Abstract
Background
The aim was to ascertain whether thrombocyte MAO (trbc-MAO) activity and depressed state are genetically associated with the MAO locus on chromosome X (Xp11.3 – 11.4). We performed novel sequencing of the MAO locus and validated genetic variants found in public databases prior to constructing haplotypes of the MAO locus in a Swedish sample (N = 573 individuals).
Results
Our results reveal a profound SNP desert in the MAOB gene. Both the MAOA and MAOB genes segregate as two distinct LD blocks. We found a significant association between two MAOA gene haplotypes and reduced trbc-MAO activity, but no association with depressed state.
Conclusion
The MAO locus seems to have an effect on trbc-MAO activity in the study population. The findings suggest incomplete X-chromosome inactivation at this locus. It is plausible that a gene-dosage effect can provide some insight into the greater prevalence of depressed state in females than males.
Background
Monoamine oxidase A (MAOA) and B (MAOB) are enzymes that deaminate monoamines such as serotonin, dopamine and noradrenaline. The genes encoding MAOA and B are located on the X chromosome in a tail-to-tail orientation and separated by approximately 20 kilobases (kb) [1,2]. Although MAOA and MAOB span 65 kb and 116 kb, respectively, both genes display a high degree of homology and most certainly have a common ancestry [3]. The frequencies of confirmed polymorphisms in the two genes vary widely among different ethnic groups [4-6]. Only two common haplotype variants of the MAOA locus were found among individuals of northern European ancestry [5].
Both enzymes are localized in the outer mitochondrial membrane [7]. They are also present in glial cells [8], although MAOA is less expressed than MAOB [9]. The enzymes differ in their expression patterns not only peripherally in the body but also in the central nervous system (CNS) [10]. MAOB is the only form that is expressed in human blood cells. MAOA is primarily expressed in catecholaminergic neurons in the human brain [10,11], whereas MAOB is expressed in serotonergic [10] and histaminergic neurons [8]. The two MAO-enzymes also differ on substrate preferences; MAOA preferentially metabolizes serotonin and norepinephrine while MAOB has a much higher affinity for phenylethylamine [12,13] and benzylamine [14].
Thrombocyte-MAO activity (Trbc-MAO) has been associated with cerebrospinal fluid (CSF) levels of serotonin metabolites in humans [15] and is higher in women than men [16-18]. This difference has been speculated to be an effect of sex steroids altering the enzyme's activity or a matter of escaped X-inactivation [19]. The proportion of variance in trbc-MAO activity explained by genetic factors (its heritability) in a Swedish population is 77% [20]. Trbc-MAO activity is weakly associated with a C/T polymorphism in intron 13 of the MAOB gene in a Swedish population [21] and is also influenced by smoking and specific medications; smokers can have a 30–40% lower trbc-MAO activity than non-smokers [22]. Trbc-MAO activity is also associated with several psychiatric syndromes [23], personality traits and mood disorders e.g. [24-28].
In the present study we address issues concerning genetic variation in MAOA and MAOB genes, activity levels of trbc-MAO, and associations with depressed state. Genetic variation was analyzed by sequencing the regulatory region of both MAOA and MAOB, and validating SNPs reported in public databases. We used multiple SNPs covering the MAO gene locus to generate haplotypes on a population level. Finally, we investigated associations between depressed state and trbc-MAO activity and genetic variants in the MAO locus in a large elderly Swedish population.
Results
Trbc-MAO activity and depressed state
We found a clearly significant difference between males (mean; 10.7) and females (mean; 12.1) (t = 4.69; p ≤ 0,0001), as well as between smokers and non-smokers in mean trbc-MAO activity (t = 5,86; p =< 0,0001). Smokers showed a 23% lower trbc-MAO activity compared to non-smokers. Females with a depressed state showed a significantly higher mean trbc-MAO activity than unaffected females (t = 2,02; p = 0,04).
Genetic variants and haplotype construction
Approximately 4.5 kb of both the MAOA and MAOB gene promotor, including the first exons, totaling 9 kb, were sequenced from a total of 148 X chromosomes. Power to discover SNPs with frequencies greater than 1% and 3% for this sample size was 77% and 100%, respectively. No variants were found in the MAOB gene. In contrast, one previously reported variation was confirmed (rs3788863) for the MAOA gene, lying within the first intron, as well as two additional variants further down stream with a minor allele frequency greater than 1%. Both the recorded and most distal variants showed complete LD with each other, therefore only the recorded variant was chosen for further analysis.
The genotyping error rate was calculated at 0,4% through males scoring as heterozygotes and from MZ twins where both twins in a pair were genotyped. These errors could not be scored differently from the sequence and therefore most likely reside in the handling of samples, e.g. contamination or labelling error.
In addition to resequencing the upstream regions, we genotyped reported SNPs in the remainder of the gene clusters by Pyrosequencing. Six of the previously reported SNPs could not be confirmed as polymorphic (rs1014876, rs3027464, rs6324, rs1040398 and two SNPs reported by Balciuniene et al.) The remaining nine polymorphic variants; one in the Norrie gene (rs766117), four SNPs in MAOB (rs1181252, rs2283729, rs3027452 and rs1799836) and four SNPs in MAOA (rs1801291, rs979605, rs6323, rs388863) were sequenced in the total sample.
