Abstract
The characterization of two distinct classes of hematopoietic stem cells based on CD34 expression and the ability of human bone marrow (BM) cells to differentiate into nonhematopoietic cells introduced new levels of complexity within the stem cell compartment. Here we report the identification and purification of a rare human stem cell population with hematopoietic and hepatic potential based on the expression of a receptor for the complement molecule C1q (C1qRp). We show that C1qRp is a positive marker of all BM-repopulating stem cells because it is expressed on both CD34− and CD34+ stem cells from umbilical cord blood and adult BM. In addition, we show that highly purified lineage-negative CD45+CD38−CD34+or−C1qR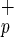 cells not only have BM-repopulating capacity but also can differentiate into human hepatocytes in vivo. The identification of human hepatocytes in mouse livers indicates that the NOD/SCID (nonobese diabetic/severe combined immunodeficient) mouse model can be a valuable tool to study the differentiation potential of adult human stem cells. These findings may have important scientific and clinical implications in the field of human stem cell biology and transplantation.
cells not only have BM-repopulating capacity but also can differentiate into human hepatocytes in vivo. The identification of human hepatocytes in mouse livers indicates that the NOD/SCID (nonobese diabetic/severe combined immunodeficient) mouse model can be a valuable tool to study the differentiation potential of adult human stem cells. These findings may have important scientific and clinical implications in the field of human stem cell biology and transplantation.
Recently, a class of murine and human hematopoietic stem cells (HSCs) was identified and characterized by its lack of CD34 expression (1–6). These studies challenged the dogma that all human repopulating cells express CD34. However, the relationship between human CD34− and CD34+ stem cells and the role of CD34− stem cells in clinical transplantation remain unclear. Xenograft repopulation assays using fetal sheep (7, 8) and immune-deficient mice (9, 10) were crucial for the identification of human CD34− stem cells because Lin−CD34− cells have little or no clonogenic [colony-forming cells (CFC)] or long-term culture-initiating cell (LTC-IC) activity. Using the sheep xenograft model, Zanjani et al. showed that Lin−CD34− cells contained stem cells capable of long-term repopulation and multilineage differentiation in vivo (5). Moreover, these cells are also able to repopulate secondary recipients, attesting to the extensive self-renewal potential of the engrafting cells. In the nonobese diabetic/severe combined immunodeficient (NOD/SCID) mouse model, SCID-repopulating cells (SRC) lacking CD34 expression (CD34neg-SRC) are found in the heterogeneous Lin−CD34− cell fraction at a very low frequency (1 in 125,000 cells; ref. 6). CD34neg-SRC have the ability to give rise to CD34+ cells in vitro and in vivo, and have different cytokine requirements from CD34pos-SRC (6, 11, 12). However, until now, the absence of a positive marker for the identification and isolation of human CD34neg-SRC has hindered their further characterization and prevented their use in clinical transplantation. Here, we investigated whether C1qRp (13–15), the human homologue of the mouse stem cell antigen AA4 (16), could be used as a marker to isolate and characterize human HSCs. AA4 has been recently identified as the antigen recognized by AA4.1 monoclonal antibody used to define murine multipotent hematopoietic precursors that can give rise to the entire spectrum of mature blood cells (16). AA4 may play a role in the homing of hematopoietic stem/progenitors to the fetal liver (16). Moreover, because recent reports have indicated that some adult human and rodent bone marrow (BM) cells have the capacity to give rise to hepatocytes (17–21), we investigated whether C1qR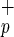 can differentiate into human hepatocytes in vivo.
can differentiate into human hepatocytes in vivo.
Experimental Procedures
Human Cells.
Umbilical cord blood (CB, more than 70 samples) and human BM from healthy donors (eight donors, median age = 35 years) were obtained from the Stem Cell and Leukemia Core at the University of Pennsylvania Cancer Center in accordance with the procedures approved by the Institutional Review Board. Three or more CB samples were pooled for each experiment. All samples were diluted 1:3 in Iscove's modified Dulbecco's medium supplemented with 5% FBS and mononuclear cells (MNC) were isolated using Ficoll/Paque (Pharmacia).
Cell Sorting and Phenotypic Analysis.
MNC expressing lineage markers (CD2, CD3, CD14, CD16, CD19, CD24, CD56, CD66b, CD41, Glycophorin A) were depleted using immunomagnetic beads (StemCell Technologies, Vancouver) according to the manufacturer's protocol. Cells lacking lineage markers (Lin−) were incubated with anti-human CD34-FITC, CD38-APC (Becton Dickinson), C1qRp-biotin (clone R3; ref. 15), and streptavidin–phycoerythrin (PE), then sorted on a Coulter EPICS Elite FACS. For phenotypic analysis, Lin− MNC from CB were incubated with CD34-PE.Cy5, CD38-APC, and C1qRp-biotin, in combination with one of the following anti-human monoclonal antibodies: AC133 (Miltenyi Biotec, Auburn, CA), CD90, CD117, HLA-DR, CD45 (Becton Dickinson), KDR (Sigma), CD162 (Immunotech, Westbrook, ME), and CXCR4 (R&D Systems).
CFC and LTC-IC Assays.
