Abstract
Agrobacterium is a unique model system as well as a major biotechnological tool for genetic manipulation of plant cells. It is still unknown, however, whether host cellular factors exist that are limiting for infection, and whether their overexpression in plant cells can increase the efficiency of the infection. Here, we examined the effect of overexpression in tobacco plants of an Arabidopsis gene, VIP1, which encodes a recently discovered cellular protein required for Agrobacterium infection. Our results indicate that VIP1 is imported into the plant cell nucleus via the karyopherin αdependent pathway and that elevated intracellular levels of VIP1 render the host plants significantly more susceptible to transient and stable genetic transformation by Agrobacterium, probably because of the increased nuclear import of the transferred-DNA.
Agrobacterium elicits neoplastic growths on many plant species. This genetic modification results from the transfer and integration into the plant genome of a single-stranded copy (T-strand) of the bacterial transferred DNA (T-DNA) from the bacterial tumor-inducing plasmid. Plant genetic transformation by Agrobacterium requires the presence of two genetic components located on the bacterial tumor-inducing plasmid: (i) T-DNA, the actual genetic element transferred into the plant cell genome, and (ii) the virulence (vir) region, encoding most components of the protein machinery mediating T-DNA transfer (recently reviewed in refs. 1–4).
One of the central processes in Agrobacterium infection is nuclear import of the T-DNA (5). Presumably, this process is mediated by two Agrobacterium proteins, VirD2 and VirE2, which are thought to directly associate with the T-strand, forming a transport (T) complex (6). Nuclear import of the T-complex is most likely assisted by a host cell protein, VIP1, that specifically interacts with VirE2 (5). The molecular mechanism by which VIP1 mediates VirE2 nuclear import, however, is unclear. Specifically, it is unknown whether the classical nuclear import machinery is involved in this process.
Another unresolved question concerning the VIP1 function is whether it represents a factor limiting for infection and whether its overexpression in plant cells can increase the efficiency of the infection. Here, we show that VIP1 is imported into the plant cell nucleus via the karyopherin α-dependent pathway and that elevated intracellular levels of VIP1 render the host plants more susceptible to transient and stable genetic transformation by Agrobacterium.
Matrials and Methods
Two-Hybrid Protein–Protein Interaction Assay.
AtKAPα (7), VirE2 (8), and VirD2 (9) were amplified by PCR as BamHI–PstI, EcoRI–BamHI, and SmaI–SalI fragments, respectively, and cloned into the corresponding sites of pSTT91 (TRP1+; ref. 10), producing fusions with LexA. High-fidelity Pfu DNA polymerase (Stratagene) was used in all PCRs. VIP1 (5) was subcloned as a PCR-amplified BamHI–SalI fragment into the BamHI–SalI sites of pGAD424 (LEU2+, CLONTECH), producing a fusion with GAL4 activation domain. All DNA constructs were verified by dideoxynucleotide sequencing (11).
For the two-hybrid assay, the potential interactors were introduced into the Saccharomyces cerevisiae strain TAT7 [L40 (12)-ura3] (13) and grown for 2 days at 30°C on a leucine-, tryptophan-, and histidine-deficient medium. Histidine prototrophy indicated protein–protein interaction (12).
Nuclear Import of VIP1 in Yeast Cells.
The srp1–31 yeast strain containing a temperature-sensitive mutation in SRP1 (14) and its parental wild-type strain were a kind gift from Gerald Fink (Whitehead Institute, Massachusetts Institute of Technology). To inactivate Srp1p, the cells were grown for 24 hr at 25°C and then shifted for 6 hr to 37°C; this period of growth at the restrictive temperature inactivates >95% of the mutant Srp1p (14).
For nuclear import assays, VIP1 fusion to the C terminus of green fluorescent protein (GFP) was first constructed by cloning a PCR-amplified VIP1 ORF into the SalI–BamHI sites of pEGFP-C1 (CLONTECH), and then the entire GFP-VIP1 cassette was subcloned into the NcoI–BamHI sites of galactose-inducible expression vector pSJ101 (URA3+; ref. 15). The resulting pSJ-GFP-VIP1 plasmid was introduced into the wild-type and srp1–31 yeast strains, and the cells were grown as described above. For induction of GFP-VIP1 expression under the restrictive temperature, the cells were washed after 4 hr of growth at 37°C, supplemented with galactose (10% final concentration), and allowed to grow for additional 2 hr at 37°C. Under the permissive temperature, the cells were simply washed and supplemented with galactose 2 hr before harvesting the cultures. Harvested cells were fixed in 3.7% formaldehyde for 30 min, washed twice with PBS, and mounted on a coverslip. For nucleus-specific staining, 2 μl of 10 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) was added to the coverslip-mounted cells. Cells were observed under a Zeiss Axioplan 2 microscope with a Zeiss Axiocam and Zeiss axiovision 3.0.6 software.
Generation of VIP1 Tobacco Plants.
