Abstract
Cited2 is a cAMP-responsive element-binding protein (CBP)/p300 interacting transcriptional modulator and a proposed negative regulator for hypoxia-inducible factor (HIF)-1α through its competitive binding with HIF-1α to CBP/p300. Disruption of the gene encoding Cited2 is embryonic lethal because of defects in the development of heart and neural tube. Morphological and Doppler echocardiographic analyses of Cited2−/− embryos reveal severe cardiovascular abnormalities, including pulmonic arterial stenosis and ventricular septal defects accompanied by high peak outflow velocities, features of the human congenital cardiac defect termed tetralogy of Fallot. The mRNA levels of several HIF-1α-responsive genes, such as vascular endothelial growth factor (VEGF), Glut1, and phosphoglycerate kinase 1, increased in the Cited2−/− hearts. The increase of VEGF levels is significant, because defects in the Cited2−/− embryos closely resemble the major defects observed in the VEGF transgenic embryos. Finally, compared with wild-type, cultured fibroblasts from Cited2−/− embryos demonstrate an enhanced expression of HIF-1α-responsive genes under hypoxic conditions. These observations suggest that functional loss of Cited2 is responsible for defects in heart and neural tube development, in part because of the modulation of HIF-1 transcriptional activities in the absence of Cited2. These findings demonstrate that Cited2 is an indispensable regulatory gene during prenatal development.
Cited2 [cAMP-responsive element-binding protein (CBP)/p300-interacting transactivators with glutamic acid (E) and aspartic acid (D)-rich tail] is one of the founding members of a new family of transcriptional activators, previously named melanocyte-specific gene-related gene (MRG)1/p35srj (1–6). Cited2 is a nuclear protein that binds directly, with high affinity to the first cysteine-histidine-rich (CH1) region of p300 and CBP. As a CBP/p300-dependent transcription factor, Cited2 competes with hypoxia-inducible factor (HIF)-1α in binding to the CH1 region of CBP/p300, thus interfering with hypoxia-driven transcription (4). Many transcription factors and factors that modulate transcription, such as RXRα, NFκB, Stat2, Mdm2, and Ets-1, also bind to the CH1 domain of CBP/p300 (7). In this fashion, Cited2 may function as a key transcriptional modulator to up- or down-regulate expression of specific genes (7). Cited2 also interacts with a Lin-11 Isl-1 Mec-3 (LIM) homeodomain-containing transcription factor, Lhx2, to enhance the recruitment of CBP/p300 and the TATA-binding protein leading to transcriptional activation of glycoprotein hormone α subunit (6). We have previously shown that Cited2 is induced by many biological stimuli, such as cytokines, serum, and lipopolysaccharide in different cell types (3). Overexpression of Cited2 in Rat1 cells results in loss of cell contact inhibition and tumor formation in nude mice, demonstrating that Cited2 is a transforming gene (3). These initial in vitro studies underscore the potential roles of Cited2 in different biological processes. Furthermore, during mouse and chicken embryo development, Cited2 transcripts are expressed at specific sites, including the precardiac mesoderm and immediately adjacent tissues such as the anterior visceral endoderm and the septum transversum. Cited2 is also expressed in the cranial neuroectoderm and adjacent tissues. In older stage mouse embryos, Cited2 has been detected homogeneously throughout the heart. This unique expression pattern suggested a potential role for Cited2 in the development of the heart and anterior head regions (2, 8).
Although previous in vitro studies have suggested Cited2 to be a modulator for HIF-1α functions through its competitive binding to the CH1 domain of CBP/p300, the role of Cited2 in regulation of HIF-1α activities in vivo has not been reported. Hypoxia induces angiogenesis, a compensatory mechanism that supplies oxygen during mouse embryogenesis. As a critical transcription factor regulating the hypoxia-induced genes, HIF-1 is one of the most potent inducers of several well-known angiogenic factors, including vascular endothelial growth factor (VEGF). HIF-1α, VEGF, and its negative regulator, Lkb1, play important roles in embryonic vascularization and neurulation (9–13). Mouse embryos deficient in these proteins exhibit abnormalities in vascular and neural tube development. To address the role of Cited2 in vivo during different cellular processes and embryonic development, we used gene targeting in embryonic stem (ES) cells to generate mice with a null mutation at the Cited2 locus. We report here that disruption of Cited2 results in cardiac malformations and neural tube defects (NTDs) in Cited2−/− embryos. The Cited2−/− embryos display increased expression of HIF-1-responsive genes and similar phenotype with VEGF transgenic embryos. These results are consistent with the model that in the absence of Cited2, HIF-1 is better able to compete for the shared binding site of CBP/p300, leading to up-regulation of HIF-1-responsive genes and the morphological defects we observe.
Methods
Generation of Cited2-Deficient Mice.
