Abstract
The amplification of DNA by LINE-1 (L1) retrotransposons has created a large fraction of the human genome. To better understand their role in human evolution we endeavored to delineate the L1 elements that have amplified since the emergence of the hominid lineage. We used an approach based on shared sequence variants to trace backwards from the currently amplifying Ta subfamily. The newly identified groups of insertions account for much of the molecular evolution of human L1s. We report the identification of a L1 subfamily that amplified both before and after the divergence of humans from our closest extant relatives. Progressively more modern groups of L1s include greater numbers of insertions. Our data are consistent with the hypothesis that the rate of L1 amplification has been increasing during recent human evolution.
The genomes of all sexually reproducing multicellular eukaryotes harbor type I (non-long terminal repeat) retrotransposons (1). These elements, which first arose over 600 million years ago (2), have amplified to very high copy numbers and constitute a major fraction of the genomic DNA of many organisms. Their activity has exerted a powerful influence on the evolution of eukaryotic species and their genomes. In humans, LINE-1 (L1; long interspersed element) sequences are by far the predominant elements of this type (3). Although more than 500,000 L1s are present in human DNA, most are ancient and predate the mammalian radiation (4, 5). Only a relatively small number are capable of undergoing transposition (6). This situation, which appears to be a characteristic of all mammalian genomes, allows L1s to be grouped into subfamilies that amplified during different periods of evolution (7–9). A complete picture of the activity of L1s and their influence on the evolution of the human genome can be derived only once the identities and sizes of all of the L1 subfamilies that amplified during human evolution have been ascertained.
Many criteria can be used to assign relative ages to L1 subfamilies. Subfamilies with members that are present in several related species are usually older than those whose members are restricted to only one of the species. A second criterion is the degree to which the members of a subfamily have become fixed in the genome of a single species. When a new L1 transposition occurs, a genomic dimorphism—i.e., the presence or absence of the insertion—is created. Over evolutionary time, the occupied allele can either be lost or become fixed in the population. Consequently, younger subfamilies have higher fractions of dimorphic elements than do older subfamilies. Also, older insertions have had time to accumulate more random mutations, therefore older subfamilies have greater average sequence divergences than younger ones (7, 10, 11). L1 subfamilies possess shared sequence variants (SSVs) that evolve in a stepwise fashion. These too can be used to ascertain the relative evolutionary order of the subfamilies.
A general classification scheme for human L1s has been proposed. In this scheme, L1Hs refers to insertions that are found only in humans. Elements that amplified during the primate radiation are designated L1PA#, with older subfamilies receiving higher numbers (4). Although this classification scheme is a solid basis for the investigation of human L1s, it was developed well before the completion of the draft human genome and therefore needs to be updated. For example, the L1PA2 subfamily should contain only the L1s that amplified just before the divergence of hominids from their last common ancestor. Recent results indicate, however, that many members of this subfamily are found as far back in the hominoid tree as gorillas (L. Mathews and G.D.S, unpublished data).
The youngest and only actively transposing subfamily of human L1s was originally named subset Ta (12). These elements are found only in the human genome. Ta elements are defined by the SSVs ACA and G at positions 5930–5932 and 6015, respectively, in their 3′ untranslated regions (numbers refer to the first identified actively transposing L1Hs element, LRE-1), whereas ancient elements typically have GAG and A nucleotides at these positions. All but one of the known de novo L1Hs insertions contains the SSVs ACA/G and therefore belong to the Ta subfamily (13). The sole exception is an ACG/G element (14). In a recent investigation of 42 Ta and 2 ACG/G full-length insertions, Boissinot et al. (15) divided the Ta subfamily into Ta-1 (younger) and Ta-0 (older) subdivisions. They concluded that ACG/G elements belong to the Ta-0 subdivision and named them “preTa.” Before the present investigation, the single de novo insertion and the two considered by Boissinot et al. (15) were the only ACG elements that had been investigated. Boissinot et al. also suggested that although Ta elements first appeared as long as 4 million years ago, most of them have amplified within the past 2 million years. In contrast, no examples of L1PA2 (GAG/A) elements that amplified after the split of humans and chimpanzees from their last common ancestor have been reported. Thus a large gap exists in our present knowledge of the amplification of human L1s between the time of the gorilla divergence (between 8 and 11 million years ago) and 2 million years ago (16). Several L1 subfamilies are likely to have amplified during this extended period of evolutionary time.
