What accounts for the great popularity and success of Drosophila melanogaster as an experimental organism? There are many contributing practical reasons, including the relative ease with which large numbers can be grown at minimal expense, its having just four pairs of chromosomes, and the efficiency with which its organs and ornately patterned exterior can be examined. But there are two additional inter-related historical factors that are no less important. First, many mutant lines have been generated that have specialized chromosomes or combinations of alleles that enable an unprecedented wealth of opportunities for genetic or developmental analysis. Most of these lines are curated and maintained in stock centers that provide for their worldwide distribution. Second, there has been a long history of cooperation that has encouraged many Drosophila geneticists to generate tools that benefit the community as a whole. Such efforts have produced specialized balancer chromosomes, chromosomes that can be used to efficiently produce genetic mosaics, collections of deletions that uncover most of the euchromatic segments of the genome, and large collections of insertional mutants. A report from Huet et al. in this issue of PNAS (1) describes the development of a new and potentially very powerful genetic technique that continues this great tradition.
Huet et al. have devised a wonderfully clever strategy to efficiently produce nested sets of deletions. The method involves a hybrid transposable element that includes components of two transposable elements, P (2) and hobo (3), as well as two Drosophila genes, yellow and white, that are used for phenotypic selection. A brief history of the role that transposable elements have played in Drosophila genetics will help to place the conception and probable utility of this novel vehicle in its proper context.
This history begins with the first mutant found—white1 (4), which has a Doc element insertion in its white gene (5). This mutant was found in a laboratory stock, but how it originated remains a mystery. The first report of highly mutable Drosophila alleles came some 15 years later when Demerec described his isolation of many new alleles of two genes in Drosophila virilis, as well as the identification of dominant enhancers of their mutability (reviewed in ref. 6). It seems reasonable to suggest that the mutability Demerec characterized was a consequence of mobilized transposable elements, but it wasn't until 1973 that another report of “innate” mutability was made. In a Drosophila Information Service volume of that year (7), R. L. Berg describes the simultaneous rise in the rate of occurrence of singed mutants in separate wild populations of Drosophila, and concludes: “This global mosaic pattern of mutability fluctuations indicates that some external cosmic agent acts through mediators—supposedly mutant microorganisms which are widely distributed but not omnipresent.”
Huet et al. have devised a wonderfully clever strategy to efficiently produce nested sets of deletions.
Work in several labs, including that of Green (8), Kidwell (9), and Rubin (2), discovered the true causal agent involved in such mutagenic episodes. They identified the P transposable element as a mutagenic agent and characterized the principal systems that regulate its transposition. These findings established the mutagenic potential of transposons in eukaryotic genomes and premiered their molecular and genomic analysis. Subsequent work has established that at least half of all spontaneous mutations in Drosophila have been caused by insertions of transposable elements (10), and that the genome of a wild-type Drosophila strain contains almost 900 transposon insertions. Together, these elements account for more than 3% of the genome (S. Celniker, personal communication). With the exception of transposable insertions that cause visible or lethal phenotypes, the consequences of harboring these invading DNA elements is not understood.
The transposons in the Drosophila genome have been characterized extensively, but the P element is the best understood in terms of regulation and mechanism of transposition. It is also the most widely used for constructing transgenic lines, engineering strains for regulated ectopic expression of genes, generating strains for making genetic mosaics, marking chromosomes, and mutagenesis. A comprehensive listing and description of the many ways P elements have been used is beyond the scope of this essay, but two that have been exploited by Huet et al. are relevant. One, Drosophila P elements preferentially transpose into genomic regions close to their starting sites (11). Two, in 1990, Cooley, Thompson, and Spradling (12) found that mitotic recombination promoted by P element transposase can delete all chromosomal DNA between two appropriately positioned P elements. These authors also pointed out that this technique could be used to delete any region of the Drosophila genome if a sufficiently large collection of lines containing P element insertions was available. Any chromosome carrying duplicated DNA segments that are both closely spaced and targets of transposase are subject to intrachromosomal recombination and therefore to deletion generation. Numerous large collections of P element insertion lines now exist, the most extensive and best characterized being the one that was organized through the Berkeley Drosophila Genome Project to generate P insertions that disrupt most Drosophila ORFs. This is an ongoing effort; and as of 1999 it had more than 1,000 lines, each carrying a P element that disrupts an essential gene (13). Each P insertion has been sequenced, providing many possible lines that potentially can be used to create deletions with precisely defined endpoints.