For generation of VIP1-transgenic plants, the Arabidopsis VIP1 ORF (5) was first inserted as a PCR-amplified SalI fragment into a plant expression vector, pCd, containing the 35S promoter of cauliflower mosaic virus, tobacco mosaic virus translational enhancer (16), and the nopaline synthase poly(A) signal. Then the entire expression cassette was subcloned as a BamHI–XbaI fragment into the binary vector pBIN19, carrying a kanamycin selection marker, to produce pBIN19-VIP1.
pBIN19-VIP1 was introduced into the disarmed Agrobacterium strain EHA105, which was then used to transform tobacco plants (Nicotiana tabacum cv. Turk) as described (17). Transgenic tobacco plants expressing VIP1 were selected on a kanamycin-containing medium and maintained and propagated in sterile conditions on an MS (Murashige and Skoog) basal medium (18) with no exogenous growth regulators. Plants were then transferred to soil in a greenhouse and allowed to set seed, and the transgenic progeny were selected by germinating the seeds on MS agar in the presence of kanamycin.
Assays for Agrobacterium-Induced Genetic Transformation.
Because the VIP1 plants are already resistant to kanamycin, to select for their genetic transformation by Agrobacterium, a binary vector with a different antibiotic resistance was required. Thus, the plasmid pBIG-HYG-GUS (19) was chosen, which carries on its T-DNA two reporter genes: hpt encoding hygromycin resistance, and an intron-containing uidA gene encoding β-glucuronidase (GUS).
For Agrobacterium infection, we used 9-mm-wide disks excised from leaves of 1-mo-old wild-type plants or VIP1 plants. For stable transformation, leaf disks were submerged in a culture of the Agrobacterium strain EHA105 (OD600 = 0.1, 0.5, or 1.0 as indicated for each individual experiment) harboring pBIG-HYG-GUS and incubated for 30 min at room temperature, followed by cocultivation for 48 hr at 25°C on tobacco regeneration medium (17). The disks were then washed three times in sterile distilled water, blotted dry, and cultured on the regeneration medium in the presence of 50 μg/ml hygromycin to select for transformed shoots and 300 μg/ml carbenicillin to eliminate Agrobacterium. Six weeks later, the developed shoots were separated from the parental leaf disk and counted under a stereoscope. For whole-shoot GUS staining, the shoots were allowed to grow for 2 more weeks, removed, and stained as described below for histochemical detection of transient GUS expression.
For transient T-DNA gene expression, the GUS activity within the leaf disks cocultivated for 48 hr at 25°C with Agrobacterium as described above for shoot regeneration was analyzed histochemically by staining with the chromogenic substrate X-Gluc (19). Individual GUS-stained areas were counted under a stereoscope (20). In control experiments, GUS activity was determined in leaf disks microbombarded with pRTL2-GUS (21), followed by incubation for 24 hr at 25°C. For biolistic delivery, 1 μg of DNA was adsorbed onto 0.24 μg of 1-μm gold particles according to the manufacturer's instructions and microbombarded into the target leaf disks at a pressure of 150 psi by using a portable Helios gene gun system (model PDS-1000/He, Bio-Rad). All transformations used at least 10–20 leaf disks per experimental system.
Northern Blot Analyses of VIP1 Plants.
Total RNA was isolated from 200 mg of leaf tissue by using the TRI-REAGENT extraction kit (Molecular Research Center, Cincinnati, OH), incubated at 37°C for 15 min in 20 mM MgCl2/2 mM DTT with 1.6 units of RNase-free RQ1-DNaseI (Promega) in the presence of ≈8 units of placental ribonuclease inhibitor (recombinant RNasin, Promega), and the reaction was terminated with 0.25 vol of DNase-stop mixture (50 mM EDTA/1.5 M sodium acetate/1% wt/vol SDS). RNA samples (10 μg per lane) were electrophoresed on a 1.7% formaldehyde/agarose gel and probed with 32P-labeled VIP1 cDNA, followed by autoradiography as described (22). rRNA within the analyzed RNA preparation was detected by ethidium bromide staining of agarose gels and served as an internal control for equal loading of the lanes.
Reverse Transcription (RT)-PCR Analysis of Early T-DNA Transcription.
Leaf disks were inoculated with the Agrobacterium strain EHA105 carrying the pBISN1 binary plasmid and analyzed by RT-PCR as described (23). In brief, total RNA was extracted from 200 mg of leaf tissue and treated with RQ1 RNase-free DNase, and 10-μg samples were reverse-transcribed with Moloney murine leukemia virus reverse transcriptase by using strand-specific reverse primers derived from uidA (23) or tobacco actin (24) gene sequences. The resulting cDNAs were PCR-amplified as described (23), by using a mixture of the corresponding forward and reverse primers. RT-PCR products were then detected by ethidium bromide staining of agarose gels. The uidA-specific forward and reverse primers, 5′-ACGATCAGTTCGCCGATGG3-′ and 5′-TCCCGCTAGTGCCTTGTCC-′, respectively, generate a 543-bp PCR product on the processed uidA transcript and a 732-bp PCR product on the unprocessed, intron-containing uidA transcript, whereas actin forward and reverse primers, 5′-TCACTGAAGCACCTCTTAACC-3′ and 5′-CAGCTTCCATTCCAATCATTG3-′, respectively, generate a 500-bp RT-PCR product. In control experiments, intron-containing T-DNA sequence within Agrobacterium cells was directly amplified by PCR, omitting the RT-PCR step.
Results
VIP1 Is Imported into the Cell Nucleus by a Karyopherin α-Dependent Pathway.
