Abstract
The velvet worms (Onychophora) are considered living fossils and are closely related to the Euarthropoda. Onychophora possess a tracheal system for respiratory function, but oxygen-transport proteins have been considered unnecessary. Here, we show that the hemolymph of the Epiperipatus sp. (Onychophora: Peripatidae) contains an arthropod-type hemocyanin, demonstrating that such protein exists outside the Euarthropoda. Thus, the evolution of oxygen carriers preceded the divergence of the Onychophora and Euarthropoda and was most likely linked to the evolution of an efficient circulatory system in a low-oxygen environment. The cDNA of the Epiperipatus hemocyanin subunit comprises 2,287 bp and encodes for a protein of 641 aa (73.6 kDa). Phylogenetic analyses of the arthropod hemocyanin sequences show that the Onychophora form a robust sister-group of the Euarthropoda, whereas the monophyly of the Tracheata is not supported.
The sudden appearance of the large multicellular animals in the Cambrian period some 500 to 550 million years ago (1, 2) has stimulated intensive paleontological, morphological, and molecular studies on the evolution of the animal body plan (2–4). The increase in size and complexity of the Metazoa required a broad range of physiological adaptations. Among these, the evolution of an efficient circulatory system was essential to sustain the large Metazoa with a sufficient amount of oxygen. In many extant taxa, respiratory proteins (hemoglobins, hemerythrins, and hemocyanins) enhance the oxygen-transport capacity of the body fluid (5). The copper-containing hemocyanins are restricted to the Mollusca and Euarthropoda (6, 7), but these proteins are not related and emerged independently from tyrosinase-like ancestors (8, 9). Arthropod hemocyanins are composed by hexamers of six similar or identical polypeptide chains of 620–650 aa that may associate to form quaternary structures containing up to 8 × 6 subunits (6). Each subunit carries one O2 bound by the virtue of two copper ions. Hemocyanins have been identified in members of all euarthropod subphyla, including the Chelicerata, Crustacea, Myriapoda, and at least one insect (7, 10). The hemocyanins belong to a large superfamily that also includes the arthropod phenoloxidases involved in immune response and cuticle sclerotization, as well as proteins of other functions such as the insect and crustacean storage proteins (8, 10, 11).
Among the first metazoan fossils, the Lobopoda are of particular interest because they are thought to be the closest relatives of the Euarthropoda (12), the world's most successful and diverse animal phylum. The (sub-)phylum Onychophora (velvet worms) includes the only extant representatives of the Lobopoda (13). The presence of respiratory proteins has been unknown among the Onychophora (14). Such proteins had been considered unnecessary because gas exchange of the Onychophora is mediated via tracheae (15). However, we report here the identification and molecular cloning of an arthropod-type hemocyanin in an onychophoran species. We discuss the implication for the evolution of respiratory proteins and the phylogeny of the Arthropoda.
Materials and Methods
Animals.
Epiperipatus sp. was originally captured March 3, 1999 in WP8, Nuevo Arenal, Costa Rica, 10°31′53" N, 84°52′50" W, 640 mNN, under rotten wood by W. Boeckeler and I. Richling. Specimens used in this study were bred in the laboratory in Hamburg (by H.R.).
Protein Biochemistry.
The hemolymph of living Epiperipatus sp. was withdrawn from the dorsal region with a glass capillary, diluted with an equal volume of 100 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/5 mM CaCl2, centrifuged for 10 min at 10,000 × g, and stored at 4°C or frozen at −30°C until use. Protein concentrations were determined according to the method of Bradford (16). Hemocyanin was partially purified by ultrafiltration using a Microcon filter with 100,000 Da cut off (Millipore). The oxygen-binding capacity of the sample was spectrometrically analyzed by reading the absorption from 260 to 380 nm on a Cary photometer. For measuring the deoxy-spectrum, the sample was reduced with sodium dithionite. For Western blotting, about 10 μg of hemolymph protein was submitted to SDS/PAGE (7.5 or 10% acrylamide). Hemocyanin was detected by various anti-hemocyanin-antibodies with antibody concentrations ranging from 1:5,000 to 1:20,000. Phenoloxidase activity after electrophoresis on a native acrylamide gel at pH 8.8 was assayed by using dopamine and thyramine as substrates (17).