Here we describe several groups of L1s that bridge this gap. These insertions account for much of the molecular evolution of L1s since the time of the human–chimpanzee last common ancestor. We also report the identification of a cluster of L1s that amplified both before and after the hominoid–great ape divergence. Our data are consistent with the hypothesis that the rate of L1 amplification has been increasing during recent human evolution. These results improve our understanding of the human genome and provide valuable tools for enhancing our knowledge of the genetic relationships among the hominoids.
Methods
Identification of L1 Insertions.
Our strategy for identifying L1Hs insertions younger than L1PA2 but older than Ta was based on the hypothesis that the four mutations that differentiate L1PA2 from Ta (5930–5932 and 6015) occurred in several steps. We reasoned that L1Hs subfamilies with combinations of nucleotides intermediate between ACA/G and GAG/A should also exist in the human genome. There are 12 different intermediate combinations of the 5930–5932 and 6015 SSVs along the possible single-step pathways leading from L1PA2 to Ta (Table 1). Accordingly, we performed a series of 12 blastn searches against the NT division of GenBank by using a query sequence derived from nucleotides 5914–6026 of LRE-1. For each of the searches the query contained a different combination of nucleotides at the diagnostic positions to reflect one of the different possible intermediates. The 200 best matches were requested for each of the searches. All of the insertions in the blast hit lists that matched the query sequences at the diagnostic positions were identified and were selected for further analysis. Positive matches were always found at the top of the list of best matches in each of the searches. Any human L1s bearing the diagnostic nucleotides that might have been missed by our method of ascertainment will be significantly diverged from the ones included and unlikely therefore to belong to the L1 groups delineated herein. The study previously reported by Boissinot et al. (15) focused only on the Ta subfamily. In their study, a search was performed only for L1s bearing the ACA/G diagnostic nucleotides, although two ACG/G insertions were included in their analyses. Thus the focuses of these two studies, and their methods of ascertainment, are significantly different. The list of accession numbers for all of the insertions analyzed in this report is available from the authors on request.
Table 1.
blast searches of the GenBank NT database for L1 insertions with different combinations of SSVs
| L1 insertions
|
No. of occurrences of SSV combination | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotides 5930–5932 | ACG | ACG | AAG | AAG | GCG | GCG | GCA | GCA | GAA | GAA | AAA | AAA | |
| Nucleotide 6015 | G | A | G | A | G | A | G | A | G | A | G | A | |
| Full-length | 23 (27%) | 8 (26%) | 0 | 5 (25%) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Truncated | 63 | 23 | 0 | 15 | 0 | 6 | 1 | 0 | 0 | 2 | 0 | 0 | |
| Total in genome | 265 | 95 | 0 | 62 | 0 | 22 | 3 | 0 | 0 | 6 | 0 | 0 | |
The percent of elements with each combination of SSVs that was full length is indicated in parentheses.
Genotyping.
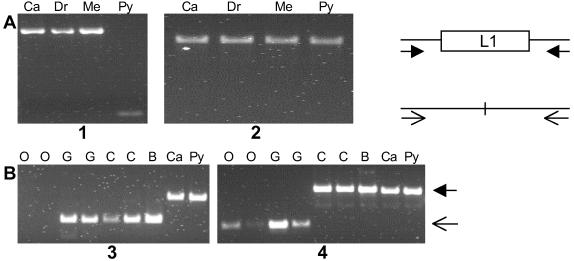
Boundaries of the L1 insertions in the GenBank entries were determined by identifying target site duplications when possible, or by alignment of the insertions with LRE-1. RepeatMasker was used to screen the flanking DNA regions for the presence of repetitive sequences. A single pair of PCR primers, located in unique sequences within the 5′ and 3′ flanking DNAs, was used to amplify both the occupied and empty alleles (Fig. 1A). The primers were designed by using the MacVector program (version 7.0). PCR amplifications were performed either with Taq DNA polymerase or with Elongase (GIBCO/BRL), depending on the length of the insertion and the expected amplification product. The reactions were performed according to the recommended conditions in a MJ Research model PTC 200 thermocycler. Genotypes were determined by direct inspection of ethidium bromide-stained agarose gels. The sequences of the primers used are available on request. Human DNA samples used for genotyping included Caucasian (GM05386), Druze (GM11522), Melanesian (GM10540), and Pygmy (GM10492) obtained from the Coriell Cell Repository (Camden, NJ). Ape DNA samples obtained from Coriell included two Pan troglodytes (GM03452 and NG06939A), one Pan paniscus (AG05253), one Gorilla gorilla (NG05251), and two Pongo pygmaeus (AG06209 and AG06105). A Gorilla gorilla sample was kindly provided by Todd Disotell, New York University.