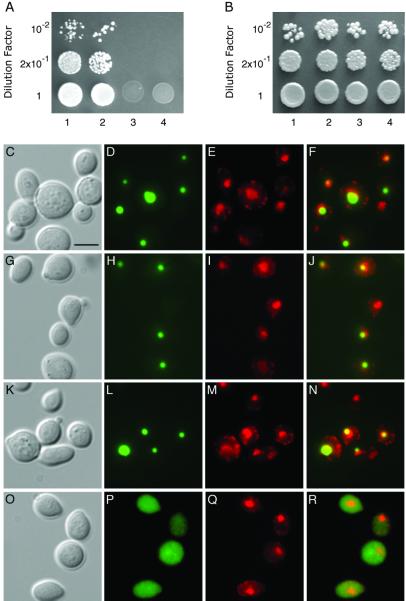
The molecular pathway by which VIP1 enters the plant cell nucleus is unknown. Because VIP1 contains a highly basic domain within its N-terminal sequence (5), it may interact with karyopherins α known to recognize basic nuclear localization signals (reviewed in refs. 25 and 26). Indeed, Fig. 1A shows that VIP1 interacted with the Arabidopsis karyopherin α, AtKAPα (7) in the yeast two-hybrid system (lane 1). In a positive control experiment, VIP1 was shown to interact with VirE2 (lane 2), whereas in negative control experiments, no interaction was observed between VIP1 and lamin C (lane 3) or between AtKAPα and VirE2 (lane 4). Also, under the nonselective conditions, i.e., in the presence of histidine, all combinations of the tested proteins resulted in efficient cell growth (Fig. 1B).
Fig 1.
Karyopherin α-dependent nuclear import of VIP1. Specific interaction between VIP1 and AtKAPα in the two-hybrid assay. (A) Cell growth on a histidine-deficient medium. (B) Cell growth in the presence of histidine. Lane 1, VIP1 + AtKAPα; lane 2, VIP1 + VirE2; lane 3, VIP1 + lamin C; lane 4, AtKAPα + VirE2. VIP1 nuclear import in srp1–31 yeast cells. (C–F) GFP-VIP1 expressed in wild-type cells grown at 25°C. (G–J) GFP-VIP1 expressed in srp1–31 cells grown at 25°C. (K–N) GFP-VIP1 expressed in wild-type cells grown at 37°C. (O–R) GFP-VIP1 expressed in srp1–31 cells grown at 37°C. (C, G, K, and O) Phase-contrast images; (D, H, L, and P) images of GFP fluorescence; (E, I, M, and Q) images of DAPI fluorescence, presented in red to facilitate image merging); (F, J, N, and R) merged GFP–DAPI images. (Bar = 5 μm.)
Next, we tested whether a karyopherin α also is involved in the VIP1 nuclear import in vivo. Because no plant knockout mutants in karyopherin α genes are available, we used a temperature-sensitive yeast mutant, srp1–31, in which its only karyopherin α protein, Srp1p, is inactivated when the cells are grown at the restrictive temperature (37°C) (14). Our previous results indicated that Srp1p and AtKAPα are homologous and functionally interchangeable (7). Fig. 1 shows that GFP-VIP1 was imported into the nuclei, often preferentially accumulating in a smaller subnuclear compartment, of both wild-type (C–F) and srp1–31 mutant cells (G–J) grown at 25°C. In contrast, GFP-VIP1 remained cytoplasmic in srp1–31 cells grown at 37°C (Fig. 1 O–R). This lack of VIP1-GFP nuclear import was not due to nonspecific effects of the elevated temperature because wild-type cells grown at 37°C still efficiently accumulated GFP-VIP1 in their nuclei (Fig. 1 K–N). In all experiments, the position of the yeast cell nucleus was confirmed by nucleus-specific staining with DAPI (Fig. 1 E, I, M, and Q) and merging of the GFP and DAPI fluorescence images (Fig. 1 F, J, L, and R). Thus, VIP1 nuclear import most likely occurs via a karyopherin α-mediated pathway.
Overexpression of VIP1 in Transgenic Plants.
To examine whether VIP1 may represent one of the limiting cellular factors during Agrobacterium infection, we constructed transgenic tobacco plants that overexpress the Arabidopsis VIP1 cDNA (5). A total of 15 independently transformed transgenic lines were produced, and two lines, designated VIP1 S1 and VIP1 S2, were analyzed in detail.
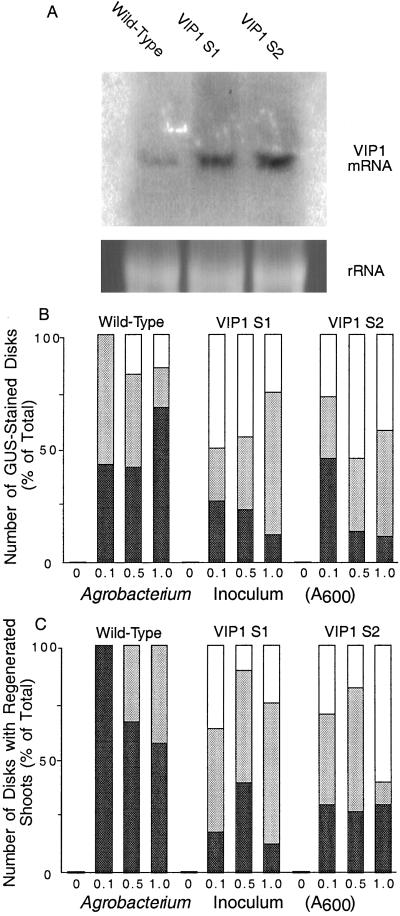
Fig. 2A shows that Northern blot analysis of total RNA obtained from both VIP1 S1 and S2 plants detected high levels of the VIP1 transcript, although the VIP1 mRNA accumulation in the S2 line was slightly higher than that in the S1 line. In contrast, the amount of the endogenous tobacco VIP1 transcript in the wild-type plants was significantly lower, suggesting that VIP1 does not represent an abundant cellular protein. Equal loading of all samples was confirmed from equal amounts of rRNA in all lanes (Fig. 2A).