Cloning of Epiperipatus Hemocyanin cDNA.
Total RNA was extracted from two adult specimens by using the guanidine hydrochloride method (18). poly(A)+RNA was purified from the total RNA by using the PolyATract kit (Promega). About 3 μg poly(A)+RNA were used for the construction of a directionally cloned cDNA expression λ-ZAP library (Stratagene). The library was amplified once. A pair of degenerated oligonucleotide primers designed according to the conserved copper-binding site A (HHWHWH) (CAYCAYTGGCAYTGGCA) and the CGWPQH/QM sequence (CATRTGNTGNGGCCANCCRCA) of the arthropod hemocyanins were used to amplify the middle region of the hemocyanin cDNA by PCR. The fragment was labeled with digoxigenin (Roche Molecular Biochemicals) and used to screen the cDNA library. Positive phage clones were converted to pBK-CMV vectors by using the material provided by Stratagene and sequenced on both strands by a commercial service (Genterprise, Mainz, Germany). We determined the sequences of two independent clones, which are identical except for the length of the 5′ ends (GenBank accession no. AJ420966).
Phylogenetic Analyses.
An alignment of 18 arthropod hemocyanin and 7 phenoloxidase amino acid sequences was constructed as described (ref. 8, and see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The data set comprises the complete alignment excluding sequences overlapping with signal peptides (707 amino acids positions); in the second data set, long gap regions and highly divergent regions were excluded (500 positions). Distances between pairs of proteins were calculated by using the PAM001 matrix implemented in PHYLIP V.3.6a2 (19). Tree constructions were performed by the neighbor-joining method. The reliability of the trees was tested by the bootstrap procedure with 100 replications (20). Maximum likelihood quartet puzzling analyses were carried out by TREE-PUZZLE V.5.0 (21). Different models of amino acid replacement were tested. The final trees were constructed by assuming the Whelan and Goldman model (22) with rate heterogeneity of eight γ categories.
Results
Identification and Analyses of Epiperipatus Hemocyanin.
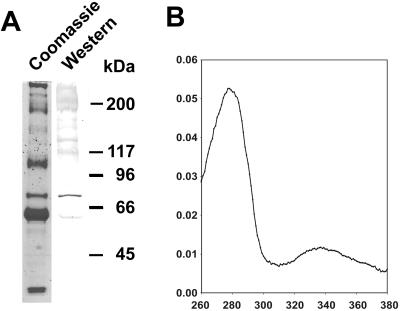
Western blot analysis of the hemolymph from the Epiperipatus sp. (Fig. 1), a member of the peripatoid Onychophora, shows the presence of a polypeptide that shares common epitopes with the arthropod hemocyanins, thus cross-reacting with antibodies raised against these proteins (Fig. 2A). The apparent molecular mass of this polypeptide of about 72 kDa is characteristic for an arthropod-type hemocyanin subunit (6). The total protein concentration in the hemolymph of Epiperipatus sp. is low (about 10–20 mg/ml), a tenth of what has been typically found in other arthropods. The hemocyanin makes up only about 5–10% of the total protein. The absorption spectrum of partially purified hemocyanin shows the presence of a peak at 340 nm (Fig. 2B), which disappears upon the addition of dithionite as reducing agent. The absorption maximum at about 340 nm is typical for the presence of an oxygenated hemocyanin. A function as a phenoloxidase can be excluded, because this protein metabolizes neither dopamine nor tyramine in a native polyacrylamide gel (data not shown).
Fig 1.
Epiperipatus sp. (Onychophora).
Fig 2.
Identification of hemocyanin in the Onychophora. (A) Hemolymph proteins (10 μg) of male and female individuals of Epiperipatus sp. were separated by SDS/PAGE and stained with Coomassie brilliant blue (lane 1). The hemocyanin was detected by using an anti-H. americanus hemocyanin antibody (lane 2). The molecular mass standard is given at Left. (B) Absorption spectrum of partially purified, oxygenated Epiperipatus hemocyanin.
Molecular Cloning of the Epiperipatus Hemocyanin cDNA.