Chimeras were produced by microinjecting targeted R19 ES cells in C57BL/6J blastocysts and transplanting the embryos into uteri of pseudopregnant females. Chimeric males were mated initially to C57BL/6J females, and agouti offspring were screened by Southern blotting of tail genomic DNA for germline transmission of the targeted allele. Two independent ES clones generated germline chimeras. Once productive heterozygous mice were identified, they were intercrossed, and the genotype of offspring was determined by Southern blotting of embryo-derived DNAs with the probe shown in Fig. 1a. All experiments were conducted in accordance with the institutional guidelines of Case Western Reserve University.
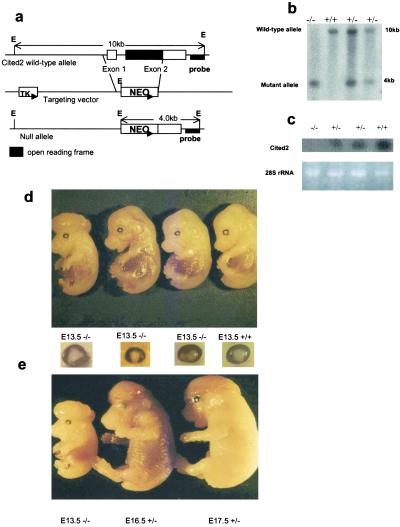
Fig 1.
Generation of Cited2-deficient mice. Cited2-deficient mice were generated by standard methods. (a) The Cited2 locus and targeting construct. Exon 1 and part of exon 2 that contains the full-length of the Cited2 coding region were deleted and replaced with a neo gene. The 3′ flanking probe used to screen ES cell clones and mice is indicated. E, EcoRI; TK, thymidine kinase. (b) Southern blot analysis of representative tail DNAs from the heterozygotic intercross. Genomic DNA was digested with EcoRI, transferred to a nylon membrane and hybridized with the 3′ flanking probe. (c) Northern blot analysis of total RNAs from the E13.5 Cited2−/−, Cited2+/−, and wild-type embryos. Ten micrograms of total RNA was loaded in each lane. (Lower) The ethidium bromide-staining pattern of 28S rRNA in each lane. (d) All of the Cited2-deficient embryos at E13.5 exhibited an eye phenotype similar to iris coloboma and a subset developed exencephaly. (e) Some of the Cited2−/− and Cited+/− embryos developed exencephaly.
Histological Analysis.
Hematoxylin/eosin-stained sections were prepared by standard methods (14). For proliferating cell nuclear antigen (PCNA) immunostaining, a mouse monoclonal antibody against PCNA and the ImmunoCruz Staining System Kit were used according to protocols provided by the manufacturer (Santa Cruz Biotechnology). BrdUrd was administered to pregnant females (100 mg/kg of body weight in PBS) on embryonic day (E)13.5 by i.p. injection. The embryos were harvested after 2 hr, fixed, and processed for immunohistochemical analysis by using protocols provided along with the anti-BrdUrd antibody (Sigma).
Heart Function of Embryos Detected by in Utero Doppler Echocardiography.
Mice were anesthetized with 2.5% tribromoethanol (0.01 ml/g of body weight, i.p.). A warm pad (Deltaphase Isothermal Pad, Braintree Scientific) maintained normothermia. After the abdomen was shaved and prewarmed ultrasound transmission gel (Parker Laboratories, Fairfield, NJ) was applied to the anterior abdominal wall, Doppler echocardiographic studies were performed by using a 15-MHz (15L8) phased array imaging and a 7-MHz Doppler transducer (Sequoia, Acuson, Lake Oswago, OR). The peak outflow velocity (Ovel) was measured, and three beats were averaged for each measurement.
Northern Blot Analysis of Cited2−/− Heart Tissue and Mouse Embryonic Fibroblast (MEF) Cultures.
Total RNA samples were collected from the heart tissue of wild-type and Cited2−/− embryos at E14.5. Total RNA samples from 24 embryos for each genotype group (Cited2−/− and Cited2+/+) were pooled to obtain enough RNA for Northern blot analysis. For hypoxia experiments, MEF cells were derived from the E13.5 embryos. Embryonic tissue was minced, passed through 18-g needles, and MEFs were cultured in Delbecco's modified Eagle's medium containing 15% FCS, glutamine, penicillin, and streptomycin for 4 days. MEFs (2.5 × 105) from individual embryos were plated on a 6-cm plate. The medium was changed once after overnight culture under normal conditions. After fresh medium was added, Set A and B of the MEFs were subjected to normoxic (21% O2) and Set C to hypoxic (1% O2) conditions. After 20 hr, plates of Set B MEF cultures grown under normoxic conditions were shifted to a hypoxic condition. Four hours later, total RNA was extracted from the three sets of the MEF cultures by using Trizol Reagent (GIBCO/BRL). For electrophoresis, 10 μg of total RNA was loaded per lane. Equal loading was demonstrated by assay of β actin mRNA levels, and mRNA levels were quantified with a densitometer and quantity one software (Bio-Rad).