Fig 2.
Northern blot analysis of VIP1 expression and in wild-type and VIP1 plants and increased Agrobacterium-mediated transient and stable genetic transformation of VIP1 plants. VIP1 S1 and VIP1 S2 represent two independent VIP1-transgenic plant lines. (A) VIP1 expression as detected by Northern blot hybridization (Upper) and amounts of rRNA in each lane as detected by the ethidium bromide staining (Lower). (B) Quantification of transient T-DNA expression. Black, gray, and white bars indicate the numbers of disks that developed 1–50, 51–99, and 100–150 GUS-stained areas per disk, respectively. Total number of GUS-stained disks for each experimental condition was defined as 100%. (C) Quantification of stable T-DNA expression. Black, gray, and white bars indicate the numbers of disks that developed 1–4, 5–9, and 10–20 shoots per disk, respectively. Total number of disks with regenerated shoots for each experimental condition was defined as 100%. All data represent three independent experiments with at least 20 disks for each experimental condition; all of these transformed disks were used for data collection.
Notably, both VIP1-transgenic lines were indistinguishable from the wild-type plants in their overall morphology. Also, no changes in seed viability were observed between the VIP1 and the wild-type plants (data not shown). Thus, overexpression of VIP1 most likely did not interfere with essential plant cellular functions.
Increased Susceptibility of VIP-Transgenic Plants to Agrobacterium Infection.
The VIP1-transgenic plants were tested for their susceptibility to Agrobacterium infection. Three fundamental criteria for the Agrobacterium-mediated genetic transformation were used: transient expression of the T-DNA, regeneration of the stably transformed shoots, and expression of the GUS reporter enzyme in the regenerated transformed plantlets. To better estimate the effects of VIp1 on the efficiency of infection, three different inocula of Agrobacterium were used in each experiment.
Early expression of the Agrobacterium T-DNA is transient, reaching its maximum ≈2 days after infection (27, 28) and occurring from the T-DNA molecules that have not yet integrated into the plant genome. To assay transient T-DNA expression, leaf disks derived from the wild-type and VIP1 S1 and S2 lines were inoculated with Agrobacterium carrying on its T-DNA a uidA gene encoding the GUS enzyme. The indigo-stained areas, representing GUS activity and thus transient T-DNA expression, on each disk were counted 48 hr after inoculation. The resulting data for each plant line and bacterial inoculum were subdivided into three groups: number of disks that developed 1–50, 51–99, and 100–150 stained areas per disk. Fig. 2B shows that while in the absence of Agrobacterium no GUS staining was observed, cocultivation of the wild-type leaf disks with the increasing amounts of Agrobacterium resulted in an increased number of GUS-stained leaf areas. Specifically, at the lowest bacterial inoculum, ≈40% of the disks were categorized to the “1–50” group, and the rest of the disks exhibited an average of 58 GUS-stained areas per disk. At higher bacterial inocula, a relatively small percentage of the wild-type disks (10–15%) developed an average of 110 and 112 GUS-stained areas per disk when cocultivated with Agrobacterium cultures of OD600 = 0.5 and 1.0, respectively (Fig. 2B). Cocultivation of the VIP1 S1 and VIP1 S2 leaf disks with Agrobacterium resulted in T-DNA expression levels significantly higher than those observed in the wild-type leaf disks. For example, even at the lowest Agrobacterium concentration (OD600 = 0.1), 49% of the disks from the VIP1 S1 plants and 25% of the disks from the VIP1 S2 plants exhibited an average of 142 and 130 GUS-stained areas per disk, respectively. At higher inocula of OD600 = 0.5 and OD600 = 1.0, 40% and 24% of the VIP1 S1 plants developed an average of 140 and 143 GUS-stained areas per disk, respectively, and 55% and 42% of the VIP1 S2 plants developed an average of 135 and 138 GUS-stained areas per disk, respectively. Statistical evaluation of the “100–150” groups for the OD600 = 0.5–1.0 inocula by using the unpaired two-tailed t test confirmed that the observed differences in T-DNA transient expression between the wild-type plants and both lines of the VIP1 plants were statistically significant (probability >95%). Because the uidA gene contained an intron (19), these results represented the GUS activity directed by the T-DNA after its transfer to the plant rather than its potentially leaky expression within Agrobacterium. Importantly, the wild-type and VIP1 plants displayed comparable levels of GUS expression (>200 GUS-stained areas per disk, data not shown) when the uidA gene was delivered biolistically, indicating that the elevated amounts of VIP1 in the S1 and S2 plants did not nonspecifically increase the degree of gene expression in the VIP1-expressing cells.