The LD map (Figure 2) displays a clear structure of the MAO locus with strong LD across the MAOA gene in a distinct block spanning approximately 65 kb. The MAOB gene also displays a similar block-like structure, although the pattern of LD is not as robust as for MAOA. This is perhaps due to the inconsistencies in allele frequencies across MAOB. Interestingly, weak LD is observed at the tail ends between the two MAO loci.
Figure 2.
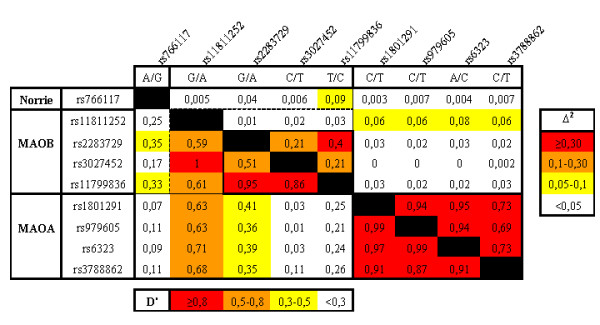
LD map. Pair-wise LD map with one individual from each female pair (N = 356). D' is shown below the diagonal and Δ2 above the diagonal. Color code D': Red: ≥0,8 Orange: 0,5–0,8 Yellow: 0,3–0,5 White: <0,3. Color code Δ2: Red: ≥0,30 Orange: 0,1–0,30 Yellow: 0,05–0,1 White: <0,05.
Furthermore, because there was no LD between the Norrie gene variant, located >66 kb upstream of MAOB, and any other variant in the MAO region, we decided not use this variant further in the haplotype assessment. Modest deviations from Hardy-Weinberg equilibrium were noted in rs766117 in the Norrie gene (p = 0,022) and rs979605 in intron 10 of the MAOA gene (p = 0,028). This could reflect the underlying LD structure [29], as demographic influences would act over larger regions [30]. However to clarify this, a denser set of SNPs would need to be genotyped.
In the male population we could identify five distinct haplotypes in the MAOB gene and four in the MAOA gene with frequencies ≥1% (Figure 1.). When analyzing the MAO locus as one large block using eight SNPs, we found ten distinct locus haplotypes with a frequencies ≥1% (data not shown). In the female population, "PHASE" assembled identical higher frequency haplotypes as were identified in the male sample, with minor discrepancies in lower frequency haplotypes due to unknown phase (Figure 1).
Figure 1.
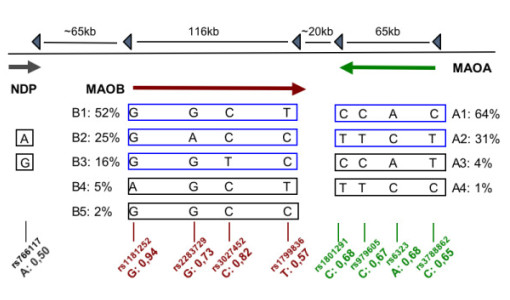
Genetic structure of the MAO locus. Haplotype and common allele frequencies in the total sample. dbSNP rs numbers for all genotyped SNPs are presented with major allele frequencies. Haplotypes frequencies illustrated for MAOA and B separately as well as the genes combined (See Text). NDP was not used in the haplotype frequency estimations.
Associations with SNPs
In the total sample, no single variant of any of the individual SNPs was associated with trbc-MAO activity. However, in females the C/C and C/T genotypes of rs979605 in the MAOA gene were associated with a significant decrease in trbc-MAO activity, (-2,9; CI 95%: -5,2 – -0,6) and (-2,4; CI 95%: -4,7 – -0,1) respectively.
Analyzed by gender, depressed state was associated with the A-allele of MAOB SNP rs1181252 in males (OR = 4,5; CI 95%: 1,0 – 21,7) and both GG and GA of rs766117 (OR = 2,2; CI 95%: 1,1 – 4,3) in females. It should be noted that the "A" allele of rs1181252 only had a population frequency of 6%.
Associations with haplotypes
There was no association between any of the MAOB haplotypes and trbc-MAO activity. Two MAOA haplotypes, A1 and A3, both sharing identical alleles at the three first haplotype positions (CCA-) (Figure 1), were associated with a significant decrease in trbc-MAO activity (Table 1). Analyses of the entire MAO locus and trbc-MAO activity did not reveal any significant findings (data not shown). We could not find any significant associations between depressed state and any specific haplotype in men or women (Table 2).
Table 1.