For the ELISA of VEGF in MEFs, 1.5 × 105 MEFs from individual embryos in passage 3 were plated on 24-well plates. The medium was changed after overnight incubation. One set of MEFs was cultured under normoxic conditions, whereas the other set was cultured under hypoxic conditions for 24 hr. The medium was removed, and the VEGF concentration was analyzed by ELISA (R&D Systems). Remaining cells were trypsinized, and numbers of viable cells in each well were counted.
Results
Loss of Cited2 Results in Embryonic Lethality.
The targeting vector was designed to replace the entire Cited2 coding region with the neomycin gene (Fig. 1a). Germline transmission of the Cited2 mutant allele was obtained from two independent targeted ES clones (Fig. 1b). The heterozygous mutant mice were intercrossed to generate Cited2−/− mice. Heterozygous pups for the Cited2 mutation (Cited2+/−) were phenotypically indistinguishable from Cited2+/+ littermates and were generally healthy. However, among 438 progeny from the heterozygous matings, 191 pups were Cited2+/+, 247 were Cited2+/−, and no Cited2−/− pups were found. Note that fewer heterozygotes were born than expected for a single Mendelian element, suggesting significant embryonic lethality among the Cited2 heterozygotes (Table 1). To determine whether the Cited2 disruption is a null mutation, total RNA of the E13.5 embryos from heterozygous crosses were evaluated by Northern blot analysis. As shown in Fig. 1c, the Cited2 mRNA was not detected in homozygous mutant embryos and was reduced by about half in heterozygous mutant embryos compared with wild-type littermates. To characterize the embryonic phenotype, timed matings between Cited2+/− mice were conducted. At E13.0 and earlier gestational periods, Cited2−/− embryos were viable and recovered at a Mendelian ratio of 1:2:1 (+/+:+/−:−/−) (Table 2). Embryonic lethality (lacking both a pulsating stream of blood within the umbilical artery/vein and a regularly contracting heart) was detected after 13 days of gestation. Most (95%) of the homozygotes died between E13.0 and E17.5, and none survived beyond E18.5. Cited2 genotype analysis revealed that all of the Cited2−/− and a proportion of the Cited2+/− embryos died in utero. All viable Cited2−/− and a small portion of the Cited2+/− (10%, 21/202) embryos recovered after E12.5 were smaller in size and displayed overall growth retardation compared with the wild-type littermates (Table 2; Fig. 1d).
Table 1.
Gene dosage effect of Cited2 disruption
| Genotype | Male (expected no., survival rate) | Female (expected no., survival rate) | Total (expected no., survival rate) |
|---|---|---|---|
| −/− | 0 (102, 0%) | 0 (89, 0%) | 0 (191, 0%) |
| +/− | 143 (204, 70%) | 104 (178, 58%) | 247 (382, 64%) |
| +/+ | 102 | 89 | 191 |
| Total | 245 | 193 | 438 |
Table 2.
Prenatal lethality of Cited2 disruption
| Age
|
No. of litters
|
No. of normal embryos
|
No. of retarded or abnormal embryos
|
No. of resorbed embryos
|
No. with Cited2 genotype | ||
|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||||
| E18.5 | 7 | 37 | 2 (2 NTD) | 15 | 15 | 24 (2 NTD) | 0 |
| E17.5 | 6 | 35 | 6 (2 NTD) | 9 | 13 | 27 (3 NTD) | 1 |
| E16.5 | 10 | 54 | 13 (11 NTD) | 13 | 21 | 36 (3 NTD) | 10 (8 NTD) |
| E14.5–15.5 | 18 | 104 | 31 (21 NTD) | 12 | 36 | 71 (1 NTD) | 28 (20 NTD) |
| E9.5–14 | 12 | 80 | 17 (17 NTD) | 2 | 23 | 44 (1 NTD) | 25 (16 NTD) |
NTD ratio in −/− embryos, 44/64 (69%); NTD ratio in +/− embryos, 10/202 (5.0%).
5 of 8 NTD embryos died.
2 of 20 NTD embryos died.
Abnormal Neural Development in Cited2 Mutant Mice.
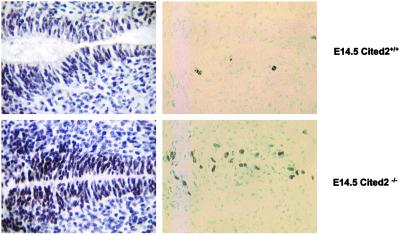
More than half of the homozygous embryos (69%) developed NTD (Table 2) consisting solely of open cranial neural tubes (exencephaly; Fig. 1d). NTD were also observed in 5% (10/202) of the heterozygous embryos (Fig. 1e). In addition, an eye phenotype referred to as iris coloboma was found in all of the Cited2−/− embryos after E12.5 (Fig. 1d). Hematoxylin/eosin-stained sections of the E12.5–14.5 embryos at the level of the ventricular outflow tract also revealed premature closure of the central canal in the spinal cord of a majority of the Cited2−/− (11/13) embryos (Figs. 2b and 3). Consistent with this observation, PCNA and BrdUrd staining in these regions demonstrated increased proliferation of ependymal layer cells surrounding the central canal in E14.5 Cited2−/− embryos compared with that of wild-type embryos (Fig. 3).