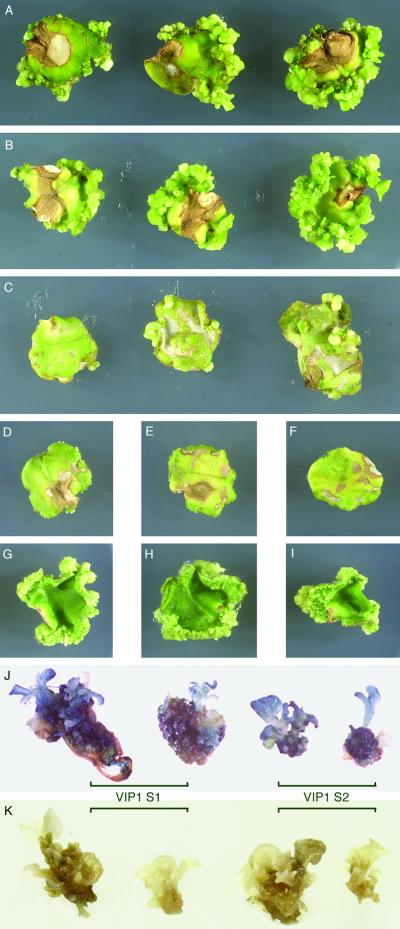
Next, we examined the ability of the VIP1 plants to regenerate transgenic shoots after genetic transformation by Agrobacterium carrying two marker genes, hpt and uidA, on its T-DNA. Leaf disks derived from the VIP1 S1 and S2 plant lines and from the wild-type tobacco plants were inoculated with increasing amounts of Agrobacterium and cultured on the regeneration medium (17) in the presence of hygromycin to allow regeneration and growth of the transgenic shoots. Fig. 3 shows that both VIP1 plant lines produced a large number of hpt-expressing, hygromycin-resistant shoots (Fig. 3 A and B). The number of shoots per disk increased with an increase in the Agrobacterium inoculum such that, at the highest bacterial concentration used, the entire circumference of the infected disk was virtually saturated with growing transformed shoots. In contrast, the infected leaf disks derived from the wild-type plants regenerated much fewer hygromycin-resistant shoots; although the number of these regenerated shoots also increased with increasing Agrobacterium inoculum (Fig. 3C), it never reached the same high density observed with the VIP1 leaf disks even at the lowest concentration of Agrobacterium (compare Fig. 3 C to A and B). Fig. 3 also shows that, in control experiments, leaf disks from the wild-type plants (Fig. 3D) and from the VIP1 S1 and VIP1 S2 lines (Fig. 3 E and F, respectively) grown on the selective medium but in the absence of Agrobacterium infection produced no shoots at all, ruling out a possibility that VIP1 expression rendered the VIP1 plants resistant to hygromycin and enabled them to regenerate untransformed shoots even on the selective medium. Another type of control experiments shown in Fig. 3 demonstrated that the regeneration capacity of the wild-type plants (Fig. 3G) in the absence of selection was comparable to that of the VIP1 S1 and VIP1 S2 plants (Fig. 3 H and I, respectively), indicating that overexpression of VIP1 in these transgenic lines did not nonspecifically increase their regeneration potential.
Fig 3.
Increased Agrobacterium-mediated stable genetic transformation of VIP1 plants. Regeneration of stably transformed shoots: (A–C) Agrobacterium-infected disks from the VIP1 S1 line, VIP1 S2 line, or wild-type plants, respectively, grown on hygromycin-containing selective medium. Left to right: disks inoculated with Agrobacterium cultures at OD600 = 0.1, 0.5, and 1.0, respectively. (D–F) Uninfected disks from the wild-type plants, VIP1 S1 line, or VIP1 S2 line, respectively, grown on hygromycin-containing selective medium. (G–I) Uninfected disks from the wild-type plants, VIP1 S1 line, or VIP1 S2 line, respectively, grown in the absence of selection. GUS-staining of transgenic shoots regenerated from Agrobacterium-infected VIP1 plants: (J) Shoots from Agrobacterium-infected VIP1 S1 and VIP1 S2 lines regenerated on hygromycin-containing medium. (K) Shoots from uninfected VIP1 S1 and VIP1 S2 lines regenerated without selection.
The increased susceptibility of the VIP1 plant lines to genetic transformation by Agrobacterium was quantified by counting the number of regenerated hygromycin-resistant shoots per each infected leaf disk tested. Similarly to quantification of transient expression (see Fig. 2B), these stable transformation data were classified into three groups: number of disks that gave rise to 1–4 shoots, 5–9 shoots, and 10–20 shoots per disk. Fig. 2C shows that while in the absence of Agrobacterium infection no shoots were formed, the wild-type plants infected with the lowest Agrobacterium inoculum produced an average of 2 shoots per disk (i.e., all disks fell into the first group of shoot numbers). At the highest inoculum used (OD600 = 1.0), only ≈40% of the disks developed an average of 5 shoots per disk, corresponding to the second group of shoot numbers. However, under no conditions were wild-type leaf disks obtained that could be classified as the third group of shoot numbers (Fig. 2C). In contrast, both VIP1 S1 and VIP1 S2 plant lines produced higher numbers of shoots per disk. Indeed, at the OD600 = 1.0 inoculum, 85% of the disks from the VIP1 S1 plants and 70% of the disks from the VIP1 S2 plants developed more than 5 shoots, falling in the second and third categories of shoot numbers per leaf disk; in the third category alone, 25% and 60% of the disks from the VIP1 S1 and VIP1 S2 lines produced an average of 16 and 19 shoots per disk, respectively. At the bacterial inoculum of OD600 = 0.5, an average of 12 and 15 shoots per disk was observed in 10% and 20% of VIP1 S1 and VIP1 S2 plants, respectively. Finally, at the lowest inoculum (OD600 = 0.1), ≈35% of disks from the VIP1 S1 plants and 40% of the disks from the VIP1 S2 plants developed an average of 13 and 14 shoots per disk, respectively. Collectively, our results suggest that overexpression of VIP1 in tobacco plants rendered them “super-susceptible” to genetic transformation by Agrobacterium.