Associations between MAO haplotypes and trbc-MAO activity, reported as unit change in mean trbc-MAO activity per allele and controlling for gender and smoking status.
| Total sample N = 340 | Males N = 156 | Females N = 184 | |
| per allele | Hemizygous | per allele | |
| Haplotypes | Estimates (Unit change in mean trbc-MAO activity per allele) | ||
| B1 | -0,38 (-1,3 – 0,5) | 0 (ref) | -0,3 (-1,1 – 0,5) |
| B2 | -0,63 (-1,8 – 0,5) | -0,08 (-1,6 – 1,5) | -0,4 (-1,5 – 0,7) |
| B3 | -0,18 (-1,6 – 1,3) | -0,8 (-2,9 – 1,2) | -0,2 (-1,5 – 1,0) |
| B4 | -1,3 (-3,5 – 0,8) | 0,7 (-2,8 – 4,3) | -1,0 (-2,8 – 0,8) |
| B5 | -1,7 (-5,1 – 1,6) | NA | -0,7 (-3,8 – 2,4) |
| Male gender | -2,1 (-3,3 – -0,9)* | ||
| Non-smokers | 2,3 (1,1 – 3,5)* | 1,5 (0,2 – 2,8) | 3,5 (2,0 – 5,0) |
| A1 | -1,1 (-1,9 – -0,3)* | -1,8 (-3,2 – -0,5)* | -1,0 (-1,7 – -0,3)* |
| A2 | 0,1 (-0,9 – 1,2) | 0 (ref) | 0,6 (-0,4 – 1,5) |
| A3 | -3,1 (-6,1 – -0,14)* | -2,3 (-5,8 – 1,2) | -4,1 (-7,4 – -0,7)* |
| A4 | -0,02 (-3,8 – 3,7) | NA | -0,5 (-3,7 – 2,7) |
| Male gender | -2,3 (-3,5 – -1,1)* | ||
| Non-smokers | 2,4 (1,1 – 3,6)* | 1,4 (0,03 – 2,8) | 3,4 (1,9 – 4,8) |
Table 2.
MAO haplotypes and depressive state, reported as odds ratios per allele. Without genetic information in the model male gender was significant [OR: 0,5 (0,3 – 0,8)] for depressive state. *Homozygote compared to heterozygote.
| MAO haplotypes and depressive state | ||||
| Total sample N = 573 | Males N = 239 | Females N = 334 | ||
| per allele | Hemizygous | per allele | Homozygotes* | |
| Odds Ratio with 95% CI | ||||
| B1 | 1,2 (0,6 – 2,5) | 1,0 (ref) | 1,4 (0,6 – 2,9) | 1,3 p = 0,57 |
| B2 | 1,5 (0,7 – 3,3) | 1,7 (0,8 – 3,6) | 1,5 (0,6 – 3,3) | 1,2 p = 0,80 |
| B3 | 1,3 (0,5 – 3,0) | 0,7 (0,3 – 1,7) | 1,7 (0,7 – 4,2) | 1,9 p = 0,51 |
| B4 | 2,0 (0,8 – 5,2) | 3,7 (0,7 – 18,7) | 2,0 (0,7 – 5,3) | 1,7 p = 0,59 |
| B5 | 0,5 (0,1 – 2,8) | 2,4 (0,4 – 14,5) | 0,3 (0,04 – 2,5) | NA |
| Male gender | 0,7 (0,3 – 1,5) | |||
| A1 | 3,0 (0,8 – 12,2) | 1,0 (ref) | 2,2 (0,6 – 8,4) | 5,5 p = 0,08 |
| A2 | 2,5 (0,6 – 10,6) | 0,9 (0,4 – 1,8) | 1,7 (0,4 – 7,1) | 1,3 p = 0,80 |
| A3 | 2,8 (0,7 – 11,4) | 1,2 (0,3 – 4,4) | 1,7 (0,4 – 6,9) | NA |
| A4 | 3,2 (0,6 – 18,6) | NA | 2,8 (0,4 – 17,3) | NA |
| Male gender | 1,4 (0,3 – 6,0) | |||
When the model was analyzed without genetic information, males have a significantly lower risk for being affected with depressed state compared to women (OR = 0,5). This gender effect may be explained by the genetic information (even though no associations were found with any of the haplotypes), because the risk for depressed state due to the male gender is differs in the analyses of the MAOA locus (OR = 1,4; non-significant) and the MAOB locus, where the estimate is similar to the model without genetic information.
Interestingly, in females all MAOB homozygote haplotypes displayed greater odds ratios with depressed state than that for heterozygotes (Table 2), indicating an additive effect. The same was true for MAOA (Table 2).
Discussion
Monoamine oxidase A and B constitute two important molecules in the human body in general and in the central nervous system (CNS) in particular. Numerous studies suggest a contribution of these two mitochondrial enzymes to complex human behaviors [26-28,31-33]. In the present study we searched the MAO locus for novel genetic variants and evaluated the genetic and haplotype structure in a Swedish population. We also assessed associations between trbc-MAO activity and depressed state, and their respective associations with the genetic structure of the MAO locus. The key findings of this study are first: the profound lack of variation at functional regions of the two MAO genes and a pattern of two distinct genetic LD blocks, one for each gene. Second: we replicated the gender differences in trbc-MAO activity and demonstrated an association between trbc-MAO activity and depressed state in women. Third: two MAOA haplotype variants were associated with decreased trbc-MAO activity although we could not replicate a previously reported genetic association between the MAOB gene and trbc-MAO activity. Fourth: we could not find any significant associations between the genetic variants and depressed state. On the other hand, there was an interesting, although not significant dose-response effect of haplotypes displayed in women, with greater odds ratios in homozygotes than heterozygotes.