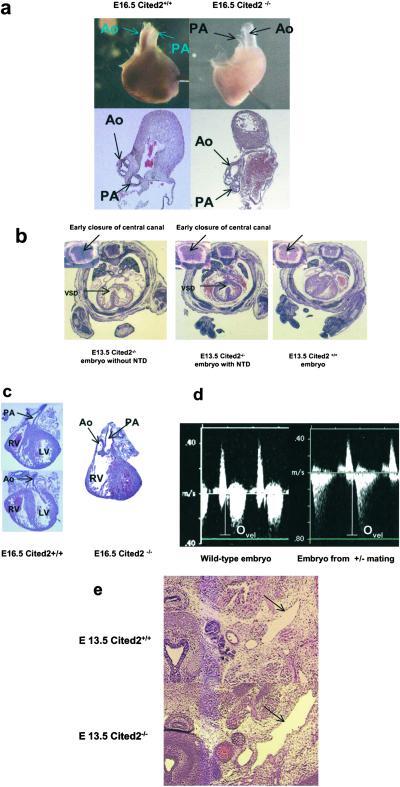
Fig 2.
Histological analysis of Cited2−/− embryonic heart. (a) Whole-mount and transverse section of an E16.5 Cited2−/− heart to demonstrate the smaller diameter of the PA. (b) Transverse sections of the E13.5 embryos stained with hematoxylin/eosin to demonstrate VSD and the early closure of the central canal of the spinal cord (Inset). (c) Transverse section of the heart from an E16.5 Cited2−/− embryo to demonstrate that both the Ao and PA connected to the RV resembling double outlet right ventricle. (d) Abnormal heart function of the Cited2-deficient embryos detected by in utero Doppler echocardiography. Cardiac blood flow seen by Doppler echocardiography in a normal E14.5 embryo derived from wild-type mouse mating. Peak systolic ejection velocity (Ovel) in this embryo is 37 cm/s (Left). An E14.5 embryo derived from the Cited2+/− mouse mating exhibited a peak systolic ejection velocity of 76 cm/s (Right). (e) Enlarged jugular lymph sac observed in an E13.5 Cited2−/− embryo (arrows). LA, left atria; LV, left ventricle; Ovel, peak systolic ejection velocity; RA, right atria; RV, right ventricle.
Fig 3.
Proliferation of E14.5 Cited2−/− spinal ependymal cells. Transverse sections of E14.5 (Upper) wild-type and (Lower) Cited2−/− embryos subjected to PCNA (Left) and BrdUrd (Right) immunostaining at the level of the ventricular outflow tract.
Abnormal Heart Development in Cited2 Mutant Mice.
All Cited2−/− embryos that survived to E16.5 exhibited conotruncal defects severe enough to be detected by stereomicroscopy of the intact heart. A consistent feature of the defects was the smaller external diameter of the pulmonary artery (PA) compared with the aorta (Ao). The Ao and PA are equal in diameter at these stages in the wild-type mouse embryo (5/5; Fig. 2a). A majority (16/19) of E13.5–16.5 Cited2−/− embryos had ventricular septal defects (VSDs) (Fig. 2b) and misalignment of the Ao and PA with respect to each other and the ventricular chambers. The range of conotruncal defects included two E13.5 and an E16.5 embryo with double outlet right ventricle (Fig. 2c) and two E12.5 and one E15.0 embryos with persistent truncus arteriosus (data not shown). Various malformations of atrioventricular valves were also observed in at least three of the E14.5–15.5 Cited2−/− embryos examined (data not shown).
To better understand the anatomy of the Cited2−/− cardiovascular system, we injected blue polymer into the left and/or right ventricles of the E14.5 wild-type or Cited2−/− embryos. The blue polymer injected into the ventricles of the embryonic heart leaked into the atria of 2/4 of the Cited−/− embryos but never into the atria of the Cited+/+ embryos (n = 4). This result supports defects in the unidirectional function of the atrioventricular valves, which were observed in some of sections from E14.5–15.5 embryos. Polymer injected into one ventricle leaked into the other ventricle (4/4) confirming the presence of functional VSDs in these animals (data not shown). The results of in utero Doppler echocardiography are consistent with the morphological findings of conotruncal defects (Fig. 2d). Three litters with only Cited2+/+ embryos (Ovel = 33.72 ± 4.65 cm/s, n = 14; Fig. 2d Left) and three litters with Cited2+/+ and Cited2+/− embryos (four Cited2+/+ and eight Cited2+/−) exhibited normal peak outflow velocities (Ovel = 36.05 ± 3.70 cm/s, n = 12). Three litters from heterozygous matings that included six Cited−/− embryos had five embryos with abnormally high peak outflow velocities (Ovel = 69.87 ± 10.65 cm/s, n = 5; Fig. 2d Right). This doubling of peak outflow velocity is consistent with a narrowing of the conotruncus in the Cited2−/− embryos, which could be the result of a normal volume of blood exiting the ventricles through the Ao and an abnormally narrowed PA (Fig. 2a).