Human CFC were assayed in semisolid methylcellulose medium in standard conditions (22). Long-term culture-initiating cell (LTC-IC) cultures were established on preformed M210B4 stroma layers by using human myeloid long-term culture medium (MyeloCult 5100, StemCell Technologies) according to described methods (23, 24). Limiting dilutions (≥3 replicates per dilution) were performed in 96-well plates in which 1,000–20,000 Lin−CD38−CD34−C1qR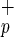 or Lin−CD38−CD34−C1qRp− per well, and 10–2,000 Lin−CD38−CD34+C1qR
or Lin−CD38−CD34−C1qRp− per well, and 10–2,000 Lin−CD38−CD34+C1qR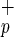 or Lin−CD38−CD34+C1qRp− per well were plated. After 5 weeks in coculture with stroma cells, the content of each well was transferred in methylcellulose medium to reveal CFC activity.
or Lin−CD38−CD34+C1qRp− per well were plated. After 5 weeks in coculture with stroma cells, the content of each well was transferred in methylcellulose medium to reveal CFC activity.
NOD/SCID Mouse Transplantation.
Eight-week-old sublethally irradiated (375 cGy) NOD/LtSz-scid (NOD/SCID) mice were transplanted with purified human cell populations by lateral tail vein injection according to a standard protocol (22). Mice received alternate-day i.p. injections of human cytokines for the first 10 days posttransplant at the following doses: 10 μg of human SCF (PeproTech, Rocky Hill, NJ), 6 μg of human IL-3 (PeproTech), and 6 μg of human G-CSF (Immunex) per mouse per injection. Mice were killed 8–10 weeks posttransplant.
Analysis of Mouse BM.
For each killed mouse, BM was harvested by flushing femurs, tibiae, and iliac crests. Red blood cells were lysed using a 6% ammonium chloride solution and the remaining BM cells were incubated with anti-human monoclonal antibodies (CD45-FITC, CD33-PE, CD19-APC) to determine the level and nature of human engraftment. In some experiments, cells were also incubated with anti-human CD2, CD3, CD4, CD8, CD20, CD34, and CD38. Human engraftment was verified by Southern blot analysis. Genomic DNA was isolated from BM by using a standard extraction protocol (22) and probed with a labeled α-satellite probe specific for human chromosome 17 (p17H8) as described (22). The limit of detection was 0.05% human DNA.
Analysis of Mouse Liver.
Livers from transplanted or control (no human cells injected) mice were collected, divided, and processed for dissociation, RNA extraction, or fixation. Liver cell suspensions were treated with ammonium chloride for red blood lysis and immunostained with anti-human CD45 and HLA-ABC (Becton Dickinson). mRNA was extracted and cDNA synthesized according to standard procedures. The cDNA was amplified by PCR using human albumin primers: 5′-CATTAGCTGCTGATTTTGTTGAAAG; 3′-TGTGCAGCAT TTTGTGACTCTG. The PCR reactions were completed using the following conditions: denaturation at 94°C for 30 s, annealing at 60°C for 75 s, and extension for 30 s for 40 cycles. A final extension step at 72°C for 10 min was included before samples were cooled at 4°C. For immunohistochemistry, multiple pieces from each liver were fixed in formalin for 24 h, dehydrated, paraffin-embedded, and serially sectioned. Sections were immunostained with anti-human monoclonal antibodies: hepatocyte-specific antigen (HSA, clone OCH15E, Dako) and c-met (clone 8F11, Vector Laboratories). The nonspecific binding of the monoclonal antibodies was assessed for each experiment by using liver section from NOD/SCID mice that were not transplanted with human cells. Antigens were unmasked using high temperature and a citric acid-based solution (Vector Laboratories). Antigens were revealed using an avidin: biotinylated alkaline phosphatase complex (Vectastain ABC-AP, Vector Laboratories) method and the Vector Red substrate. Sections were counter stained with methyl green (Vector Laboratories).
Results and Discussion
C1qR Is Expressed on Lin−CD38−CD34+ and Lin−CD38−CD34− Cells.
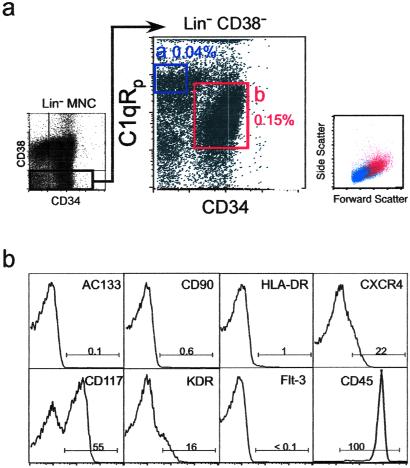
Human C1qRp is expressed on myeloid and endothelial cells (13). Because human HSC are restricted to the Lin−CD38− fraction (6, 22), we examined the cell surface expression of C1qRp and CD34 in Lin−CD38− mononuclear cells isolated from CB or BM. We show here that C1qRp is expressed on two primitive cell subsets: Lin−CD38−CD34− (Fig. 1a, gate a) and Lin−CD38−CD34+ cells (Fig. 1a, gate b). Our results indicate that C1qRp is expressed on >99% of all Lin−CD38−CD34+ cells (Fig. 1a, gate b), a population highly enriched for CD34pos-SRC (22). Lin−CD38−CD34−C1qR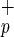 cells are smaller than Lin−CD38−CD34+C1qR
cells are smaller than Lin−CD38−CD34+C1qR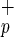 , as determined by forward light scatter (Fig. 1a). Phenotypic analysis indicates that Lin−CD38−CD34−C1qR
, as determined by forward light scatter (Fig. 1a). Phenotypic analysis indicates that Lin−CD38−CD34−C1qR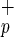 cells do not express stem cell-associated markers such as Thy-1 (CD90) or AC133 (Fig. 1b). However, some of these cells express the cytokine receptors c-kit (CD117, SCF receptor) or KDR [high-affinity vascular endothelial growth factor (VEGF) receptor], and molecules involved in BM homing, such as CXCR4 (Fig. 1b) and CD162 (data not shown). All Lin−CD38−CD34−C1qR
cells do not express stem cell-associated markers such as Thy-1 (CD90) or AC133 (Fig. 1b). However, some of these cells express the cytokine receptors c-kit (CD117, SCF receptor) or KDR [high-affinity vascular endothelial growth factor (VEGF) receptor], and molecules involved in BM homing, such as CXCR4 (Fig. 1b) and CD162 (data not shown). All Lin−CD38−CD34−C1qR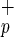 cells expressed CD45, confirming their hematopoietic origin (Fig. 1b). Reverse transcription–PCR analysis of purified Lin−CD38−CD34+or−C1qR