Considering the size and importance of the MAO locus, relatively few polymorphic sites have been verified. We observed two new variants through sequence screening a partial region of MAOA intron 1, but in MAOB neither the previously reported nor any novel variants were found in the areas sequenced. It is surprising that so few SNPs were discovered given our power to detect variants with very low frequencies. SNP deserts have been previously noted on the q arm of the X chromosome [34]. Gilad and colleagues [4] have described similar features across MAOA, where extensive LD and low nucleotide diversity suggest recent action by population structure forces and perhaps a recent positive selection sweep [35]. Although we could not evaluate the influence of such forces, evidence of strong LD and the lack of decay across MAOA in our Swedish sample complement these previous findings. Linkage disequilibrium decays rapidly between the two MAO genes (separated by approximately 20 kb). Perhaps selection is in action much more locally than would be expected in each MAO gene, both separated by regions of higher recombination than that within each gene.
Previous studies have indicated that the MAOA gene may harbour relatively few haplotypes within a block structure [5]. We observed similar results here with two haplotypes encompassing 95% of the haplotypic variation. We found similar results for the MAOB gene, with a distinct block structure in which three haplotypes explain 93% of the variation. So few haplotypes over such long distances have been observed previously (McCarthy et al, manuscript) and are proposed signatures of selection and population substructure on the X chromosome [36,37].
A previous Swedish association between the MAOB gene and trbc-MAO activity [21] could not be replicated nor distinctly refuted, as we found a small non-significant effect of the same allele in males. However, none of the haplotype blocks carrying this allele could strengthen or support this effect, suggesting that this allele is not in high LD with a larger region of the MAOB gene.
Two MAOA haplotypes (A1 and A3) showed a significant association with reduced trbc-MAO activity. Both haplotypes shared the initial sequence variants [CCA], but varied at the fourth allele [T/C]. Given that only MAOB is expressed in platelets there is no clear explanation for this finding. Given the minor kinetic differences between platelet and brain MAO-B [38] and the correlation of MAOB and MAOA levels in regions of the brain [39], this association may reflect MAOA activity in the brain. On the other hand, it is possible that the MAOA locus holds cis-acting regulatory elements affecting MAOB expression. Another possible explanation could be that one or several single-base variants affected by methylation cause changes in the expression pattern [40].
Our study is based on a relatively large population-based sample of normally aging adults, although it is not without its limitations. We have controlled for smoking, but were unable to do so for intake of certain medications. The study sample was included in a larger study where associations between depressed state and the serotonin receptor 2A and the serotonin transporter were evaluated [41]. The influence of these genes has not been corrected for in the analysis.
Conclusion
Good et al [19] demonstrated that trbc-MAO activity is related to the number of X chromosomes. We replicated a significant difference in trbc-MAO activity between males and females reported by others e.g. [17]. The findings suggest incomplete X chromosome inactivation at this locus and are consistent with other findings of genes escaping inactivation on the X chromosome [42,43]. It has been hypothesized that this dosage imbalance between males and females might be crucial for gender characteristics [19,44]. Recently it was demonstrated that a number of genes, including MAOA, escape X-inactivation [45]. Furthermore, the X-inactivation pattern, which shows a substantial heritability [46], increases in the elderly. Although we could not find a significant association between variants of MAOB or MAOA and depressed state in this population, we found an interesting dose-response effect in women, with a higher risk for depressed state with homozygosity. Whether levels of trbc-MAO activity are correlated with the number of X chromosomes and whether this might be linked to the higher prevalence of depressive symptoms in females deserves further investigation. Nevertheless it is plausible that a partially doubled gene activity on the X chromosome can explain difference in prevalence of depressive state in men and women.
Methods
Participants
The participants were taken from a longitudinal twin study of aging, the Swedish Adoption/Twin Study of Aging (SATSA) with up to five occasions of measurement [47]. SATSA is a sub-sample of the population based Swedish Twin Registry [48]. All participants are Caucasian and born in Sweden. For the present analyses we selected all individuals who participated in an in-person testing session during which questionnaires were administered and a blood sample was drawn. The mean age of the sample was 61,3 years at the time of testing. Twenty two percent of the participants were current smokers; 35% of the males and 15% of the females.
Zygosity was initially based on self-reports of similarity and confirmed by serological analyses and comparisons of up to 10 DNA markers.
For preliminary screening of the promoter, the first exon and intron regions for novel variants, 94 Swedish male blood donors were randomly selected from a larger sample set collected to study MAOB regulation. All were between the ages of 20 to 40 years and non-smokers.
This study was reviewed and approved by the Ethics Committee of the Karolinska Institute, the Swedish Data Inspection Board, and the IRBs at the University of Southern California and the Pennsylvania State University. All subjects provided informed consent.