Disruption of Cited2 Enhanced HIF-1α-Responsive Gene Expressions.
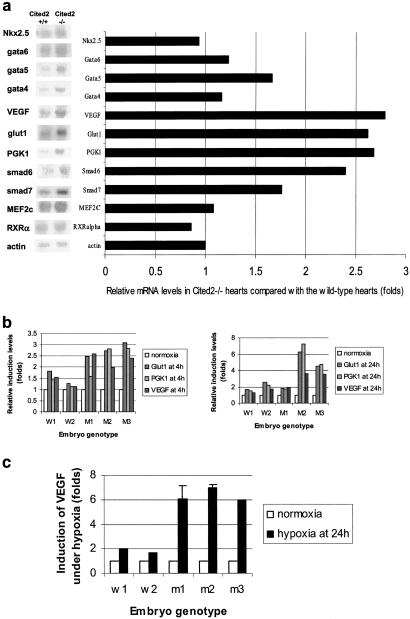
Cited2 has been proposed to be a negative regulator for HIF-1 through its competitive binding to the CH1 domain of CBP/p300, where HIF-1α also binds (4). The consequences of Cited2 disruption supported its proposed role as a regulator of HIF-1 function. As shown in Fig. 4a, PGK1, Glut1, and VEGF mRNAs were highly expressed in the E14.5 Cited2−/− embryonic heart. The transcript levels of all three HIF-1-responsive genes (VEGF, Glut1, and phosphoglycerate kinase (PGK)1 assayed were also increased in the Cited2−/− MEFs compared with MEFs from the wild-type littermate controls under hypoxic conditions, especially at the 4-hr hypoxia time point (Fig. 4b). The difference in induction kinetics among MEF cultures could be due to variations in cell population and genetic background of different primary MEF cultures. VEGF protein levels in the supernatant were also much higher in mutant compared with the wild-type MEFs under hypoxia at 24 hr (Fig. 4c). This is significant because the heart defects and the enlarged jugular lymph sac (Fig. 2e) of the Cited−/− embryos are similar to those observed in mouse embryos overexpressing VEGF (12). No significant differences between mutant and wild-type embryonic heart tissues were observed in the transcript levels of genes known to be important for heart development (MEF2, Nkx2.5, GATA4–6, RXRα, Smad 6 and 7) except for a modest increase in Smad6 mRNA in Cited2−/− hearts (15).
Fig 4.
Northern blot and ELISA analysis of Cited2−/− heart tissue and MEF cultures. (a) Northern blot analysis and PhosphorImager analysis of RNA levels from the E14.5 wild-type and Cited2−/− hearts. Five micrograms of total RNA isolated from the pool of 24 E14.5 Cited2+/+ or Cited2−/− embryonic heart was loaded per lane and hybridized with cDNA probes for Nkx2.5, Gata4, Gata5, Gata6, Smad6, Smad7, MEF2C, VEGF, Glut1, PGK1, RXRα, and β actin (control for sample loading). The data represent duplicate results. (b) PhosphorImager analysis of HIF-1α target genes, Glut1, PGK1, and VEGF, in the wild-type (W1, W2) and Cited2−/− (M1, M2, M3) MEF cultures, which were derived from independent littermate embryos, under normoxia or hypoxia (1% O2) at 4 (b-1) or 24 hr (b-2). (c) VEGF levels in the supernatants of the wild-type (W1, W2) and mutant (M1–3) cultures, each generated from independent littermate embryos. The levels of VEGF detected were normalized with the viable cell numbers in each well for comparison. For hypoxia induction, one set of the MEF cultures under normoxia was compared with another set subjected to 1% O2 for 24 hr. Error bars indicate the standard deviation in independent VEGF ELISA measurements.
Discussion
In the current study, we used the gene knockout approach and physiological measurements to characterize the consequences of Cited2 disruption. In addition, we provide a mechanistic interpretation for the observed phenotype. We observed malformations in the heart and neural tube regions of Cited2−/− embryos, where Cited2 expressed during embryonic development (2). The morphological defects in the cardiovascular system are supported by in utero Doppler echocardiography (Fig. 2d Right). The results from this physiological measurement strongly support that malfunction in outflow tracts may cause the embryonic lethality of the Cited2 null embryos. The defects of Cited2−/− embryos described in this study are similar to those found in embryos overexpressing the VEGF gene (12). In addition, we demonstrate a higher expression of several HIF-1 target genes including VEGF in the Cited2−/− heart and fibroblasts. These data strongly suggest that the morphological and functional defects in Cited2−/− embryos are mediated by the dysregulated expression of HIF-1 target genes.