cells expressed CD45, confirming their hematopoietic origin (Fig. 1b). Reverse transcription–PCR analysis of purified Lin−CD38−CD34+or−C1qR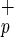 cells showed that these cells do not express LIF receptor or c-mpl (thrombopoietin receptor) and confirmed the expression of KDR in both populations (data not shown). To determine whether Lin−CD38−CD34−C1qR
cells showed that these cells do not express LIF receptor or c-mpl (thrombopoietin receptor) and confirmed the expression of KDR in both populations (data not shown). To determine whether Lin−CD38−CD34−C1qR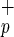 cells are also present in adult hematopoietic tissues, we evaluate the expression of C1qRp on eight different adult BM donors. Although the frequency of Lin−CD38−CD34−C1qR
cells are also present in adult hematopoietic tissues, we evaluate the expression of C1qRp on eight different adult BM donors. Although the frequency of Lin−CD38−CD34−C1qR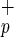 cells appears to be lower in adult BM compared with CB (0.01% versus 0.04%), a similar pattern of C1qRp expression was observed (data not shown).
cells appears to be lower in adult BM compared with CB (0.01% versus 0.04%), a similar pattern of C1qRp expression was observed (data not shown).
Fig 1.
Identification of a novel stem cell population: Lin−CD38−CD34−C1qR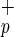 . (a) Mononuclear cells from umbilical CB were lineage-depleted (Lin−) and the expression of CD34 and CD38 was evaluate (Left). Cells lacking CD38 expression (Lin−CD38−) were further analyzed and subdivided into CD34−C1qR
. (a) Mononuclear cells from umbilical CB were lineage-depleted (Lin−) and the expression of CD34 and CD38 was evaluate (Left). Cells lacking CD38 expression (Lin−CD38−) were further analyzed and subdivided into CD34−C1qR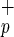 and CD34+C1qR
and CD34+C1qR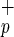 populations (Center, regions a and b). The percentage of each subpopulation relative to total CB MNC is shown next to each sort gate. The forward and side scatter of Lin−CD38−CD34−C1qR
populations (Center, regions a and b). The percentage of each subpopulation relative to total CB MNC is shown next to each sort gate. The forward and side scatter of Lin−CD38−CD34−C1qR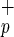 (blue) and Lin−CD38−CD34+C1qR
(blue) and Lin−CD38−CD34+C1qR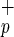 (red) are shown on the Right. (b) Phenotypic analysis of Lin−CD38−CD34−C1qR
(red) are shown on the Right. (b) Phenotypic analysis of Lin−CD38−CD34−C1qR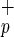 cells. Each panel is a representative flow cytometry histogram representing fluorescence intensity (x axis) as a function of cell number (y axis) for various markers associated with stem cells.
cells. Each panel is a representative flow cytometry histogram representing fluorescence intensity (x axis) as a function of cell number (y axis) for various markers associated with stem cells.
Functional Activity of C1qR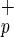 Cells in Vitro.
Cells in Vitro.
For functional evaluation, Lin−CD38− cells were subdivided according to CD34 and C1qRp expression and each subpopulation was assayed for the presence of committed (CFC) and primitive hematopoietic progenitors (LTC-IC) by using standard colony assays. Both Lin−CD38−CD34−C1qR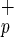 and Lin−CD38−CD34−C1qRp− have very low frequencies of CFC and LTC-IC (Table 1). In these in vitro assays, fractionation of Lin−CD38−CD34− based on C1qRp expression does not increase clonogenic activity indicating that the use of C1qRp as a positive marker of stem cells does not enrich for hematopoietic progenitors. The high frequency of CFC and LTC-IC normally found in Lin−CD38−CD34+ cells was unaffected when cells were further divided into C1qR
and Lin−CD38−CD34−C1qRp− have very low frequencies of CFC and LTC-IC (Table 1). In these in vitro assays, fractionation of Lin−CD38−CD34− based on C1qRp expression does not increase clonogenic activity indicating that the use of C1qRp as a positive marker of stem cells does not enrich for hematopoietic progenitors. The high frequency of CFC and LTC-IC normally found in Lin−CD38−CD34+ cells was unaffected when cells were further divided into C1qR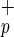 and C1qRp− fractions (Table 1). Interestingly, myeloid (CFU-G, -M, and -GM) and erythroid (BFU-E) colonies were present in both fractions (data not shown), indicating that purification based on C1qRp expression does not skew the differentiation pattern of Lin−CD38−CD34+ cells. Our results also indicate that Lin−CD38−CD34+C1qR
and C1qRp− fractions (Table 1). Interestingly, myeloid (CFU-G, -M, and -GM) and erythroid (BFU-E) colonies were present in both fractions (data not shown), indicating that purification based on C1qRp expression does not skew the differentiation pattern of Lin−CD38−CD34+ cells. Our results also indicate that Lin−CD38−CD34+C1qR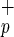 cells contain almost all multipotent and committed hematopoietic progenitors found in the Lin−CD38−CD34+ population.
cells contain almost all multipotent and committed hematopoietic progenitors found in the Lin−CD38−CD34+ population.