DNA and trbc-MAO activity
DNA samples were available from 573 twins. Trbc-MAO activity measures were available from 565 twins. The trbc-MAO activity is expressed as nmoles of 2-phenylethylamine oxidized per minute and per 1010 platelets. Trbc-MAO activity measures have previously been described in detail [20].
Depressed state
Depressive symptoms were measured with the Center for Epidemiologic Studies Depression Scale (CES-D), a 20-item self-report instrument developed for use in the community and well established for use with older adults [49,50]. The scale has been shown to have minimal overlap with physical illness [51] and assesses current symptoms during the past week. Respondents scoring 16 or higher on the CES-D scale are considered to have a clinically relevant depressed state. In this study population of 574 participants, 144 were classified as having a depressed state, 17.9% of the males and 30.2% of females.
Genotyping & sequencing
Approximately 4.5 kb of each gene was initially sequenced in search of novel SNPs in both MAOA and MAOB, first in 94 Swedish males and later 45 twins with CES-D scores (36 males and 9 females). Power to detect minor allele frequencies (q) between 1 and 5% was determined as by Glatt et al. [52], 1-(1-q)N where N is the number of chromosomes. Amplification and nested sequencing primers were designed with the CPrimers programme from Genbank entry GI:8671203 containing the promoter, coding exon 1 and flanking intronic sequence of MAOA (~5.0 kb, nucleotides 46490–51454) and Genbank entry GI:2440066 spanning the same characterized sequences of MAOB (~4 kb nucleotides 35033–39021).
Direct sequencing reactions were performed using DYEnamic ET Dye Terminator Cycle Sequencing Kit (Amersham Biosciences) and separated using a Megabace 1000. Reads were base called with Phred [53], assembled using Phrap and viewed using Consed Version 13 [54]. All SNPs were documented and cross validated with dbSNP at NCBI.
Twelve SNPs identified from public databases (dbSNP at NCBI) and two novel SNPs previously reported (introns 3 and 10 of MAOB) a Swedish sample [5] were sequenced in 95 participants (142 chromosomes) by Pyrosequencing to confirm their presence in this population. For Pyrosequencing, either the forward or the reverse primer in each primer pair was biotinylated. Sequencing primers with a length of 14 and 18 bases were placed within one base of the SNP. The PCR reaction was performed in a 50 μl reaction volume, containing 5 ng of genomic DNA, 10 pmoles of each primer, 0.2 mM of each dNTP, 1.5 mM MgCl2 and 1.5 U of Taq. Thermal cycling was performed in a PTC-225 DNA machine (MJ Research Inc., Cambridge, MA, USA) at 95°C for 5 min followed by 50 cycles of 95°C for 30 s, 45 s of annealing at an optimized temperature, followed by 72°C for 30 s and a final extension of 5 min at 72°C. The biotinylated PCR product was immobilized onto streptavidin-coated sepharose beads and DNA strands were separated by denaturation with 0.2 M NaOH. The pyrosequencing reaction was performed on a PSQ96™ Instrument from Pyrosequencing AB (Uppsala, Sweden) as described by [55,56]. Detailed primer and assay information are available upon request.
Statistical analysis
Male haplotypes could be extrapolated directly since the MAO locus is located on the X chromosome and males are thereby hemizygous. Female bi-allelic haplotypes were estimated using an EM algorithm (Sham 1998) and the pair-wise LD measures D' [57] and Δ2 [58]. We used "PHARE" (by David G Cox, available at http://bioinformatics.org/macroshack/programs/PHARE) to create input files for "PHASE" [59,60] to construct female haplotypes.
We used linear regression to estimate the association between trbc-MAO activity and genotypic information using a generalized estimating equation (GEE) approach and alternating logistic regression (ALR) [61] to estimate the association between depressed state and genotypic information. We first modeled the association between single SNPs and each of the two outcomes and then modeled the association between haplotype constructs and the two outcomes. All estimates were adjusted for current smoking status. We estimated both dominance and co-dominance models. Explanatory variables in the dominance models were binary whereas in the co-dominance models they were coded as the number of reference alleles (i.e., 0, 1, or 2 for females and 0 or 1 for males). The parameter estimates for the co-dominance models represent the change in the outcome (trbc-MAO activity or odds of being in a depressed state) per affected allele. Due to the continuous nature of the trbc-MAO measure, only one individual from each complete twin pair and single participating individuals were analyzed (N = 340). Among females we also estimated the effect of being homozygote compared to heterozygote. If the co-dominance model is a good fit to the data then these estimates should be similar to the "per allele" estimates from the co-dominance model. All statistical analyses were performed in SAS 8.01 using GENMOD procedure (SAS Institute Inc. Cary, NC).
Authors' contributions
MJ: Design of the study, performed data analysis and interpretation of data. Carried out the molecular genetic studies (genotyping) and drafted the manuscript.
SM: Participated in the design of the study. Carried out the molecular genetic studies (sequencing), sequence alignment and critically revised the manuscript.