Fig 1.
Genotyping L1 insertions. The occupied and empty alleles at the sites of the insertions were detected by PCR amplification of genomic DNA. The diagram on the right illustrates that the same pair of primers located in DNA flanking the insertion site can amplify both alleles. (A) Fixed and dimorphic insertions were detected in the genomic DNA isolated from four ethnically diverse humans. Results for insertions AC016986 (ACG/A) and AL359703 (AAG/A) are shown in gels 1 and 2, respectively. Ca, Caucasian; Dr, Druze; Me, Melanesian; Py, Pygmy. (B) Species distribution of the AC007705 (ACG/A) and AL353663 (AAG/A) insertions (gels 3 and 4). Primers for AC007705 failed to amplify either the occupied or the empty allele in orangutans. O, orangutan; G, gorilla; C, chimpanzee; B, bonobo; Ca, human Caucasian, Py, human Pygmy.
Sequence Alignments and Comparisons.
Sequence alignments and pairwise sequence comparisons were performed with the Clustal W program in paup version 4.0b4a (17) and MacVector, followed by manual refinement. Several different alignments were performed. A complete alignment of all of the full-length insertions (Table 1) was constructed. To the insertions identified in the blast searches we added five L1Hs-Ta sequences (three Ta-0, one Ta-1nd, and one Ta-1d) selected from Boissinot et al. (15), and five L1PA2 sequences selected from Ovchinnikov et al. (18). Pairwise sequence comparisons were calculated by MacVector. Insertion AC005820, an ACG/A element that is located on the Y chromosome, was excluded from the calculations of average sequence divergence because insertions on the Y chromosome accumulate mutations faster than insertions located on the autosomes (5). Insertions HS15D23, AC003667, and HS101G11 were also excluded from the calculations for reasons that are described in Results. Consensus sequences were also constructed from the aligned full-length elements belonging to each of the individual groups. SSVs were identified by inspection of the group consensus sequences. A nucleotide position was considered to be an SSV if a single base at that position was characteristic of one or more, but not all, of the groups of insertions. No additional SSVs were identified within the blast query sequence. The presence of intact ORFs was determined by using the MacVector program. Poly(A) tail lengths were calculated by counting the number of A residues that followed the presumed polyadenylation signal that is located at the 3′ end of the L1 3′ untranslated region.
Phylogenetic Analysis.
Maximum parsimony (MP), maximum likelihood (ML), and neighbor-joining (NJ) phylogenetic analyses were performed with paup or with MacVector. NJ analyses were performed with the Clustal W output files (described above), uncorrected p values, and 1,000 bootstraps. Separate analyses were performed with either all of the full-length L1 insertions described in this report or the consensus sequences of the Ta (ACA/G), ACG/G, ACG/A, AAG/A, and L1PA2 groups of sequences, plus the individual sequences AC003667 (the only full-length GCG/A element identified) and HS15D23 (an unusual AAG/A element; see Results). The resulting phylogenetic trees were drawn with TreeView version 1.6.2 (19).
Results
Identification of Older Groups of Human L1 Insertions.
To identify human L1 insertions with ages intermediate between L1PA2 and Ta we performed 12 independent blast searches of the NT division of GenBank. A total of 147 insertions that matched the query sequences at the 5930–5932 and 6015 SSVs were identified (Table 1). In 6 cases, no insertions bearing the SSVs of the query were found. In 2 cases (GCA/G and GAA/A) no full-length and few truncated insertions were found. These insertions were not considered further because only limited information can be derived from a small number of truncated elements. In the remaining 4 cases, several insertions, including one or more full-length elements, were identified. We estimated the number of insertions belonging to each group in the haploid human genome by accounting for the fact that the NT division of GenBank contained only 32.5% of the human genome at the time of the blast searches. A previous study estimated that between 560 and 700 L1Hs-Ta elements were present in the human genome (15).
Previous work indicates that as many as 34% of the Ta-1 insertions but fewer than 5–10% of the GAG L1 insertions in the human genome are full-length (15, 20). The fractions of full-length ACG/G, ACG/A, and AAG/A insertions were all close to 25% (Table 1). These results are consistent with the hypothesis that these insertions occurred between the times of peak L1PA2 and Ta retrotransposition. No estimate for the fraction of full-length GCG/A elements was derived because very few of these elements were identified.
Poly(A) Tail Shortening.