Table 1.
Colony-forming activity of human Lin−CD38− as a function of CD34 and C1qRp expression
| Phenotype | Frequency of CFC | Frequency of LTC-IC |
|---|---|---|
CD34−C1qR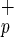
|
1/2,000 | 1/3,500 |
CD34−C1qR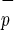
|
1/3,125 | 1/4,000 |
CD34+C1qR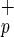
|
1/3 | 1/16 |
CD34+C1qR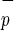
|
1/4 | 1/24 |
CFC frequency measured after 2 weeks in culture.
LTC-IC frequency determined after 5 weeks in culture on M210B4 stroma layers followed by 2 weeks in methylcellulose medium.
Repopulating Activity of C1qR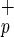 Cells.
Cells.
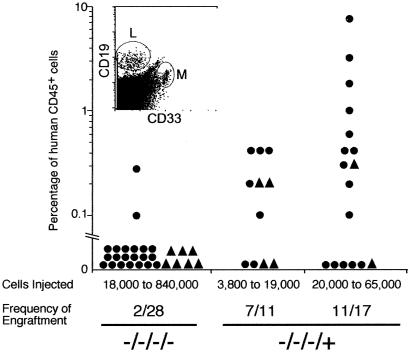
We used the NOD/SCID xenotransplantation model to determine whether subfractionation of Lin−CD38−CD34+or− cells based on C1qRp expression can be used to further enrich for human HSC activity. Various doses of purified cells were transplanted into NOD/SCID mice by using described protocols (22), to determine the frequency of SRC within each cell population. Mouse BM was analyzed for the presence of human cells 8–10 weeks post transplantation. Levels of human cell engraftment in NOD/SCID mice transplanted with CB (Fig. 2, circles) or BM (Fig. 2, triangles) Lin−CD38−CD34−C1qR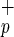 or Lin−CD38−CD34−C1qRp− cells were quantified by flow cytometry and Southern blot analysis (Fig. 2). Only 2 mice of 21 transplanted with CB Lin−CD38−CD34−C1qRp− cells (doses ranging from 20,000 to 840,000 cells) and none of the seven mice injected with BM cells engrafted (limit of detection: <0.05% human cells). In contrast, 18 of 28 mice had detectable human cell engraftment when transplanted with CB or BM Lin−CD38−CD34−C1qR
or Lin−CD38−CD34−C1qRp− cells were quantified by flow cytometry and Southern blot analysis (Fig. 2). Only 2 mice of 21 transplanted with CB Lin−CD38−CD34−C1qRp− cells (doses ranging from 20,000 to 840,000 cells) and none of the seven mice injected with BM cells engrafted (limit of detection: <0.05% human cells). In contrast, 18 of 28 mice had detectable human cell engraftment when transplanted with CB or BM Lin−CD38−CD34−C1qR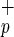 cells, indicating that the C1qR
cells, indicating that the C1qR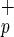 subfraction is highly enriched in SRC. Lin−CD38−CD34−C1qR
subfraction is highly enriched in SRC. Lin−CD38−CD34−C1qR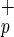 cells gave rise to both myeloid and lymphoid cells in engrafted mice (Fig. 2 Inset). As previously described, we observed the presence of CD34+CD38− cells and, in some cases, T cells in the BM of mice transplanted with Lin−CD38−CD34−C1qR
cells gave rise to both myeloid and lymphoid cells in engrafted mice (Fig. 2 Inset). As previously described, we observed the presence of CD34+CD38− cells and, in some cases, T cells in the BM of mice transplanted with Lin−CD38−CD34−C1qR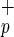 cells (data not shown; ref. 6). Transplantation with as few as 5,000 Lin−CD38−CD34−C1qR
cells (data not shown; ref. 6). Transplantation with as few as 5,000 Lin−CD38−CD34−C1qR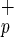 cells resulted in engraftment, whereas as many as 840,000 Lin−CD38−CD34−C1qRp− cells were incapable of BM repopulation (Fig. 2). Regression analysis obtained from all of the CB indicated that only 7,500 Lin−CD38−CD34−C1qR
cells resulted in engraftment, whereas as many as 840,000 Lin−CD38−CD34−C1qRp− cells were incapable of BM repopulation (Fig. 2). Regression analysis obtained from all of the CB indicated that only 7,500 Lin−CD38−CD34−C1qR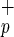 cells were necessary to obtain BM repopulation compared with 125,000 Lin−CD34− cells, as described by Bhatia et al. (6). The levels of human engraftment obtained with Lin−CD38−CD34−C1qR
cells were necessary to obtain BM repopulation compared with 125,000 Lin−CD34− cells, as described by Bhatia et al. (6). The levels of human engraftment obtained with Lin−CD38−CD34−C1qR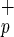 cells were comparable to those reported by Bhatia et al. (6). As the percentage of C1qR
cells were comparable to those reported by Bhatia et al. (6). As the percentage of C1qR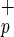 in Lin−CD38−CD34− cells varies from 20 to 35% (more than 70 CB examined), we could estimate that the use of C1qRp provides a 3- to 5-fold enrichment in CD34neg-SRC compared with Lin−CD38−CD34− cells. C1qRp represents the first positive marker to isolate the CD34neg-SRC present in Lin−CD38−CD34− population. Furthermore, as shown in Fig. 1, more than 99% of Lin−CD38−CD34+ cells express C1qRp. We confirmed the presence of CD34pos-SRC activity in Lin−CD38−CD34+C1qR
in Lin−CD38−CD34− cells varies from 20 to 35% (more than 70 CB examined), we could estimate that the use of C1qRp provides a 3- to 5-fold enrichment in CD34neg-SRC compared with Lin−CD38−CD34− cells. C1qRp represents the first positive marker to isolate the CD34neg-SRC present in Lin−CD38−CD34− population. Furthermore, as shown in Fig. 1, more than 99% of Lin−CD38−CD34+ cells express C1qRp. We confirmed the presence of CD34pos-SRC activity in Lin−CD38−CD34+C1qR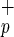 at a frequency similar to what we reported previously (22)—i.e., 1 SRC in approximately 600 Lin−CD38−CD34+ cells (data not shown). Thus, positive selection of human stem cells based on the expression of C1qRp represents a new means of simultaneously isolating CD34neg and CD34pos-SRC. Patients transplanted with C1qR
at a frequency similar to what we reported previously (22)—i.e., 1 SRC in approximately 600 Lin−CD38−CD34+ cells (data not shown). Thus, positive selection of human stem cells based on the expression of C1qRp represents a new means of simultaneously isolating CD34neg and CD34pos-SRC. Patients transplanted with C1qR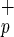 cells instead of CD34+ cells may clinically benefit from receiving both CD34+ and CD34− BM-repopulating cells.