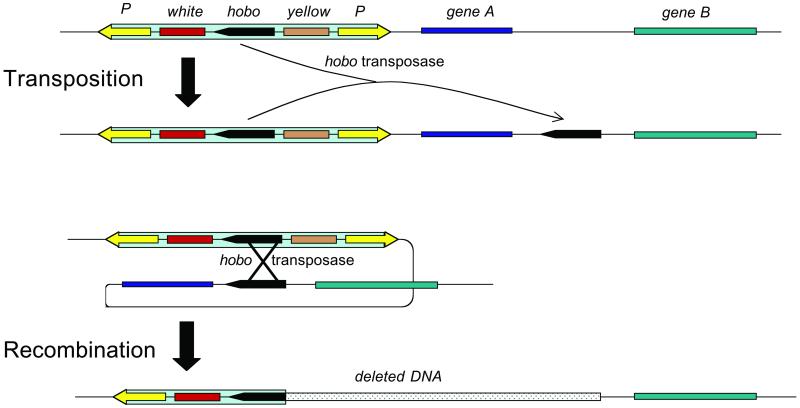
Enter Huet et al. Building on the Gelbart lab's extensive characterization and deep understanding of the hobo transposable element, these workers created a hybrid element (P{wHy}) that contains ingredients of both the P and hobo elements (Fig. 1). Its ends derive from the P element LTRs and render the construct a target of P-mediated transposition. Huet et al. created insertion lines containing P{wHy} by using standard P-based methods. Adjacent to the P ends of P{wHy} are two genes whose functions are easily scored in adult flies: white+ (w), without which the fly's eyes develop without pigment, and yellow+ (y), without which the fly's body color fails to darken. Between w+ and y+, the authors placed a segment of hobo that is a target of the hobo transposase and that when mobilized, efficiently moves to sites in its the immediate vicinity. Lines carrying P{wHy} were shown to efficiently generate deletions in the presence of hobo transposase.
Figure 1.
Deletions in lines carrying the hybrid P-hobo element P{wHy} are generated by hobo transposase in two steps. Deletions extend precisely to the point where the duplicated element inserted.
This remarkable property of P{wHy} lines is best understood in the context of the deletion-generating strategy engineered by Cooley et al. (12). Introduction of hobo transposase to a P{wHy} line can cause a local duplication of the hobo segment of P{wHy}, and if the duplicated element is in the same orientation as the original one, transposase-mediated recombination deletes DNA between the two elements. Depending on the relative orientation of the duplicated element, one of the two visible markers, w+ or y+, will be deleted. Flies expressing only one of the markers are easily scored; the authors show that most flies in these phenotypic classes have such deletions. Because all of the deletions share one common endpoint, the site of the insertion of P{wHy}, they constitute a nested set.
This method of generating deletions is marvelously efficient. In their study, Huet et al. created two such nested sets (100 and 113 deletions), starting with two independent P{wHy} lines. These deletions extended from 216 bp to 400 kbp, with smaller deletions being the most common. Genetic tests suggest that 60 kbp was the range within which the density of deletions was sufficiently high that deletions can distinguish every transcription unit in the region adjacent to original transposon. This elegant and efficient production of so extensive a repertory of deletions is a remarkable achievement.
Saturation and high resolution deletion analysis has no precedent in metazoans, and it is a tantalizing prospect to contemplate what its impact will be in Drosophila. Huet et al. suggest that sets of nested deletions will be useful to resolve complementation of extant alleles, to create null conditions by generating trans-heterozygous pairs of deletions, and to analyze genetic functions that are refractory to standard genetic approaches. And following on with the tradition that has long nurtured this field, they offer that they have initiated an effort to develop a collection of P{wHy} insertions that are regularly spaced over the entire genome. This collection will undoubtedly be a useful resource for many in the community to use, for it has the potential to arm Drosophila geneticists with a uniquely powerful weapon to attack many of the fascinating unresolved mysteries of chromosome organization and function.
Footnotes
See companion article on page 9948.
References
- 1.Huet F, Lu J T, Myrick K V, Baugh L R, Crosby M A, Gelbart W M. Proc Natl Acad Sci USA. 2002;99:9948–9953. doi: 10.1073/pnas.142310099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingham P M, Kidwell M G, Rubin G M. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- 3.Blackman R K, Grimaila R, Koehler M M, Gelbart W M. Cell. 1987;49:497–505. doi: 10.1016/0092-8674(87)90452-1. [DOI] [PubMed] [Google Scholar]
- 4.Morgan T H. Science. 1910;31:201–210. doi: 10.1126/science.31.789.201. [DOI] [PubMed] [Google Scholar]
- 5.Zachar Z, Bingham P M. Cell. 1982;30:529–541. doi: 10.1016/0092-8674(82)90250-1. [DOI] [PubMed] [Google Scholar]
- 6.Demerec M. Bot Rev. 1935;1:233–248. [Google Scholar]
- 7.Berg R L. Drosophila Inf Serv. 1974;51:100–102. [Google Scholar]
- 8.Green M M. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 1b. London: Academic; 1977. pp. 929–946. [Google Scholar]
- 9.Kidwell M G. Genet Res. 1979;33:105–117. [Google Scholar]
- 10.Finnegan D J. In: The Genome of Drosophila melanogaster. Lindsley D L, Zimm G G, editors. New York: Academic; 1992. pp. 1096–1107. [Google Scholar]
- 11.Tower J, Karpen G H, Craig N, Spradling A C. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooley L, Thompson D, Spradling A C. Proc Natl Acad Sci USA. 1990;87:3170–3173. doi: 10.1073/pnas.87.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spradling A C, Stern D, Beaton A, Rhem E J, Laverty T, Mozden N, Misra S, Rubin G M. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]