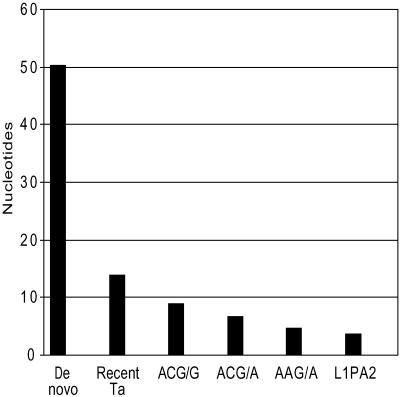
In a previous study we reported that the average length of the poly(A) tails of dimorphic Ta insertions was intermediate between the average lengths of the tails of de novo and GAG/A insertions (18). Fig. 2 depicts our previous data combined with the average lengths of the poly(A) tails of the ACG/G, ACG/A, and AAG/A groups of insertions. As shown, the poly(A) tails of the three new groups of L1 insertions were intermediate in length between the dimorphic Ta and GAG/A groups. This finding further supports the hypothesis that these groups of insertions arose after the peak amplification of the GAG/A insertions but before the amplification of the dimorphic Ta elements.
Fig 2.
Average length of L1 poly(A) tails. For each insertion, the number of pure adenine residues following the presumed polyadenylation signal at the 3′ end of each L1 insertion was determined. The figure depicts the average value for each group of L1s. The data for the de novo, dimorphic-Ta, and L1PA2 groups were previously reported (18).
Genotyping the Insertions in Modern Humans and the Great Apes.
The fraction of insertion loci in each of the groups that was dimorphic was determined by performing PCR genotyping on a panel of four geographically diverse humans (Fig. 1A). Five of the 19 (26%) ACG/G and 3 of the 21 (14%) ACG/A elements that were tested were dimorphic in our test population. In contrast, none of the 6 GCG/A or 17 AAG/A elements that were tested was dimorphic. The dimorphic fractions of the Ta-1 and Ta-0 elements were previously estimated to be 68% and 22%, respectively (15). Thus the dimorphic fractions of the Ta-0 and ACG/G groups are similar and higher than for the other groups. We also examined the full-length insertions for the presence of two intact ORFs. Neither the single full-length GCG/A nor any of the full-length AAG/A insertions had intact ORFs, indicating that they are unlikely to be capable of retrotransposition (21). One of the ACG/A insertions did have intact ORFs and is therefore potentially functional.
The same PCR genotyping assay was used to determine the species distribution of the elements (Fig. 1B). L1s that amplified after the hominid–ape divergence should be present only in the human genome, whereas older elements should also be found in the genomes of the great apes. We tested 18 ACG/G, 16 ACG/A, 5 GCG/A, and 12 AAG/A insertions for their presence in the various species. None of the ACG/G, ACG/A, and GCG/A elements were found in the genomes of chimpanzees, bonobos, gorillas, or orangutans. In contrast, 5 AAG/A elements were present in humans, chimps, and bonobos, and 2 of these were also present in gorillas. None were found in orangutans. We examined the sequences of the 2 insertions present in the gorilla genomes (AL359703 and HS21C002) to determine whether they were bona fide members of the AAG/A group (data not shown). As described below, this group of insertions was distinguished by 22 SSVs. Although both insertions were truncated, they contained the AAG/A-specific SSVs at all possible positions (8 and 7 respectively, including the nucleotides AAG/A). These data indicate that all of the tested ACG and GCG elements transposed after the hominid–great ape divergence but that many of the AAG/A elements inserted before this time. Interestingly, 7 of the 12 AAG/A insertions were specific for humans and were absent from the ape genomes.
Sequence-Based Analyses.
To determine the sequence divergences of the elements identified in the blast searches we performed all possible pairwise comparisons for the groups that contained more than a single full-length insertion. Elements with the ACG/G sequence had the lowest average divergence (0.0094 ± 0.0013), a result that is similar to the divergence of Ta-0 elements. In contrast, the average divergence of the ACG/A elements (0.0141 ± 0.0034) was intermediate, and the divergence of the AAG/A elements was greatest (0.039 ± 0.003) and similar to the divergence of the L1PA2 subfamily as a whole. These data indicate that the L1PA2 and AAG/A elements inserted first, during the same period of human evolution, and were followed later by the ACG/A and finally the ACG/G and Ta elements.