That the shoots regenerated on the leaf disks derived from the VIP1 plants were indeed transgenic, i.e., resulted from the Agrobacterium-mediated genetic transformation, was inferred from their growth in the presence of hygromycin. However, besides the hpt gene coding for this antibiotic resistance, the transforming T-DNA also carried the uidA gene encoding GUS enzymatic activity. Thus, in addition to hpt, the regenerated shoots are expected to carry the uidA transgene. To confirm the presence of uidA in the transgenic tissues, the shoots from VIP1 S1 and VIP1 S2 disks (see Fig. 3 A and B, respectively) were allowed to grow further into small plantlets, removed, and analyzed for the GUS activity by histochemical staining of the entire shoot and its cognate callus. Fig. 3J shows that the hygromycin-resistant shoots regenerated from Agrobacterium-infected VIP1 S1 and VIP1 S2 plant lines efficiently expressed the uidA transgene, resulting in the blue staining of the entire shoot. In control experiments, shoots regenerated from uninfected VIP1 plants in the absence of selection did not express GUS (Fig. 3K), indicating that the staining of the transformed shoots (Fig. 3J) was indeed due to the GUS activity and not to the presence of VIP1 in these tissues.
Overexpression of VIP1 Enhances Early Steps of the Agrobacterium Infection Process.
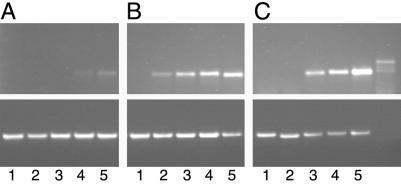
Potential involvement of VIP1 in the T-DNA nuclear import implies that it affects early stages of the Agrobacterium–plant cell T-DNA transfer. The efficiency of this process can be assessed from RT-PCR detection of the corresponding transcript shortly after inoculation (23). Indeed, Fig. 4A shows that, in Agrobacterium-infected wild-type plants, the T-DNA-specific transcripts were detected 12 hr after inoculation. Importantly, this RT-PCR analysis detected the T-DNA transcripts produced within the host plant cells and not within Agrobacterium because they did not contain the 189-bp intron sequence introduced into the T-DNA region to distinguish between these two types of T-DNA-specific PCR products (23, 29) (compare lanes 1–5 to lane 6 in Fig. 4).
Fig 4.
RT-PCR analysis of early T-DNA expression in VIP1 plants. (A) Wild-type plants. (B) VIP1 S1 line. (C) VIP1 S2 line. (Upper) T-DNA-specific products. (Lower) Actin-specific products. Lanes 1–5, RT-PCR analysis of leaf samples collected 0, 4, 8, 12, and 24 hr after inoculation, respectively; lane 6, PCR analysis of Agrobacterium alone control.
RT-PCR analysis of the infected VIP1 plant lines detected the T-DNA-specific products significantly earlier, already 4–8 hr after inoculation, and they continued to accumulate thereafter (Fig. 4 B and C). In control experiments, analysis of actin-specific transcripts generated similar amounts of PCR products in all plant samples (lanes 1–5, but obviously not in Agrobacterium alone sample, lane 6), indicating equal efficiencies of the RT-PCRs (Fig. 4). Thus, the onset of the enhanced susceptibility of VIP1 plants to Agrobacterium infection occurs early in the infection process and is most likely due to the elevated levels of T-DNA nuclear import mediated by overexpressed VIP1.
Discussion
The ability of Agrobacterium to infect eukaryotic cells is not limited to plants, and, in laboratory conditions, Agrobacterium has been shown to genetically transform yeast (30, 31), filamentous fungi and cultivated mushrooms (32), and human cells (33). Thus, Agrobacterium has been used as a model organism capable of a wide-range transkingdom DNA transfer. Furthermore, disarmed Agrobacterium strains that lack the wild-type T-DNA are widely used in plant genetic engineering (reviewed in ref. 34). One of the long-standing goals of these basic scientific and applied aspects of Agrobacterium research is the increase of the transformation efficiency. To date, this objective has been addressed by modifying the Agrobacterium itself, e.g., introducing multiple copies of various vir genes (35–37), or by optimizing tissue culture and inoculation techniques (38). Conversely, no endogenous host factors have been described that improve the Agrobacterium-mediated gene transfer. We hypothesized that VIP1, a recently discovered Arabidopsis protein, which is required for Agrobacterium infection (5), but which is not an abundant cellular protein, may be one of such limiting host factors.