PFS: Participated in the design of the study and critically revised the manuscript for important intellectual content.
PD: Planed and performed the statistical analysis.
BA: Participated in the design of the study and critically revised the manuscript.
LO: Substantially revised the manuscript for important intellectual content.
MS: Participated in the design of the study and critically revised the manuscript.
NLP: Participated in the design of the study and substantially revised the manuscript for important intellectual content.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
SATSA is supported by grants AG 04563, AG 10175, the Swedish Council for Social Research, and the MacArthur Foundation Research Network on Successful Aging. The work herein is also supported by Kapten Artur Erikssons fund, Organons stiftelse för stöd till forskning inom gynekologi-obstertik och psykiatri, VR 10909 and 4145, the Söderström Königska Stiftelsen and funds from Karolinska Institutet and the Karolinska Hospital. Sequencing of the genetic loci was supported by the Genome Program of the Swedish Foundation for Strategic research.
Contributor Information
Mårten Jansson, Email: marten.jansson@cmm.ki.se.
Shane McCarthy, Email: Shane.McCarthy@cgb.ki.se.
Patrick F Sullivan, Email: pfsulliv@med.unc.edu.
Paul Dickman, Email: Paul.Dickman@meb.ki.se.
Björn Andersson, Email: Bjorn.Andersson@cgb.ki.se.
Lars Oreland, Email: lars.oreland@neuro.uu.se.
Martin Schalling, Email: Martin.Schalling@cmm.ki.se.
Nancy L Pedersen, Email: Nancy.Pedersen@meb.ki.se.
References
- Kochersperger LM, Parker EL, Siciliano M, Darlington GJ, Denney RM. Assignment of genes for human monoamine oxidases A and B to the X chromosome. J Neurosci Res. 1986;16:601–616. doi: 10.1002/jnr.490160403. [DOI] [PubMed] [Google Scholar]
- Grimsby J, Chen K, Wang LJ, Lan NC, Shih JC. Human monoamine oxidase A and B genes exhibit identical exon-intron organization. Proc Natl Acad Sci U S A. 1991;88:3637–3641. doi: 10.1073/pnas.88.9.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Denney RM, Breakefield XO. Norrie disease and MAO genes: nearest neighbors. Hum Mol Genet. 1995;4 Spec No:1729–1737. doi: 10.1093/hmg/4.suppl_1.1729. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rosenberg S, Przeworski M, Lancet D, Skorecki K. Evidence for positive selection and population structure at the human MAO-A gene. Proc Natl Acad Sci U S A. 2002;99:862–867. doi: 10.1073/pnas.022614799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J, Syvanen AC, McLeod HL, Pettersson U, Jazin EE. The geographic distribution of monoamine oxidase haplotypes supports a bottleneck during the dispersion of modern humans from Africa. J Mol Evol. 2001;52:157–163. doi: 10.1007/s002390010144. [DOI] [PubMed] [Google Scholar]
- Tivol EA, Shalish C, Schuback DE, Hsu YP, Breakefield XO. Mutational analysis of the human MAOA gene. Am J Med Genet. 1996;67:92–97. doi: 10.1002/(SICI)1096-8628(19960216)67:1<92::AID-AJMG16>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Greenawalt JW, Schnaitman C. An appraisal of the use of monoamine oxidase as an enzyme marker for the outer membrane of rat liver mitochondria. J Cell Biol. 1970;46:173–179. doi: 10.1083/jcb.46.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlund KN, Denney RM, Rose RM, Abell CW. Localization of distinct monoamine oxidase A and monoamine oxidase B cell populations in human brainstem. Neuroscience. 1988;25:439–456. doi: 10.1016/0306-4522(88)90250-3. [DOI] [PubMed] [Google Scholar]
- Saura J, Bleuel Z, Ulrich J, Mendelowitsch A, Chen K, Shih JC, Malherbe P, Da Prada M, Richards JG. Molecular neuroanatomy of human monoamine oxidases A and B revealed by quantitative enzyme radioautography and in situ hybridization histochemistry. Neuroscience. 1996;70:755–774. doi: 10.1016/S0306-4522(96)83013-2. [DOI] [PubMed] [Google Scholar]
- Thorpe LW, Westlund KN, Kochersperger LM, Abell CW, Denney RM. Immunocytochemical localization of monoamine oxidases A and B in human peripheral tissues and brain. J Histochem Cytochem. 1987;35:23–32. doi: 10.1177/35.1.3025289. [DOI] [PubMed] [Google Scholar]
- Arai R, Kimura H, Nagatsu I, Maeda T. Preferential localization of monoamine oxidase type A activity in neurons of the locus coeruleus and type B activity in neurons of the dorsal raphe nucleus of the rat: a detailed enzyme histochemical study. Brain Res. 1997;745:352–356. doi: 10.1016/S0006-8993(96)01239-5. [DOI] [PubMed] [Google Scholar]