Because the site of hemocyanin synthesis in the Onychophora is unknown, a cDNA library was constructed from poly(A)+ RNA of the complete animal. Screening with various anti-hemocyanin antibodies did not give rise to a hemocyanin cDNA clone. Therefore, a set of degenerated oligonucleotides were designed according to the conserved arthropod hemocyanin sequences (8). These primers were used to amplify an 1,128-bp fragment of the hemocyanin from the cDNA library. The complete cDNA-sequence was obtained by screening the library with this fragment as a probe. A single clone-type of 2,287 bp was obtained that covers the complete coding region plus the 5′ and 3′ untranslated regions (Fig. 5). The deduced amino acid sequence of the Epiperipatus hemocyanin covers 641 aa with an estimated molecular mass of 73.6 kDa, which is in excellent agreement with that obtained by SDS/PAGE. The protein contains all amino acids that are required for the function of a true oxygen carrier, in particular the residues that are required for oxygen binding and cooperativity (23). These residues include the six copper-binding His in the second domain (Fig. 3) and three Phe in the first (Phe-54) and second domain (Phe-204 and Phe-375; Fig. 5). Phe-54 is implied to be a key residue in regulation of oxygen affinity (23). The calcium-binding site, which is present in the third domain of most hemocyanins (23), is formed by the conserved residues Ser-517, Thr-520, and Asp-589. As in the chelicerate and myriapod hemocyanins (24), an α-helix of the first domain (1.2) present in the crustacean hemocyanins and insect hexamerins is missing in the Epiperipatus hemocyanin. The protein does not contain any signal peptide required for excretion via the endoplasmic reticulum, suggesting that, similar to the chelicerate hemocyanins (6), it is released into the hemolymph via holocrine secretion.
Fig 3.
Alignment of copper-binding sites A (CuA) and B (CuB) of selected arthropod hemocyanins. Amino acids conserved between Epiperipatus and other hemocyanins are shaded in gray; the copper-binding histidines are shaded in black. Hc, hemocyanin.
Sequence Comparison and Phylogenetic Analyses.
Comparison of the Epiperipatus hemocyanin and other members of the arthropod hemocyanin superfamily (refs. 8 and 11; Fig. 6, which is published as supporting information on the PNAS web site) shows the highest identity with the chelicerate hemocyanins (39.0–44.8% identity), whereas lower scores were observed with the myriapod (34.1 to 37.9%), the crustacean (30.1–34.9%), and the insect hemocyanins (33.6%). The phenoloxidases share 33.9 to 38.7% of the amino acids with the onychophoran hemocyanin.
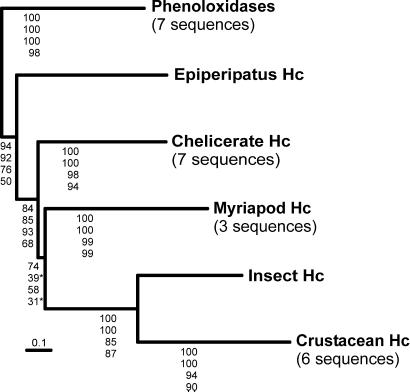
Phylogenetic analyses were carried out by distance-matrix and maximum likelihood methods by using two different amino acid sequence alignment data sets constructed from 25 hemocyanin and phenoloxidase sequences. The first data set of 707 aa positions comprises the complete sequences without the regions overlapping with the signal peptides; in the second data set (500 positions), highly divergent regions and long gaps were excluded from the calculation. The arthropod phenoloxidases most likely constitute the most ancient branch of the arthropod hemocyanin superfamily and thus were considered as the outgroup (8). Except for the position of the myriapod hemocyanins, the general topologies of the resulting trees are identical (Fig. 4; Fig. 7, which is published as supporting information on the PNAS web site). The 707-aa data set supports a sister group position of the myriapod hemocyanins with the monophyletic crustacean and hexapod proteins, whereas the reduced data set favors an association of the myriapod and the chelicerate hemocyanins. The Epiperipatus hemocyanin is in basal position with respect to all other hemocyanins, with modest to high bootstrap and quartet puzzling support values (50–94%).
Fig 4.