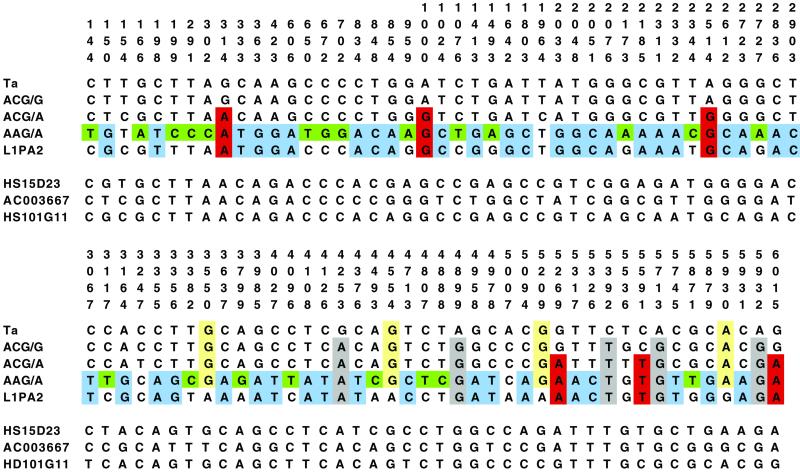
To better determine the relationships among the different groups of insertions we aligned all of the full-length sequences and searched for SSVs. Several different classes of SSVs were identified (Fig. 3). The Ta insertions were differentiated from all of the other groups at 5 positions (gray) and L1PA2 insertions were unique at 4 positions (yellow). In contrast, ACG/G and ACG/A elements were not unique at any positions, suggesting that they represent intermediate steps in the evolution of L1s from L1PA2 to Ta. The 6 positions highlighted in red indicate the relative order of the ACG/G and ACG/A groups along the L1 evolutionary pathway. At these locations, ACG/A elements are similar to older AAG/A and L1PA2 insertions but different from the younger ACG/G and Ta insertions. Surprisingly, AAG/A elements were unique at 22 positions (green). The presence of so many SSVs unique to the AAG/A elements makes it unlikely that they represent a transitional step between L1PA2 and Ta and more likely that they represent a side branch in the evolution of hominoid L1s. At an additional 43 sites (blue) the AAG/A elements were similar only to the L1PA2 insertions and different from the other groups. This finding is further evidence supporting the relatively close evolutionary relationship between the AAG/A and L1PA2 insertions.
Fig 3.
Alignment of full-length L1 insertions. Consensus sequences for the Ta, ACG/G, ACG/A, AAG/A, and L1PA2 insertions described in this report were aligned. Only the SSVs that distinguished among the different groups of insertions are shown. The position numbers refer to the actively transposing element LRE-1 (22). Three individual sequences are also shown. AC003667 was the only full-length GCG/A insertion, and HS15D23 was an atypical AAG/A insertion (see text). Insertion HS101G11 is a composite element with an older nucleotide at positions 5′ of 3356, and a younger nucleotide at positions 3′ of 3356.
Fig. 3 also includes the sequences of three individual L1 insertions. Although element HS15D23 was identified by the blast search using the AAG/A query sequence, close inspection of its sequence makes its inclusion in the AAG/A group problematic. HS15D23 shares only 5 of the 22 (green) SSVs that are unique to the remaining AAG/A elements. This element also possesses the younger SSV at 22 of the 43 (blue) sites that are shared only by the AAG/A and the L1PA2 insertions. Thus HS15D23 appears to occupy a position intermediate between L1PA2 or AAG/A and ACG/A. Insertion AC003667 also appears to occupy an intermediate evolutionary position. This element, which was the only full-length GCG/A insertion identified in our GenBank searches, possesses 30 older, 12 younger, and 1 unique nucleotide at the blue-highlighted positions. In addition, insertion AC003667 possesses none of the green-highlighted nucleotides that are unique to the AAG/A elements. Neither HS15D23 nor AC003667 was detected in any of the ape genomes. Altogether, element AC003667 represents the best candidate for bridging the evolutionary gap between the L1PA2 and the ACG/A subfamilies.
The third individual sequence listed in Fig. 3, insertion HS101G11, was identified in our search for ACG/G insertions. Close inspection of this sequence reveals that it possesses a younger nucleotide at nearly all positions 3′ of 3637 and an older nucleotide at the positions 5′ of 3356. Thus element HS101G11 likely resulted from a recombination or extended gene conversion event between an ACG/G and an ACG/A (or older) element.
Phylogenetic Analyses.