HIF-1α is an important transcription factor that is required for cardiovascular development and O2 homeostasis (9–11). It activates transcription of erythropoietin and VEGF to provide metabolic adaptation under reduced O2 conditions (16). It also activates expression of glucose transporters and glycolytic enzymes, such as PGK1 and Glut1 (9–11, 15). Mice lacking HIF-1α die of abnormal mesenchymal cell death at midgestation (10, 11). Cited2 is transcriptionally induced by hypoxia and has been suggested to be a regulator for HIF-1α in vitro through their competitive binding to CBP/p300 (4). In this study, we use gene targeting to provide supporting evidence that Cited2 is a negative regulator for HIF-1α in vivo. VEGF, which is overexpressed in Cited2−/− heart and fibroblasts under hypoxia, plays an important role in coronary vasculogenesis and angiogenesis (11, 16). The development of the cardiovascular system is exquisitely dependent on normal levels and appropriately timed expression of VEGF. Haploinsufficiency in mice carrying one functional VEGF allele results in early embryonic lethality. Conversely, a modest (2- to 3-fold) increase in VEGF during development results in lethality of VEGF transgenic embryos at E12.5–14.5 of severe VSD and abnormal outflow tracts in the heart and enlargement of jugular lymph sac (12). The lethal heart phenotype associated with VEGF overexpression suggested that VEGF is crucial for remodeling of this developing organ (12). Interestingly, similar heart defects were observed in the Cited2−/− embryos, which expressed higher levels of VEGF (Fig. 4). The dilatation of the jugular lymph sacs in Cited2−/− embryos is also consistent with the increased permeability of vessels as a consequence of elevated VEGF levels (Fig. 2e; ref. 11). Similar to the VEGF overexpressed transgenic model, we did not observe obvious vascular defects in the Cited2−/− embryos. This observation could be due to a higher localized concentration of VEGF, because HIF-1α is mainly expressed in the head and heart regions during gestation at E14.5 (9). Alternatively, the heart may be more sensitive than the vasculature to a modest elevation of VEGF expression in a hypoxic environment (11).
Hypoxia not only plays an important role in embryonic heart morphogenesis but is also involved in the development of many tissues (9). At E12.5, hypoxic regions are limited mainly to neural tubes, cardiac myocytes, metanephric masses, and intervertebrae regions (9). The development of NTD in HIF-1α null mice suggests that response to oxygen tension may influence neural tube closure (9). Furthermore, p53 targets HIF-1α for ubiquitination, and the loss of p53 activity primarily leads to increased expression of HIF-1α under hypoxia (17). Interestingly, a majority of p53 null embryos developed normally, and 20% of female null mice exhibited NTD (18). Compared with the p53 null phenotype, deletion of Cited2 resulted in more severe NTD, because more than half of the Cited2-deficient embryos and 5% of the Cited2+/− embryos exhibited NTD. Consistent with the NTD in Cited2−/− embryos, disruption of Lkb1, which leads to overexpression of VEGF, results in NTD in Lkb1−/− embryos (13). We also observed overproliferation of the ependymal layer in the spinal cord during early development (Fig. 3), which may result in the early obliteration of the central canal in all of the Cited2−/− embryos at E12.5–14.5 (Fig. 2a). Extensive cell death in cephalic mesenchyme was demonstrated in HIF-1α null embryos (10). However, the correlation between the overproliferation in the ependymal cells and the physiological status of mesenchyme in its adjacent region is unclear, as are the mechanisms for the NTD in some of the Cited2−/− embryos.
Our results show that loss of the Cited2 allele is a completely penetrant recessive lethal mutation, even on a segregating genetic background. Like many other p300-dependent transcription factors, such as p300 itself, HIF-1α, p53, AP-2, RXRα, and Pax-3, Cited2 is required for proper neural tube morphogenesis. Among these transcription factors, targeted disruptions of p300, HIF-1α, RXRα, and Pax-3 genes contribute to heart defects (19, 20). In fact, the striking similarities in the knockout phenotype among p300-dependent transcription factors also provide a possible explanation for the gene dosage effects of Cited2 (4). Previous studies have suggested that proper integration of transcription signaling by CBP/p300 requires a balanced interaction among CBP/p300-interacting proteins, including Cited2 (4, 6). Bhattacharya et al. (4) demonstrated that endogenous Cited2 can be induced by hypoxia and inhibits HIF-1 transactivation by competitively blocking the HIF-1α/CBP/p300 interaction (4). Our in vitro experiments and gene-targeting results support crosscoupling or interference between CBP/p300 and transcription factors, such as HIF-1α. Thus, a reduction in the dose of Cited2 might disturb a critical balance of some transcription factors during early gestation and cause dysfunction of other factors. Death and NTD phenotype observed in a fraction of the heterozygous embryos (Tables 1 and 2) also support a requirement for the precise control of the Cited2 levels in vivo.