VIP1 has been shown to specifically interact with VirE2 and facilitate its import into the host cell nucleus (5) because VirE2 is a protein component of the Agrobacterium T-complex (6), and because VIP1 is capable of forming ternary VIP1–VirE2–single-stranded DNA complexes (5), VIP1 has been proposed to assist nuclear uptake of the invading T-complexes during the Agrobacterium infection process (2, 3, 5). This active role of VIP1 in Agrobacterium infection suggests that elevating its expression levels within the host cells may enhance the transformation efficiency. Indeed, our data indicate that transgenic tobacco plants that overexpress VIP1 represent better, more susceptible hosts for Agrobacterium. Thus, Agrobacterium-mediated transformation of tobacco, one of the natural hosts of Agrobacterium, apparently does not occur at its maximal possible efficiency, and it can be improved by raising the intracellular levels of VIP1.
Consistent with the proposed function of VIP1 early in infection (5), VIP1 overexpression improved not only the stable but also the earlier, transient expression of T-DNA. One possible scenario for this stimulating effect of increased cellular levels of VIP1 is that, in wild-type plants, the cytoplasmic pool of the de novo synthesized VIP1 is very limited because this protein is relatively not abundant and because it accumulates in the cell nucleus (5). However, it is this cytoplasmic VIP1 that most likely has to associate with the transported T-complex to help its nuclear uptake. Thus, the relatively low amounts of VIP1 in the host cell cytoplasm may be insufficient to saturate multiple T-complexes, each of which is coated with numerous molecules of VirE2; for example, one 22-kb molecule of the nopaline-type T-strand has been calculated to contain 1,176 molecules of VirE2 (39). Overexpression from the strong cauliflower mosaic virus 35S promoter likely increases the cytoplasmic pool of VIP1. As more VIP1 binds to the T-complexes, their nuclear import, and potentially subsequent intranuclear transport, would become more efficient, resulting in higher frequencies of transient and stable genetic transformation. An alternative explanation that VIP1 simply increases the overall capacity to regenerate shoots from leaf disks and/or enhances the levels of foreign gene expression is unlikely because the VIP1 plants were indistinguishable from the wild-type tobacco in their ability to form shoots in the absence of selection and in the expression of GUS activity after biolistic delivery of the uidA gene.
How does VIP1 enhance the transformability of the host plant? Our results suggest that, in plants that overexpress VIP1, the T-DNA molecules are transcribed much sooner after inoculation than in the wild-type plants. This increase in transcription is specific because it was not observed with a cellular gene coding for actin. That VIP1 mediates nuclear import of VirE2 and, by implication, the entire T-complex (5) suggests that the elevated levels of T-DNA transcription are due to enhanced T-DNA nuclear import. Our data indicate that this nuclear import process occurs via a karyopherin α-dependent pathway. Thus, VIP1 may function as an “adapter” molecule between VirE2 and karyopherin α, “piggy-backing” VirE2 into the host cell nucleus. A similar mechanism has been reported for several other protein complexes (reviewed in ref. 40).
Besides helping us to better understand the Agrobacterium-mediated genetic transformation of plants and the role the host cellular factors in this process, our results may ultimately contribute to the design of new strategies for improving genetic transformability of agronomically important plants that are normally difficult to stably transform by using Agrobacterium. Obviously, it is still unknown whether insufficient amounts of a VIP1-like protein restrict the transformability of such plant species; however, our findings indicate that “transformation-limiting” host cellular factors exist and that their overexpression can significantly enhance the degree of the Agrobacterium-mediated genetic transformation.
Acknowledgments
We thank Aaron Neiman for guidance with fluorescence microscopy experiments and Katherine Aguila for her help with the experiments. This work was supported by grants from National Institutes of Health, National Science Foundation Functional Genomic Initiative, U.S. Department of Agriculture, U.S.–Israel Binational Agricultural Research and Development, and Binational Science Foundation (to V.C.), and, in part, by a U.S.–Israel Binational Agricultural Research and Development Postdoctoral Fellowship (to T.T.) and a U.S.–Israel Binational Agricultural Research and Development Research Fellow Award (to V.C.).