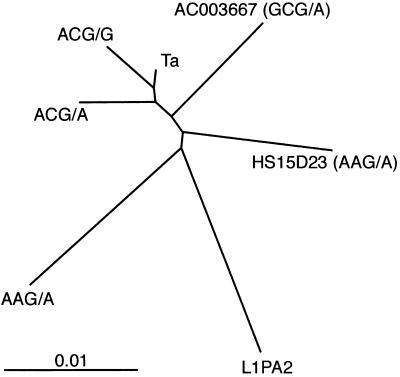
The evolutionary relationships among the insertions were further explored by phylogenetic analyses. First, all of the full-length elements described in this report, including five L1PA2 elements (see Methods), were used in NJ, MP, and ML calculations. All three methods derived the same overall topology for the relationships among the sequences—i.e., separate large groupings containing (i) the Ta and ACG/G, (ii) the ACG/A, and (iii) the L1PA2 and AAG/A insertions. A NJ tree is included in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org. None of the methods provided strong statistical support for these relationships. This lack probably resulted from the fact that the L1 sequences had low levels of divergence, and the inter- and intragroup divergence levels were similar. To provide a global view of the relationships among the L1 groups, we repeated the NJ analysis after substituting consensus sequences for the individual Ta (ACA/G), ACG/G, ACG/A, AAG/A, and L1PA2 insertions. The resulting tree (Fig. 4) had the same overall topology as the ML, MP, and NJ trees calculated by using the individual sequences. The L1PA2 and AAG/A sequences are most distant from the actively transposing Ta elements. L1PA2 and AAG/A also have the longest branch lengths, indicating that they are the most diverged from the other elements. Insertions HS15D23 and AC003667 occupy intermediate positions and also have intermediate branch lengths. Insertion groups that have dimorphic members in the modern human genome (Ta, ACG/G, and ACG/A) cluster at one end of the phylogram. Finally, Ta and ACG/G sequences are joined at a terminal branch point, indicating that they are closely related.
Fig 4.
NJ phylogeny of the full-length group consensus sequences and individual insertions described in Fig. 2. Insertion HS101G11 was not included. The scale indicates nucleotide substitutions per site.
Discussion
The goal of this study was to fill a large gap in our knowledge of the evolution of L1 elements in the human genome. This gap stretched from the time of divergence of the gorilla and human lineages, usually estimated to have occurred between 8 and 11 million years ago, till the rapid Ta expansion ≈2 million years ago. Our approach relied on the use of SSVs. In total we analyzed 147 L1 insertions isolated from GenBank. Only 3 insertions belonging to the groups investigated here had previously been examined. Our results indicate that these elements largely fill the gap between the L1PA2 and Ta subfamilies. We propose that the likely order of the molecular evolution of human L1s (from oldest to youngest) is L1PA2 (GAG/A)–GCG/A–ACG/A–ACG/G–ACA/G. This conclusion is based on many lines of evidence, including structural analyses, genotyping, species distribution, and phylogenetic studies. Our data also suggest that the L1PA2 and AAG/A groups expanded during a similar period of hominoid evolution.
The proposed evolutionary step from GCG/A to ACG/A is the least well supported by the data. Only 7 GCG/A insertions, including a single full-length element, were present in the NT division of GenBank at the time that we performed our searches (June 2001). Also, both the HS15D23 (AAG/A) and AC003667 (GCG/A) insertions were positioned between the L1PA2 and ACG/A groups by genotyping, sequence, and phylogenetic analyses. Thus either one could theoretically have been the ACG/A progenitor. On the other hand, the presence of several AAG/A-specific SSVs in the AC003667 insertion makes it an unlikely intermediate. Most probably, neither of these insertions is the actual precursor of the ACG/A insertions. Instead, it is more plausible that the actual progenitor elements are either not yet in the database or have been lost from the human genome. Our inability to be certain about this link in the evolutionary chain points to one of the interesting results from this study, namely that the groups of L1s that inserted around the time or shortly after the hominid–great ape divergence have relatively few members.
Our data indicate that progressively more modern L1 groups include greater numbers of insertions (Table 1 and Results). This observation is consistent with the hypothesis that the rate of L1 amplification has been increasing during recent human evolution; however, other explanations are also possible. The apparent rate of L1 amplification is the result of two independent rates, the rates of transposition and fixation. A change in either of these rates could give rise to an apparent increase in the rate of amplification. An increase in the transposition rate could have important implications for human health. Previous studies have shown that the amplification of all types of transposable elements has declined during the last 35–50 million years of human evolution (3, 5). It will be important to confirm whether this long-term trend has been reversed during recent human evolution and why. A better understanding of the size, distribution, and molecular evolution of L1s in the genomes of humans and great apes may shed more light on this important question.
