) Nat. Genet. 26, 44-50. [DOI] [PubMed] [Google Scholar]
- 19.Satyal S. H., Schmidt, E., Kitagawa, K., Sondheimer, N., Lindquist, S., Kramer, J. M. & Morimoto, R. I. (2000) Proc. Natl. Acad. Sci. USA 97, 5750-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fire A., Harrison, S. W. & Dixon, D. (1990) Gene 93, 189-198. [DOI] [PubMed] [Google Scholar]
- 21.Morris J. Z., Tissenbaum, H. A. & Ruvkun, G. (1996) Nature (London) 382, 536-539. [DOI] [PubMed] [Google Scholar]
- 22.Lin K., Dorman, J. B., Rodan, A. & Kenyon, C. (1997) Science 278, 1319-1322. [DOI] [PubMed] [Google Scholar]
- 23.Timmons L., Court, D. L. & Fire, A. (2001) Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- 24.Brenner S. (1974) Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis J. A. & Fleming, J. T. (1995) in Caenorhabditis elegans: Modern Biological Analysis of an Organism, eds. Epstein, H. F. & Shakes, D. C. (Academic, San Diego), pp. 4–29.
- 26.Swaminathan R., Hoang, C. P. & Verkman, A. S. (1997) Biophys. J. 72, 1900-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman D. B. & Johnson, T. E. (1988) Genetics 118, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tissenbaum H. A. & Ruvkun, G. (1998) Genetics 148, 703-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogg S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A. & Ruvkun, G. (1997) Nature (London) 389, 994-999. [DOI] [PubMed] [Google Scholar]
- 30.Feany M. B. & Bender, W. W. (2000) Nature (London) 404, 394-398. [DOI] [PubMed] [Google Scholar]
- 31.Krobitsch S. & Lindquist, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9, 13-25. [DOI] [PubMed] [Google Scholar]
- 33.Faber P. W., Alter, J. R., MacDonald, M. E. & Hart, A. C. (1999) Proc. Natl. Acad. Sci. USA 96, 179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker J. A., Connolly, J. B., Wellington, C., Hayden, M., Dausset, J. & Neri, C. (2001) Proc. Natl. Acad. Sci. USA 98, 13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffan J. S., Kazantsev, A., Spasic-Boskovic, O., Greenwald, M., Zhu, Y. Z., Gohler, H., Wanker, E. E., Bates, G. P., Housman, D. E. & Thompson, L. M. (2000) Proc. Natl. Acad. Sci. USA 97, 6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nucifora F. C., Sasaki, M., Peters, M. F., Huang, H., Cooper, J. K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, V. L., et al. (2001) Science 291, 2423-2428. [DOI] [PubMed] [Google Scholar]
- 37.Michelitsch M. D. & Weissman, J. S. (2000) Proc. Natl. Acad. Sci. USA 97, 11910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai Y., Onodera, O., Chun, J., Strittmatter, W. J. & Burke, J. R. (1999) Exp. Neurol. 155, 195-203. [DOI] [PubMed] [Google Scholar]
- 39.Scherzinger E., Sittler, A., Schweiger, K., Heiser, V., Lurz, R., Hasenbank, R., Bates, G. P., Lehrach, H. & Wanker, E. E. (1999) Proc. Natl. Acad. Sci. USA 96, 4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S., Berthelier, V., Yang, W. & Wetzel, R. (2001) J. Mol. Biol. 311, 173-182. [DOI] [PubMed] [Google Scholar]
- 41.Dalley B. K. & Golomb, M. (1992) Dev. Biol. 151, 80-90. [DOI] [PubMed] [Google Scholar]
- 42.Cherkasova V., Ayyadevara, S., Egilmez, N. & Reis, R. S. (2000) J. Mol. Biol. 300, 433-448. [DOI] [PubMed] [Google Scholar]
- 43.Furuyama T., Nakazawa, T., Nakano, I. & Mori, N. (2000) Biochem. J. 349, 629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lithgow G. J., White, T. M., Melov, S. & Johnson, T. E. (1995) Proc. Natl. Acad. Sci. USA 92, 7540-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen P. L. (1993) Proc. Natl. Acad. Sci. USA 90, 8905-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenyon C. (2001) Cell 105, 165-168. [DOI] [PubMed] [Google Scholar]
- 47.Fargnoli J., Kunisada, T., Fornace, A. J., Jr., Schneider, E. L. & Holbrook, N. J. (1990) Proc. Natl. Acad. Sci. USA 87, 846-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fawcett T. W., Sylvester, S. L., Sarge, K. D., Morimoto, R. I. & Holbrook, N. J. (1994) J. Biol. Chem. 269, 32272-32278. [PubMed] [Google Scholar]
- 49.Cummings C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T. & Zoghbi, H. Y. (1998) Nat. Genet. 19, 148-154. [DOI] [PubMed] [Google Scholar]
- 50.Warrick J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L. & Bonini, N. M. (1999) Nat. Genet. 23, 425-428. [DOI] [PubMed] [Google Scholar]
- 51.Fernandez-Funez P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature (London) 408, 101-106. [DOI] [PubMed] [Google Scholar]
- 52.Auluck P. K., Chan, H. Y., Trojanowski, J. Q., Lee, V. M. & Bonini, N. M. (2002) Science 295, 865-868. [DOI] [PubMed] [Google Scholar]