cells instead of CD34+ cells may clinically benefit from receiving both CD34+ and CD34− BM-repopulating cells.
Fig 2.
Human cell engraftment of NOD/SCID mice transplanted with various doses of Lin−CD38−CD34−C1qR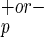 cells. Purified Lin−CD38−CD34−C1qR
cells. Purified Lin−CD38−CD34−C1qR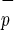 (−/−/−/−) or Lin−CD38−CD34−C1qR
(−/−/−/−) or Lin−CD38−CD34−C1qR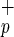 (−/−/−/+) cells isolated from CB (circle) or BM (triangle) were transplanted into NOD/SCID mice at the indicated dose ranges. The percentage of human CD45+ cells present in the murine BM 8–10 weeks posttransplant was determined by flow cytometry and confirmed by Southern blot analysis. For each cell phenotype and dose range, the frequency of engraftment represents the number of mice with detectable levels of human cells in the BM over the total number of mice transplanted. The multilineage potential of the transplanted cells was evidenced by the presence of human myeloid (M, CD33+) and lymphoid (L, CD19+) cells in the murine BM (Inset).
(−/−/−/+) cells isolated from CB (circle) or BM (triangle) were transplanted into NOD/SCID mice at the indicated dose ranges. The percentage of human CD45+ cells present in the murine BM 8–10 weeks posttransplant was determined by flow cytometry and confirmed by Southern blot analysis. For each cell phenotype and dose range, the frequency of engraftment represents the number of mice with detectable levels of human cells in the BM over the total number of mice transplanted. The multilineage potential of the transplanted cells was evidenced by the presence of human myeloid (M, CD33+) and lymphoid (L, CD19+) cells in the murine BM (Inset).
C1qR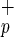 Cells Can Differentiate into Human Hepatocytes.
Cells Can Differentiate into Human Hepatocytes.
Recent reports have indicated that the BM of adult rodents contains cells with the capacity to give rise to hepatocytes (17–19). Lagasse et al. (19) established that some hematopoietic stem cells present in adult BM copurified with stem cells that gave rise to hepatocytes, supporting the new concept that somatic stem cells can change cell fate. However, these reports were based on adult rodent stem cell populations, which may differ from human stem cells in their capacity to give rise to multiple tissues. Nevertheless, the studies by Theise et al. (20) and Alison et al. (21), based on patients who received BM transplants, indicate that some human adult stem cells present in BM also have the ability to give rise to hepatocytes. Those two studies did not distinguish whether HSCs, mesenchymal stem cells, or hepatic stem cells circulating in the BM were responsible for the observed liver engraftment. Thus, we investigated the potential of human C1qR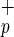 stem cells to differentiate in vivo into hepatocytes.
stem cells to differentiate in vivo into hepatocytes.