During the submission of this manuscript, Bamforth et al. (21) reported similar heart and neural-tube defects resulting from Cited2 disruption and demonstrated that AP-2 (TFAP2), a transcription factor for neural tube and NCC development, is one of the potential targets for Cited2 in vivo. The authors suggested that abnormal migration of NCCs could explain the heart defects in the Cited2 null mice (21). Neural crest cell abnormalities could result in cardiac defects, including those of the Cited2−/− embryos (19, 22), and the expression pattern of Cited2 during development also suggests its role in the migration of NCCs (2). Our in situ hybridization data with Snail, a molecular marker for NCC (23), demonstrated a decrease in the number of NCCs in the circumpharyngeal region of the E10.5 Cited2−/− embryos (data not shown). However, this mechanism alone may not explain all of the common features displayed in the Cited2−/− embryos. The expression of Pax3 and RXRα, which are p300-dependent transcription factors involved in NCC migration and OFT development (19, 22), appears to be normal in our Cited2−/− embryos (data not shown). Unlike the completely penetrant defect of the fourth pharyngeal arch arteries (PAAs) observed in the E10.5 Tbx1-deficient embryos (24), the Cited2−/− embryos did not show an obvious defect in the fourth PAAs at the E10.5 stage (data not shown). In agreement with our assessment, a recent study using a sensitive episcopic fluorescence image-capturing technique detected only a very subtle defect in PAAs in one of the Cited2−/− embryos examined at a much later stage, E14.5 (25). Interestingly, Martinez-Barbera et al. detected little or no difference in the expression patterns of many neural crest markers, such as Crabp-1, Wnt1, Wnt3, and Pax3, in Cited2 null embryos in their most recent report (26). In addition, immunostaining of neural crest derivatives revealed no significant defects in cranial and dorsal root ganglia in Cited2−/− embryos (26). Consistent with these recent independent data, our results suggest that defects in NCC alone may not be sufficient to explain all of the phenotypes in the Cited2 null embryos. Cited2, if involved, may play a different role from Pax3 and RXRα in NCC migration. In addition, we propose that the global effects of Cited2 on hypoxia-mediated events may also be involved. This global effect may encompass effects on NCCs, as well as other cell types.
Congenital cardiac malformations represent the largest group of congenital defects in humans, with 1 of 200 live births affected. The defects we observed in the Cited2−/− embryos, such as VSD and conotruncal defects, represent the most common congenital malformations of human infants (19). Human exencephaly or spina bifida occurs in approximately 1 in 1,000 births (27). The etiology of both sets of malformations is poorly understood. The human Cited2 gene has been mapped to 6q23.3 (5). Interestingly, a wide variety of congenital heart malformations have been reported in patients with 6q deletion, with 50% of the patients containing the deletion between 6q15 and 6q25 exhibited cardiac anomalies (28). It will be of interest to determine whether the Cited2 gene is mutated in patients with ToF and other forms of congenital heart diseases.
Acknowledgments
We thank Dr. D. Yang at Eli Lilly Company for providing the targeting vector pGT-N29/tk and for helpful discussions; Drs. D. Sallee, C. Colmenares, and D. Donner for insightful suggestions; Ms. N. Liu, Ms. M. Hitomi, Ms. Y. Qiu, and Dr. G. Adhikary for technical assistance; and Duke University Medical Center for producing Cited2 chimeric mice. This study was supported by National Institutes of Health Grants CA78433, DK50570, HL48819 (to Y.-C.Y.), and NSF0074882 (to M.W.).
Abbreviations
CH1, high affinity to the first cysteine-histidine-rich
CBP, cAMP-responsive element-binding protein
ES, embryonic stem
PCNA, proliferating cell nuclear antigen
Ovel, outflow velocity
MEF, mouse embryonic fibroblast
En, embryonic day n
NTD, neural tube defect
PA, pulmonary artery
Ao, aorta
VSD, ventricular septal defect
HIF, hypoxia-inducible factor
To whom reprint requests should be addressed. E-mail: yxy36@po.cwru.edu.