- Fowler CJ, Oreland L. Substrate- and stereoselective inhibitor of human brain monoamine oxidase by 4-dimethylamino-alpha, 2-dimethylphenethylamine (FLA 336) J Pharm Pharmacol. 1981;33:403–406. doi: 10.1111/j.2042-7158.1981.tb13819.x. [DOI] [PubMed] [Google Scholar]
- Arai Y, Kinemuchi H, Hamamichi N, Satoh N, Tadano T, Kisara K. Inhibition of rat brain monoamine oxidase by some analogues of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine and 1-methyl-4-phenylpyridinium ion. Neurosci Lett. 1986;66:43–48. doi: 10.1016/0304-3940(86)90163-1. [DOI] [PubMed] [Google Scholar]
- White HL, Glassman AT. Multiple binding sites of human brain and liver monoamine oxidase: substrate specificities, selective inhibitions, and attempts to separate enzyme forms. J Neurochem. 1977;29:987–997. doi: 10.1111/j.1471-4159.1977.tb06502.x. [DOI] [PubMed] [Google Scholar]
- Oreland L, Wiberg A, Asberg M, Traskman L, Sjostrand L, Thoren P, Bertilsson L, Tybring G. Platelet MAO activity and monoamine metabolites in cerebrospinal fluid in depressed and suicidal patients and in healthy controls. Psychiatry Res. 1981;4:21–29. doi: 10.1016/0165-1781(81)90004-4. [DOI] [PubMed] [Google Scholar]
- Bridge TP, Soldo BJ, Phelps BH, Wise CD, Francak MJ, Wyatt RJ. Platelet monoamine oxidase activity: demographic characteristics contribute to enzyme activity variability. J Gerontol. 1985;40:23–28. doi: 10.1093/geronj/40.1.23. [DOI] [PubMed] [Google Scholar]
- Harro M, Eensoo D, Kiive E, Merenakk L, Alep J, Oreland L, Harro J. Platelet monoamine oxidase in healthy 9- and 15-years old children: the effect of gender, smoking and puberty. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25:1497–1511. doi: 10.1016/S0278-5846(01)00212-3. [DOI] [PubMed] [Google Scholar]
- Sandler M, Reveley MA, Glover V. Human platelet monoamine oxidase activity in health and disease: a review. J Clin Pathol. 1981;34:292–302. doi: 10.1136/jcp.34.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Lawrence K, Thomas NS, Price CJ, Ashburner J, Friston KJ, Frackowiak RS, Oreland L, Skuse DH. Dosage-sensitive X-linked locus influences the development of amygdala and orbitofrontal cortex, and fear recognition in humans. Brain. 2003;126:2431–2446. doi: 10.1093/brain/awg242. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Oreland L, Reynolds C, McClearn GE. Importance of genetic effects for monoamine oxidase activity in thrombocytes in twins reared apart and twins reared together. Psychiatry Res. 1993;46:239–251. doi: 10.1016/0165-1781(93)90092-U. [DOI] [PubMed] [Google Scholar]
- Garpenstrand H, Ekblom J, Forslund K, Rylander G, Oreland L. Platelet monoamine oxidase activity is related to MAOB intron 13 genotype. J Neural Transm. 2000;107:523–530. doi: 10.1007/s007020070075. [DOI] [PubMed] [Google Scholar]
- Berlin I, Anthenelli RM. Monoamine oxidases and tobacco smoking. Int J Neuropsychopharmacol. 2001;4:33–42. doi: 10.1017/S1461145701002188. [DOI] [PubMed] [Google Scholar]
- Oreland L. Platelet monoamine oxidase, personality and alcoholism: the rise, fall and resurrection. Neurotoxicology. 2004;25:79–89. doi: 10.1016/S0161-813X(03)00115-3. [DOI] [PubMed] [Google Scholar]
- Stalenheim EG, von Knorring L, Oreland L. Platelet monoamine oxidase activity as a biological marker in a Swedish forensic psychiatric population. Psychiatry Res. 1997;69:79–87. doi: 10.1016/S0165-1781(96)03056-9. [DOI] [PubMed] [Google Scholar]
- Verkes RJ, Van der Mast RC, Kerkhof AJ, Fekkes D, Hengeveld MW, Tuyl JP, Van Kempen GM. Platelet serotonin, monoamine oxidase activity, and [3H]paroxetine binding related to impulsive suicide attempts and borderline personality disorder. Biol Psychiatry. 1998;43:740–746. doi: 10.1016/S0006-3223(97)00317-X. [DOI] [PubMed] [Google Scholar]
- Kirk KM, Whitfield JB, Pang D, Heath AC, Martin NG. Genetic covariation of neuroticism with monoamine oxidase activity and smoking. Am J Med Genet. 2001;105:700–706. doi: 10.1002/ajmg.1555. [DOI] [PubMed] [Google Scholar]
- Oreland L, Damberg M, Hallman J, Garpenstrand H. Smoking only explains part of the associations between platelet monoamine oxidase activity and personality. J Neural Transm. 2002;109:963–975. doi: 10.1007/s007020200079. [DOI] [PubMed] [Google Scholar]
- Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet. 1998;63:1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Hill WG, Cardon LR. Allelic association patterns for a dense SNP map. Genet Epidemiol. 2004;27:442–450. doi: 10.1002/gepi.20038. [DOI] [PubMed] [Google Scholar]