Previous work indicated that the fraction of full-length insertions is much higher for the Ta-1 group than for ancient GAG insertions. Our results indicate that L1 groups with intermediate ages have intermediate fractions of full-length insertions. Several potential mechanisms can account for these results. Boissinot et al. (15) suggested that full-length insertions are selected against. If this phenomenon takes place over a period of time, the fraction of full-length insertions will be related to the age of the elements. Alternatively, a series of mutations in the L1 reverse transcriptase that progressively increased the processivity of the enzyme could also give rise to our observations. This later mechanism can be tested by in vitro transposition experiments performed with L1s containing various amino acid substitutions. Because an increase in the processivity of the L1 reverse transcriptase should increase the length of all L1 insertions it will also be helpful to determine whether the average length of truncated elements has increased. Another possibility is that mutations in non-L1 genes that are required for transposition, or influence its outcome, account for the increasing fraction of full-length insertions.
Currently, the method of choice for dating L1 subfamilies relies on a comparison of the subfamily's average sequence diversity with the L1 mutation rate. Unfortunately, a refined measure of the hominoid L1 mutation rate is not available. Boissinot et al. (15) used a value of 0.25% per million years that they based on unpublished data in their recent study of the Ta subfamily. They concluded that the peak of Ta-0 insertion activity occurred around 2 million years ago. Using the same measure, we estimate the ages of the ACG/G and ACG/A and AAG/A groups to be 1.9, 2.8, and 7.8 million years, although the estimate for the AAG/A elements is based on relatively little data. Future studies may improve upon these estimates by providing a refined estimate of the L1 mutation rate. In addition, other methods for dating human L1 insertions may be developed. For example, compared with older L1 insertions, the DNA flanking recent L1 insertions has a higher G+C content and fewer microsatellites (18).
The poly(A) tails of the ACG/G, ACG/A, and AAG/A groups of L1s were shorter than the tails belonging to polymorphic Ta insertions and longer than GAG/A insertions. The results depicted in Fig. 2 suggest that poly(A) shortening may proceed monotonically with time. Thus the length of the pure-A portion of the L1 poly(A) tail may be a useful method for establishing the relative age of groups of L1s in the genomes of other primate and mammalian species. Although shortening of the poly(A) tail may occur in part as a result of random mutations in the poly(A) tail, other mechanisms must be involved because the rate of shortening is faster than the L1 mutation rate (18). It will be interesting to determine whether the rate of shortening is similar in different species. If so it may be a useful method for determining the age of groups of L1s that is at least partially independent of the nucleotide mutation rate.
None of the GCG/A or ACG/A insertions that we tested were present in the genomes of the great apes. We cannot exclude the possibility that some insertions belonging to these groups are present in apes, especially considering the anthropocentric bias inherent in our method of discovery. Nevertheless, the data support the first occurrence of these groups in hominids. In contrast, elements belonging to the AAG/A group were found in the genomes of humans, chimpanzees, bonobos, and gorillas. This group of elements clearly arose before the hominid–great ape divergence. Conversely, several of the AAG/A insertions were specific for humans and were absent from the ape genomes. The most likely explanation for this observation is that these elements transposed after the hominid–ape divergence. An alternative explanation is that these elements were lost from the ape genomes but maintained in the human genome. We consider this unlikely. Currently we have genotyped over 50 L1PA2 and older elements in the genomes of humans and the great apes and have not found any that have been selectively deleted from only one lineage (L. Mathews and G.D.S., unpublished data). Thus the loss of 7 AAG/A elements only from the ape genomes is unlikely. To our knowledge, the data reported here represent the first identification of a L1 subfamily that amplified both before and after the divergence of humans from our closest extant relatives. It will be interesting to determine to what extent this group of elements has amplified in the great ape genomes. Further analysis of this group of elements may also provide an independent verification for the time of the hominid–great ape divergence.
Finally, we turn our attention to the issue of L1 nomenclature and subfamily designation. Data presented here and previously show that the ACG/G elements cannot be meaningfully distinguished from the other Ta-0 insertions. Thus the previous decision to include them within this subfamily is supported (15). On the other hand, the ACG/A elements can be distinguished by several SSVs and are, on the average, older than the Ta-0 insertions. Also, the AAG/A group of elements, although small, has diverged from its contemporaneous L1PA2 insertions by a fairly large set of SSVs. Both of these groups may merit subfamily designation. No formal set of rules governs the decision of when to call a group of insertions a subfamily. As our understanding of the molecular evolution of L1s deepens, the number of elements in each group decreases. Therefore, we have avoided giving names to the groups we have analyzed and refrained from calling them subfamilies. Instead we have referred to them by the SSVs that were used to identify them. Although this may not be an ideal naming system, we prefer to defer this issue until a better nomenclature can be agreed upon. More important is an improved understanding of the evolution of L1s and their influence on the evolution of humans and all eukaryotes.