References
- 1.Shioda T., Fenner, M. H. & Isselbacher, K. J. (1997) Gene 204, 235-241. [DOI] [PubMed] [Google Scholar]
- 2.Dunwoodie S. L., Rodriguez, T. A. & Beddington, R. S. P. (1998) Mech. Dev. 72, 27-40. [DOI] [PubMed] [Google Scholar]
- 3.Sun H. B., Zhu, Y., Sledge, G. & Yang, Y. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya S., Michels, C. L., Leung, M. K., Arany, Z. P., Kung, A. L. & Livingston, D. M. (1999) Genes Dev. 13, 64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung M. K., Jones, T., Michels, C. L., Livingston, D. M. & Bhattacharya, S. (1999) Genomics 61, 307-313. [DOI] [PubMed] [Google Scholar]
- 6.Glenn D. J. & Maurer, R. A. (1999) J. Biol. Chem. 274, 36159-36167. [DOI] [PubMed] [Google Scholar]
- 7.Giles R. H., Peters, D. J. M. & Breuning, M. H. (1998) Trends Genet. 14, 178-183. [DOI] [PubMed] [Google Scholar]
- 8.Schlange T., Andree, B., Arnold, H. & Brand, T. (2000) Mech. Dev. 98, 157-160. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y. M., Jeong, C. H., Koo, S. Y., Son, M. J., Song, H. S., Bae, S. K., Raleigh, J. A., Chung, H. Y., Yoo, M. A. & Kim, K. W. (2001) Dev. Dyn. 220, 175-186. [DOI] [PubMed] [Google Scholar]
- 10.Iyer N. V., Kotch, L. E., Agani, F., Leung, S., W., Laughner, E., Wenger, R. H., Gassmann, M., Gearhart, J. D., Lawler, A. M., Yu, A. Y., et al. (1998) Genes Dev. 12, 149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan H. E., Lo, J. & Johnson, R. S. (1998) EMBO J. 17, 3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miquerol L., Langille, B. L. & Nagy, A. (2000) Development (Cambridge, U.K.) 127, 3941-3946. [DOI] [PubMed] [Google Scholar]
- 13.Ylikorkala A., Rossi, D. J., Korsisaari, N., Luukko, K., Alitalo, K., Henkemeyer, M. & Makela, T. P. (2001) Science 293, 1323-1326. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman M. H., (1995) The Atlas of Mouse Development (Academic, London), pp. 1–15.
- 15.Zhu H. & Bunn, H. F. (1999) Respir. Physiol. 115, 239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dor Y., Camenisch, T. D., Itin, A., Fishman, G. I., McDonald, J. A., Carmeliet, P. & Keshet, E. (2001) Development (Cambridge, U.K.) 128, 1531-1538. [DOI] [PubMed] [Google Scholar]
- 17.Ravi R., Mookerjee, B., Bhujwalla, Z. M., Sutter, C. H., Artemov, D., Zeng, Q., Dillehay, L. E., Madan, A., Semenza, G. L. & Bedi, A. (2000) Genes Dev. 14, 34-44. [PMC free article] [PubMed] [Google Scholar]
- 18.Sah V. P., Attardi, L. D., Mulligan, G. J., Williams, B. O., Bronson, R. T. & Jacks, T. (1995) Nat. Genet. 10, 175-180. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava D. & Olson, E. N. (2000) Nature (London) 407, 221-226. [DOI] [PubMed] [Google Scholar]
- 20.Yao T. P., Oh, S. P., Fuchs, M., Zhou, N. D., Ch'ng, L. E., Newsome, D., Bronson, R. T., Li, E., Livingston, D. M. & Eckner, R. (1998) Cell 93, 361-372. [DOI] [PubMed] [Google Scholar]
- 21.Bamforth S. D., Braganca, J., Eloranta, J. J., Murdoch, J. N., Marques, F. R., Kranc, K. R., Farza, H., Henderson, D. J., Hurst, H. C. & Bhattacharya, S. (2001) Nat. Genet. 29, 469-474. [DOI] [PubMed] [Google Scholar]
- 22.Conway S. J., Henderson, D. J., Kirby, M. L., Anderson, R. H. & Copp, A. J. (1997) Cardiovasc. Res. 36, 163-173. [DOI] [PubMed] [Google Scholar]
- 23.Sefton M., Sanchez, S. & Nieto, M. A. (1998) Development (Cambridge, U.K.) 125, 3111-3121. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay E. A., Vitelli, F., Su, H., Morishima, M., Huynh, T., Pramparo, T., Jurecic, V., Ogunrinu, G., Sutherland, H. F., Scambler, P. J., et al. (2001) Nature (London) 410, 97-101. [DOI] [PubMed] [Google Scholar]
- 25.Weninger W. J. & Mohun, T. (2002) Nat. Genet. 30, 59-65. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Barbera J. P., Rodriguez, T. A., Greene, N. D. E., Weninger, W. J., Simeone, A., Copp, A. J., Beddington, R. S. & Dunwoodie, S. (2002) Hum. Mol. Genet. 11, 283-293. [DOI] [PubMed] [Google Scholar]
- 27.Juriloff D. M. & Harris, M. J. (2000) Hum. Mol. Genet. 9, 993-1000. [DOI] [PubMed] [Google Scholar]
- 28.Hopkin R. J., Schorry, E., Bofinger, M., Milatovich, A., Stern, H. J., Jayne, C. & Saal, H. M. (1997) Am. J. Med. Genet. 70, 377-386. [PubMed] [Google Scholar]