Livers from NOD/SCID mice transplanted with Lin−CD45+CD38−CD34+or−C1qR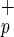 cells were assessed by flow cytometry, immunohistochemistry, and RNA expression analysis. Paraffin-embedded liver sections were immunostained for human HSA and human c-met (hepatocyte growth factor receptor). Liver sections from non-injected NOD/SCID mice were used as negative controls. Both Lin−CD38−CD34−C1qR
cells were assessed by flow cytometry, immunohistochemistry, and RNA expression analysis. Paraffin-embedded liver sections were immunostained for human HSA and human c-met (hepatocyte growth factor receptor). Liver sections from non-injected NOD/SCID mice were used as negative controls. Both Lin−CD38−CD34−C1qR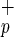 and Lin−CD38−CD34+C1qR
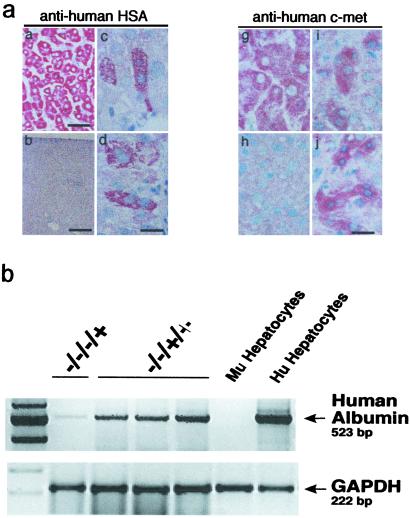
and Lin−CD38−CD34+C1qR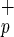 cells gave rise to individual and/or clusters of human hepatocytes in mouse livers based on their morphology and the presence human-specific positive staining for HSA (Fig. 3a, c–f) or c-met (Fig. 3a, i–j). To not solely rely on immunohistochemistry and morphology, we also used reverse transcription–PCR to examine the presence human albumin transcripts in the liver of transplanted mice (Fig. 3b). We detected human-specific albumin transcripts in mice transplanted with Lin−CD38−CD34+or−C1qR
cells gave rise to individual and/or clusters of human hepatocytes in mouse livers based on their morphology and the presence human-specific positive staining for HSA (Fig. 3a, c–f) or c-met (Fig. 3a, i–j). To not solely rely on immunohistochemistry and morphology, we also used reverse transcription–PCR to examine the presence human albumin transcripts in the liver of transplanted mice (Fig. 3b). We detected human-specific albumin transcripts in mice transplanted with Lin−CD38−CD34+or−C1qR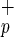 , confirming the presence of human hepatocytes and demonstrating their ability to express a human hepatocyte-specific gene in the context of mouse liver tissue (Fig. 3b). Furthermore, using flow cytometry on mouse liver suspensions, we detected a rare population (≤0.1%) of human (HLA-ABC+) non-hematopoietic (CD45−) cells that could represent human hepatocytes (data not shown). The lack of CD45 expression suggests that these cells are not simply residual cells from the transplanted populations (CD45+).
, confirming the presence of human hepatocytes and demonstrating their ability to express a human hepatocyte-specific gene in the context of mouse liver tissue (Fig. 3b). Furthermore, using flow cytometry on mouse liver suspensions, we detected a rare population (≤0.1%) of human (HLA-ABC+) non-hematopoietic (CD45−) cells that could represent human hepatocytes (data not shown). The lack of CD45 expression suggests that these cells are not simply residual cells from the transplanted populations (CD45+).
Fig 3.
Identification of human hepatocytes in livers from NOD/SCID mice transplanted with Lin−CD38−CD34+C1qR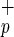 or Lin−CD38−CD34−C1qR
or Lin−CD38−CD34−C1qR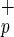 cells (ranging from 500 to 1,500 and 8,000 to 70,000 cells, respectively). (a) Photomicrographs of NOD/SCID mouse liver sections from mice transplanted with purified human Lin−CD38−CD34−or+C1qR
cells (ranging from 500 to 1,500 and 8,000 to 70,000 cells, respectively). (a) Photomicrographs of NOD/SCID mouse liver sections from mice transplanted with purified human Lin−CD38−CD34−or+C1qR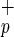 cells isolated from umbilical CB. Livers from transplanted mice were recovered 8–10 weeks posttransplant, paraffin-embedded, and processed for immunocytochemistry. Serial sections were immunostained for human HSA (a–d) and c-met (g–j). Human liver sections were used as a positive control (a and g) and liver sections from noninjected NOD/SCID mice were used as a negative control for each monoclonal antibody (b and h). No HSA+ or c-met+ cells were found in liver sections from noninjected mice (b and h). Human hepatocytes were identified as single cells (c and d) or small clusters of two to eight cells (i and j) of HSA+ or c-met+ cells in mice transplanted with Lin−CD38−CD34−or+C1qR
cells isolated from umbilical CB. Livers from transplanted mice were recovered 8–10 weeks posttransplant, paraffin-embedded, and processed for immunocytochemistry. Serial sections were immunostained for human HSA (a–d) and c-met (g–j). Human liver sections were used as a positive control (a and g) and liver sections from noninjected NOD/SCID mice were used as a negative control for each monoclonal antibody (b and h). No HSA+ or c-met+ cells were found in liver sections from noninjected mice (b and h). Human hepatocytes were identified as single cells (c and d) or small clusters of two to eight cells (i and j) of HSA+ or c-met+ cells in mice transplanted with Lin−CD38−CD34−or+C1qR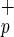 . [Scale bars = 100 μm (a), 200 μm (b), 10 μm (c–j).] (b) Expression of human albumin in livers from NOD/SCID mice transplanted with Lin−CD38−CD34−C1qR
. [Scale bars = 100 μm (a), 200 μm (b), 10 μm (c–j).] (b) Expression of human albumin in livers from NOD/SCID mice transplanted with Lin−CD38−CD34−C1qR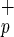 (−/−/−/+) or Lin−CD38−CD34+C1qR
(−/−/−/+) or Lin−CD38−CD34+C1qR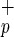 (−/−/+/+) cells. RNA extracted from noninjected mice was used as a negative control and from human hepatocytes as a positive control.
(−/−/+/+) cells. RNA extracted from noninjected mice was used as a negative control and from human hepatocytes as a positive control.