Supplementary Material
Acknowledgments
We thank Susan Chi and Dr. Lauren Mathews for critical reading of the manuscript. We also thank the anonymous reviewers for their helpful suggestions. This work was supported in part by Grant 5R21CA87356 from the National Cancer Institute.
Abbreviations
L1, LINE-1 (long interspersed element)
SSV, shared sequence variant
MP, maximum parsimony
ML, maximum likelihood
NJ, neighbor-joining
References
- 1.Arkhipova I. & Meselson, M. (2000) Proc. Natl. Acad. Sci. USA 97, 14473-14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik H. S., Burke, W. D. & Eickbush, T. H. (1999) Mol. Biol. Evol. 16, 793-805. [DOI] [PubMed] [Google Scholar]
- 3.Smit A. F. (1999) Curr. Opin. Genet. Dev. 9, 657-663. [DOI] [PubMed] [Google Scholar]
- 4.Smit A. F. A., Toth, G., Riggs, A. D. & Jurka, J. (1995) J. Mol. Biol. 246, 401-417. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium (2001) Nature (London) 409, 860-921.11237011 [Google Scholar]
- 6.Sassaman D. M., Dombroski, B. A., Moran, J. V., Kimberland, M. L., Naas, T. P., DeBerardinis, R. J., Gabriel, A., Swergold, G. D. & Kazazian, H. H., Jr. (1997) Nat. Genet. 16, 37-43. [DOI] [PubMed] [Google Scholar]
- 7.Casavant N. C. & Hardies, S. C. (1994) J. Mol. Biol. 241, 390-397. [DOI] [PubMed] [Google Scholar]
- 8.Furano A. V. (2000) Prog. Nucleic Acid Res. Mol. Biol. 64, 255-294. [DOI] [PubMed] [Google Scholar]
- 9.Goodier J. L., Ostertag, E. M., Du, K. & Kazazian, H. H., Jr. (2001) Genome Res. 11, 1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascale E., Liu, C., Valle, E., Usdin, K. & Furano, A. V. (1993) J. Mol. Evol. 36, 9-20. [DOI] [PubMed] [Google Scholar]
- 11.Adey N. B., Schichman, S. A., Graham, D. K., Peterson, S. N., Edgell, M. H. & Hutchison, C. A., 3rd (1994) Mol. Biol. Evol. 11, 778-789. [DOI] [PubMed] [Google Scholar]
- 12.Skowronski J., Fanning, T. G. & Singer, M. F. (1988) Mol. Cell. Biol. 8, 1385-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazazian H. H. & Moran, J. V. (1998) Nat. Genet. 19, 19-24. [DOI] [PubMed] [Google Scholar]
- 14.Kazazian H. H. J., Wong, C., Youssoufian, H., Scott, A. F., Phillips, D. G. & Antonarakis, S. (1988) Nature (London) 332, 164-166. [DOI] [PubMed] [Google Scholar]
- 15.Boissinot S., Chevret, P. & Furano, A. V. (2000) Mol. Biol. Evol. 17, 915-928. [DOI] [PubMed] [Google Scholar]
- 16.Ruvolo M., Disotell, T. R., Allard, M. W., Brown, W. M. & Honeycutt, R. L. (1991) Proc. Natl. Acad. Sci. USA 88, 1570-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swofford D. L., (2000) paup*: Phylogenetic Analysis Using Parsimony (and Other Methods) (Sinauer, Sunderland, MA).
- 18.Ovchinnikov I., Troxel, A. B. & Swergold, G. D. (2001) Genome Res. 11, 2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page R. D. (1996) Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- 20.Grimaldi G., Skowronski, J. & Singer, M. F. (1984) EMBO J. 3, 1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W., Gilbert, N., Ooi, S. L., Lawler, J. F., Ostertag, E. M., Kazazian, H. H., Boeke, J. D. & Moran, J. V. (2001) Mol. Cell. Biol. 21, 1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dombroski B. A., Mathias, S. L., Nanthakumar, E., Scott, A. F. & Kazazian, H. H., Jr. (1991) Science 254, 1805-1808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.