Abbreviations
GFP, green fluorescent protein
T-DNA, transferred DNA
DAPI, 4′,6diamidino-2-phenylindole
GUS, β-glucuronidase
RT, reverse transcription
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gelvin S. B. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223-256. [DOI] [PubMed] [Google Scholar]
- 2.Tzfira T., Rhee, Y., Chen, M.-H. & Citovsky, V. (2000) Annu. Rev. Microbiol. 54, 187-219. [DOI] [PubMed] [Google Scholar]
- 3.Tzfira T. & Citovsky, V. (2000) Mol. Plant Pathol. 1, 201-212. [DOI] [PubMed] [Google Scholar]
- 4.Zupan J., Muth, T. R., Draper, O. & Zambryski, P. C. (2000) Plant J. 23, 11-28. [DOI] [PubMed] [Google Scholar]
- 5.Tzfira T., Vaidya, M. & Citovsky, V. (2001) EMBO J. 20, 3596-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zupan J. & Zambryski, P. C. (1997) Crit. Rev. Plant Sci. 16, 279-295. [Google Scholar]
- 7.Ballas N. & Citovsky, V. (1997) Proc. Natl. Acad. Sci. USA 94, 10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirooka T., Rogowsky, P. M. & Kado, C. I. (1987) J. Bacteriol. 169, 1529-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard E., Zupan, J., Citovsky, V. & Zambryski, P. C. (1992) Cell 68, 109-118. [DOI] [PubMed] [Google Scholar]
- 10.Sutton A., Heller, R. C., Landry, J., Choy, J. S., Sirko, A. & Sternglanz, R. (2001) Mol. Cell. Biol. 21, 3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft R., Tardiff, J., Kranter, K. S. & Leinwand, L. A. (1988) BioTechniques 6, 544-547. [PubMed] [Google Scholar]
- 12.Hollenberg S. M., Sternglanz, R., Cheng, P. F. & Weintraub, H. (1995) Mol. Cell. Biol. 15, 3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SenGupta D. J., Zhang, B., Kraemer, B., Pochart, P., Fields, S. & Wickens, M. (1996) Proc. Natl. Acad. Sci. USA 93, 8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb J. D. J., Schlenstedt, G., Pellman, D., Kornitzer, D., Silver, A. P. & Fink, G. R. (1995) Proc. Natl. Acad. Sci. USA 92, 7647-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kironmai K. M., Muniyappa, K., Friedman, D. B., Hollingsworth, N. M. & Byers, B. (1998) Mol. Cell. Biol. 18, 1424-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallie D. R., Lucas, W. J. & Walbot, V. (1989) Plant Cell 1, 303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsch R. B., Fry, J. E., Hoffman, N. L., Eichholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229-1231.17757866 [Google Scholar]
- 18.Murashige T. & Skoog, F. (1962) Physiol. Plant. 15, 473-497. [Google Scholar]
- 19.Tzfira T., Jensen, C. S., Wangxia, W., Zuker, A., Altman, A. & Vainstein, A. (1997) Plant Mol. Biol. Rep. 15, 219-235. [Google Scholar]
- 20.Schöpke C., Taylor, N. J., Cárcamo, R., Beachy, R. N. & Fauquet, C. (1997) Plant Cell Rep. 16, 526-530. [DOI] [PubMed] [Google Scholar]
- 21.Carrington J. C., Freed, D. D. & Leinicke, A. J. (1991) Plant Cell 3, 953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ausubel F. M., Brent, R., Kingston, R. E., Moore, D. D., Smith, J. A., Seidman, J. G. & Struhl, K., (1987) Current Protocols in Molecular Biology (Greene-Wiley, New York).
- 23.Narasimhulu S. B., Deng, X.-B., Sarria, R. & Gelvin, S. B. (1996) Plant Cell 8, 873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thangavelu M., Belostotsky, D., Bevan, M. W., Flavell, R. B., Rogers, H. J. & Lonsdale, D. M. (1993) Mol. Gen. Genet. 240, 290-295. [DOI] [PubMed] [Google Scholar]
- 25.Chook Y. M. & Blobel, G. (2001) Curr. Opin. Struct. Biol. 11, 703-715. [DOI] [PubMed] [Google Scholar]
- 26.Powers M. A. & Forbes, D. J. (1994) Cell 79, 931-934. [DOI] [PubMed] [Google Scholar]
- 27.Janssen B. J. & Gardner, R. C. (1990) Plant Mol. Biol. 14, 61-72. [DOI] [PubMed] [Google Scholar]
- 28.Nam J., Mysore, K. S., Zheng, C., Knue, M. K., Matthysse, A. G. & Gelvin, S. B. (1999) Mol. Gen. Genet. 261, 429-438. [DOI] [PubMed] [Google Scholar]
- 29.Tzfira T., Jensen, C. S., Vainstein, A. & Altman, A. (1997) Physiol. Plant. 99, 554-561. [Google Scholar]
- 30.Piers K. L., Heath, J. D., Liang, X., Stephens, K. M. & Nester, E. W. (1996) Proc. Natl. Acad. Sci. USA 93, 1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bundock P., den Dulk-Ras, A., Beijersbergen, A. & Hooykaas, P. J. J. (1995) EMBO J. 14, 3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Groot M. J., Bundock, P., Hooykaas, P. J. & Beijersbergen, A. G. (1998) Nat. Biotechnol. 16, 839-842. [DOI] [PubMed] [Google Scholar]
- 33.Kunik T., Tzfira, T., Kapulnik, Y., Gafni, D., Dingwall, C. & Citovsky, V. (2001) Proc. Natl. Acad. Sci. USA 98, 1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelvin S. B. (1998) Curr. Opin. Biotechnol. 9, 227-232. [DOI] [PubMed] [Google Scholar]
- 35.Liu C. N., Li, X. Q. & Gelvin, S. B. (1992) Plant Mol. Biol. 20, 1071-1087. [DOI] [PubMed] [Google Scholar]
- 36.Wang K., Herrera-Estrella, A. & Van Montagu, M. (1990) J. Bacteriol. 172, 4432-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin S. G., Komari, T., Gordon, M. P. & Nester, E. W. (1987) J. Bacteriol. 169, 4417-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newell C. A. (2000) Mol. Biotechnol. 16, 53-65. [DOI] [PubMed] [Google Scholar]
- 39.Citovsky V., Guralnick, B., Simon, M. N. & Wall, J. S. (1997) J. Mol. Biol. 271, 718-727. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Bustos J., Heitman, J. & Hall, M. N. (1991) Biochim. Biophys. Acta 1071, 83-101. [DOI] [PubMed] [Google Scholar]