Two recent reports (25, 26) have indicated the possibility of fusion in vitro between embryonic stem cells and BM or neuronal stem cells. It is unlikely that the human hepatocytes observed in our study are in fact the product of a fusion between human HSCs and mouse liver cells. Indeed, if such human HSC/mouse liver hybrids were formed, we would expect the hybrid to have both human HSC markers and mouse liver markers. It seems very unlikely that human HSC/mouse hepatocyte hybrids would acquire human liver markers, down-regulate CD45 expression, and furthermore be able to express the transcripts for human albumin. It is more plausible, indeed, to speculate that human HSCs circulating into the liver of the mice “transdifferentiated” within the mouse liver microenvironment. This last hypothesis is indirectly supported by a recent study of C. Verfaillie's group which indicates that multipotent adult progenitor cells (MAPCs) derived from adult human BM can differentiate in vitro into functional hepatocyte-like cells (27). In this study, fusion cannot be responsible for the hepatic “transdifferentiation” observed. Finally, if human HSCs have the capacity to spontaneously fuse with other cell types in mature organs, recipients of BM transplantation should have polyploid cells in many organs. The organs of sex-mismatched BM recipients have been extensively studied for the presence of donor-derived cells (20, 21, 28), but there has been no report so far of male–female polyploid cells.
In conclusion, this study identifies C1qRp as a novel human stem cell marker expressed on both CD34neg and CD34pos-SRC. The presence of C1qRp on CD34neg-SRC provides a new means of defining and studying the most primitive population of human BM-repopulating cells. These data also provide the first direct demonstration that a highly purified and phenotypically defined human hematopoietic stem cell population can repopulate the BM and differentiate in vivo into hepatocytes by using the NOD/SCID mouse model. Although the frequency of human hepatocytes detected was low (0.05–0.1%), this is the first demonstration that a highly purified population of human HSCs can provide hepatic engraftment in absence of overt tissue damage—i.e., sublethal irradiation. Present experiments are attempting to increase the hepatocyte generation by inducing liver damage. Because the purified human HSC populations used in this study may have contained two stem cell fractions responsible for BM repopulation and hepatocyte differentiation, our data gives no indication as to whether hematopoietic and hepatic engraftment can be provided by the same human stem cell. So far, only Krause et al. (29) have provided evidence that a single mouse stem cell can provide multitissue engraftment. Gene-marking/clonal analysis experiments will be needed in the future to address whether a same human stem cell has the capacity to repopulate the entire blood system and at the same time generate hepatocytes. Despite the lack of a definitive proof of the bipotentiality of Lin−CD38−CD34−or+C1qR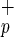 cells, the use of HSCs for the potential treatment of human liver diseases represents an exciting new therapeutic strategy. HSCs represent a safe and accessible source of human stem cells that can be genetically manipulated and may thus prove to be an ideal vehicle for delivering therapeutic genes to the liver. The NOD/SCID xenotransplant model will play an important role in evaluating this potential. Moreover, in light of the recent identification of a single mouse BM-derived stem cell with multiorgan and multilineage engraftment (28), additional experiments will be required to determine the full developmental capacity of these primitive human C1qR
cells, the use of HSCs for the potential treatment of human liver diseases represents an exciting new therapeutic strategy. HSCs represent a safe and accessible source of human stem cells that can be genetically manipulated and may thus prove to be an ideal vehicle for delivering therapeutic genes to the liver. The NOD/SCID xenotransplant model will play an important role in evaluating this potential. Moreover, in light of the recent identification of a single mouse BM-derived stem cell with multiorgan and multilineage engraftment (28), additional experiments will be required to determine the full developmental capacity of these primitive human C1qR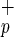 populations. In conclusion, our results suggest that C1qR
populations. In conclusion, our results suggest that C1qR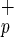 cells may become a valuable source of stem cells for both BM repopulation and treatment of liver disease.
cells may become a valuable source of stem cells for both BM repopulation and treatment of liver disease.
Acknowledgments
We thank Mark Iezzi for maintaining the NOD/SCID mouse colony, Dr. Martin Carroll and Qing Xu for providing us with CB and BM, and Dr. Marisa Fonseca for purifying and biotinylating the R3 monoclonal antibody. We thank Drs. Curt Civin, Stephen Emerson, and Fiona Watt for their critical reading of the manuscript. This work was supported by grants from the Association for International Cancer Research (to D.A.B.) and the National Institutes of Health (to A.J.T.). G.H.D. was supported by a fellowship and funds to M.C.S. from the Howard Hughes Medical Institute.