- Buchsbaum MS, Coursey RD, Murphy DL. The biochemical high-risk paradigm: behavioral and familial correlates of low platelet monoamine oxidase activity. Science. 1976;194:339–341. doi: 10.1126/science.968488. [DOI] [PubMed] [Google Scholar]
- von Knorring AL, Bohman M, von Knorring L, Oreland L. Platelet MAO activity as a biological marker in subgroups of alcoholism. Acta Psychiatr Scand. 1985;72:51–58. doi: 10.1111/j.1600-0447.1985.tb02570.x. [DOI] [PubMed] [Google Scholar]
- Eensoo D, Paaver M, Pulver A, Harro M, Harro J. Low platelet MAO activity associated with high dysfunctional impulsivity and antisocial behavior: evidence from drunk drivers. Psychopharmacology (Berl) 2003. [DOI] [PubMed]
- Miller RD, Taillon-Miller P, Kwok PY. Regions of low single-nucleotide polymorphism incidence in human and orangutan xq: deserts and recent coalescences. Genomics. 2001;71:78–88. doi: 10.1006/geno.2000.6417. [DOI] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A. Adjusting the focus on human variation. Trends Genet. 2000;16:296–302. doi: 10.1016/S0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- Fowler CJ, Ekstedt B, Egashira T, Kinemuchi H, Oreland L. The interaction between human platelet monoamine oxidase, its monoamine substrates and oxygen. Biochem Pharmacol. 1979;28:3063–3068. doi: 10.1016/0006-2952(79)90614-2. [DOI] [PubMed] [Google Scholar]
- Fowler CJ, Wiberg A, Oreland L, Marcusson J, Winblad B. The effect of age on the activity and molecular properties of human brain monoamine oxidase. J Neural Transm. 1980;49:1–20. doi: 10.1007/BF01249185. [DOI] [PubMed] [Google Scholar]
- Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- Jansson M, Gatz M, Berg S, Johansson B, Malmberg B, McClearn GE, Schalling M, Pedersen NL. Association between depressed mood in the elderly and a 5-HTR2A gene variant. Am J Med Genet. 2003;120B:79–84. doi: 10.1002/ajmg.b.20016. [DOI] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci U S A. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci U S A. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Escapees on the X chromosome. Proc Natl Acad Sci U S A. 1999;96:14180–14182. doi: 10.1073/pnas.96.25.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Kristiansen M, Knudsen GP, Bathum L, Naumova AK, Sorensen TI, Brix TH, Svendsen AJ, Christensen K, Kyvik KO, Orstavik KH. Twin study of genetic and aging effects on X chromosome inactivation. Eur J Hum Genet. 2005;13:599–606. doi: 10.1038/sj.ejhg.5201398. [DOI] [PubMed] [Google Scholar]
- Finkel D, Pedersen NL. Processing Speed and Longitudinal Trajectories of Change for Cognitive Abilities: The Swedish Adoption / Twin Study of Aging. Aging, Neuropsychology and Cognition. 2004;In press [Google Scholar]
- Lichtenstein P, De Faire U, Floderus B, Svartengren M, Svedberg P, Pedersen NL. The Swedish Twin Registry: a unique resource for clinical, epidemiological and genetic studies. J Intern Med. 2002;252:184–205. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- Radloff LS. The CES-D scale: a self report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- Blazer DG. Depression in late life: review and commentary. J Gerontol A Biol Sci Med Sci. 2003;58:249–265. doi: 10.1093/gerona/58.3.m249. [DOI] [PubMed] [Google Scholar]
- Berkman LF, Berkman CS, Kasl S, Freeman DHJ, Leo L, Ostfeld AM, Cornoni-Huntley J, Brody JA. Depressive symptoms in relation to physical health and functioning in the elderly. Am J Epidemiol. 1986;124:372–388. doi: 10.1093/oxfordjournals.aje.a114408. [DOI] [PubMed] [Google Scholar]
- Glatt CE, DeYoung JA, Delgado S, Service SK, Giacomini KM, Edwards RH, Risch N, Freimer NB. Screening a large reference sample to identify very low frequency sequence variants: comparisons between two genes. Nat Genet. 2001;27:435–438. doi: 10.1038/86948. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Nordfors L, Jansson M, Sandberg G, Lavebratt C, Sengul S, Schalling M, Arner P. Large-scale genotyping of single nucleotide polymorphisms by Pyrosequencingtrade mark and validation against the 5'nuclease (Taqman((R))) assay. Hum Mutat. 2002;19:395–401. doi: 10.1002/humu.10062. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363, 365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The Interaction of Selection and Linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effects of inbreeding at loci with heterozygote advantage. Genetics. 1968;60:615–628. doi: 10.1093/genetics/60.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey VJ, Zeger SL. Modelling multivariate binary data with alternating logistic regressions. Biometrica. 1993. pp. 517–526.