Abbreviations
BM, bone marrow
CB, cord blood
CFC, colony-forming cells
HSA, hepatocyte-specific antigen
HSC, hematopoietic stem cell
Lin−, lineage-negative
LTC-IC, long-term culture-initiating cell
NOD/SCID, nonobese diabetic/LtSz-scid
SRC, SCID-repopulating cells
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Osawa M., Hanada, K., Hamada, H. & Nakauchi, H. (1996) Science 273, 242-245. [DOI] [PubMed] [Google Scholar]
- 2.Goodell M. A., Rosenzweig, M., Kim, H., Marks, D. F., DeMaria, M., Paradis, G., Grupp, S. A., Sieff, C. A., Mulligan, R. C. & Johnson, R. P. (1997) Nat. Med. 3, 1337-1345. [DOI] [PubMed] [Google Scholar]
- 3.Jones R. J., Collector, M. I., Barber, J. P., Vala, M. S., Fackler, M. J., May, W. S., Griffin, C. A., Hawkins, A. L., Zehnbauer, B. A., Hilton, J., et al. (1996) Blood 88, 487-491. [PubMed] [Google Scholar]
- 4.Donnelly D. S., Zelterman, D., Sharkis, S. & Krause, D. S. (1999) Exp. Hematol. 27, 788-796. [DOI] [PubMed] [Google Scholar]
- 5.Zanjani E. D., Almeida-Porada, G., Livingston, A. G., Flake, A. W. & Ogawa, M. (1998) Exp. Hematol. 26, 353-360. [PubMed] [Google Scholar]
- 6.Bhatia M., Bonnet, D., Murdoch, B., Gan, O. I. & Dick, J. E. (1998) Nat. Med. 4, 1038-1045. [DOI] [PubMed] [Google Scholar]
- 7.Zanjani E. D., Almeida-Porada, G., Ascensao, J. L., MacKintosh, F. R. & Flake, A. W. (1997) Stem Cells 15, 79-93. [DOI] [PubMed] [Google Scholar]
- 8.Srour E. F., Zanjani, E. D., Cornetta, K., Traycoff, C. M., Flake, A. W., Hedrick, M., Brandt, J. E., Leemhuis, T. & Hoffman, R. (1993) Blood 82, 3333-3342. [PubMed] [Google Scholar]
- 9.Dick J. E. (1996) Semin. Immunol. 8, 197-206. [DOI] [PubMed] [Google Scholar]
- 10.Dao M. A., Tsark, E. & Nolta, J. A. (1999) Curr. Opin. Mol. Ther. 1, 553-557. [PubMed] [Google Scholar]
- 11.Fujisaki T., Berger, M. G., Rose-John, S. & Eaves, C. J. (1999) Blood 94, 1926-1932. [PubMed] [Google Scholar]
- 12.Nakamura Y., Ando, K., Chargui, J., Kawada, H., Sato, T., Tsuji, T., Hotta, T. & Kato, S. (1999) Blood 94, 4053-4059. [PubMed] [Google Scholar]
- 13.Nepomuceno R. R. & Tenner, A. J. (1998) J. Immunol. 160, 1929-1935. [PubMed] [Google Scholar]
- 14.Kim T. S., Park, M., Nepomuceno, R. R., Palmarini, G., Winokur, S., Cotman, C. A., Bengtsson, U. & Tenner, A. J. (2000) Mol. Immunol. 37, 377-389. [DOI] [PubMed] [Google Scholar]
- 15.Nepomuceno R. R., Henschen-Edman, A. H., Burgess, W. H. & Tenner, A. J. (1997) Immunity 6, 119-129. [DOI] [PubMed] [Google Scholar]
- 16.Petrenko O., Beavis, A., Klaine, M., Kittappa, R., Godin, I. & Lemischka, I. R. (1999) Immunity 10, 691-700. [DOI] [PubMed] [Google Scholar]
- 17.Theise N. D., Badve, S., Saxena, R., Henegariu, O., Sell, S., Crawford, J. M. & Krause, D. S. (2000) Hepatology 31, 235-240. [DOI] [PubMed] [Google Scholar]
- 18.Petersen B. E., Goff, J. P., Greenberger, J. S. & Michalopoulos, G. K. (1998) Hepatology 27, 433-445. [DOI] [PubMed] [Google Scholar]
- 19.Lagasse E., Connors, H., Al-Dhalimy, M., Reitsma, M., Dohse, M., Osborne, L., Wang, X., Finegold, M., Weissman, I. L. & Grompe, M. (2000) Nat. Med. 6, 1229-1234. [DOI] [PubMed] [Google Scholar]
- 20.Theise N. D., Nimmakayalu, M., Gardner, R., Illei, P. B., Morgan, G., Teperman, L., Henegariu, O. & Krause, D. S. (2000) Hepatology 32, 11-16. [DOI] [PubMed] [Google Scholar]
- 21.Alison M. R., Poulsom, R., Jeffery, R., Dhillon, A. P., Quaglia, A., Jacob, J., Novelli, M., Prentice, G., Williamson, J. & Wright, N. A. (2000) Nature (London) 406, 257. [DOI] [PubMed] [Google Scholar]
- 22.Bhatia M., Wang, J. C. Y., Kapp, U., Bonnet, D. & Dick, J. E. (1997) Proc. Natl. Acad. Sci. USA 94, 5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogge D. E., Lansdorp, P. M., Reid, D., Gerhard, B. & Eaves, C. J. (1996) Blood 88, 3765-3773. [PubMed] [Google Scholar]
- 24.Sutherland H. J., Eaves, C. J., Lansdorp, P. M., Thacker, J. D. & Hogge, D. E. (1991) Blood 78, 666-672. [PubMed] [Google Scholar]
- 25.Terada N., Hamazaki, T., Oka, M., Hoki, M., Mastalerz, D. M., Nakano, Y., Meyer, E. M., Morel, L., Petersen, B. E. & Scott, E. W. (2002) Nature (London) 416, 542-545. [DOI] [PubMed] [Google Scholar]
- 26.Ying Q. L., Nichols, J., Evans, E. P. & Smith, A. G. (2002) Nature (London) 416, 545-548. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz R. E., Reyes, M., Koodie, L., Jiang, Y., Blackstad, M., Lund, T., Lenvik, T., Johnson, S., Hu, W. S. & Verfaillie, C. M. (2002) J. Clin. Invest. 109, 1291-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korbling M., Katz, R. L., Khanna, A., Ruifrok, A. C., Rondon, G., Albitar, M., Champlin, R. E. & Estrov, Z. (2002) N. Engl. J. Med. 346, 738-746. [DOI] [PubMed] [Google Scholar]
- 29.Krause D. S., Theise, N. D., Collector, M. I., Henegariu, O., Hwang, S., Gardner, R., Neutzel, S. & Sharkis, S. J. (2001) Cell 105, 369-377. [DOI] [PubMed] [Google Scholar]