Phylogeny of the arthropod hemocyanin superfamily. The simplified tree was deduced from the analyses of an amino acid alignment of the Epiperipatus hemocyanin with seven chelicerate, three myriapod, six crustacean, and one insect hemocyanins (Fig. 6). The phenoloxidases (seven sequences) were considered as the outgroup (8). The complete tree may be found in Fig. 7. The numbers at the nodes are support values (from top to bottom): bootstrap numbers estimated by a PAM-based neighbor-joining from the (i) 707- and (ii) 500-aa data sets or estimated by a maximum likelihood quartet puzzling approach (21) with the (iii) 707- and (iv) 500-aa data sets.
Discussion
Most morphologists consider the Onychophora as a taxon closely related to or included in the Arthropoda (15, 25, 26). The presence of an arthropod-type hemocyanin in the Onychophora, which is absent in other animal phyla, confirms this assumption. Molecular phylogenetic studies tend to agree on the view that the Onychophora are allied with the Euarthropoda but generally do not resolve the exact position of this taxon (27–29). Phylogenetic reconstruction of the evolution of arthropod hemocyanins using the phenoloxidases as the outgroup results in a robust tree, in which the main nodes are well supported in both neighbor-joining and maximum likelihood analyses (Fig. 4). The trees show that the onychophoran hemocyanin is in a sister-group position with the hemocyanins of the Euarthropoda (Chelicerata, Myriapoda, Crustacea, and Hexapoda). This arrangement supports the notion that the Onychophora are, in fact, an early offshoot of the arthropod clade. As already demonstrated in earlier studies using hemocyanins (24) or other molecular markers (30–33), the hexapod and crustacean hemocyanins form a well supported common clade. The sister group of these pancrustacean proteins are the hemocyanins of the Myriapoda in this analysis, but this topology does not reach the significance level (24).
Although the Onychophora possess a unique tracheal system, we show that at least some of these species possess a hemocyanin that is most likely used for oxygen transport. Similar to the Myriapoda (34, 35), the evolution of trachea for air breathing in the Onychophora was, therefore, not accompanied by a loss of respiratory proteins. Phylogenetic analyses indicate that the emergence of the hemocyanins preceded the divergence of the Onychophora (Proarthropoda) and Euarthropoda. Thus, the evolution of the hemocyanins occurred in the (pan-)arthropod stem line in the Cambrian or even an earlier period. The emergence of such respiratory proteins was most likely linked with a rise in the concentration of atmospheric oxygen during the late Proterozoic period (36). At that time, the animals commenced to increase in size (37), which required the formation of an efficient circulatory system and specific oxygen-transport proteins. The common ancestor of Euarthropoda and Onychophora recruited the copper-based phenoloxidases for this task, which were transformed into hemocyanins (8). The fact that different animal phyla use different types of respiratory proteins suggests that protein-mediated oxygen transport was invented several times independently during the evolution of the Metazoa.
Supplementary Material
Acknowledgments
We thank J. Markl for excellent working facilities, N. Hellmann and H. Decker for the oxygen-binding studies, J. Bonaventura for discussions, W. Gebauer, U. Hoeger, and E. Jaenicke for their help with the experiments, and J. R. Harris for correcting the language. This work is supported by the Deutsche Forschungsgemeinschaft (Bu956/3 and 956/5).
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ420966).
References
- 1.Convey Morris S. (1993) Nature (London) 361, 219-225. [Google Scholar]
- 2.Valentine J. W., Jablonski, D. & Erwin, D. E. (1999) Development (Cambridge, U.K.) 126, 851-859. [DOI] [PubMed] [Google Scholar]
- 3.Grenier J. K., Garber, T. L., Warren, R., Whitington, P. M. & Carroll, S. B. (1997) Curr. Biol. 7, 547-553. [DOI] [PubMed] [Google Scholar]
- 4.Panganiban G., Irvine, S. M., Lowe, C., Roehl, H., Corley, L. S., Sherbon, B., Grenier, J. K., Fallon, J. F., Kimble, J., Walker, M., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 5162-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtz D. M. (1999) Essays Biochem. 34, 85-100. [DOI] [PubMed] [Google Scholar]
- 6.Markl J. & Decker, H. (1992) Adv. Comp. Environ. Physiol. 13, 325-376. [Google Scholar]
- 7.van Holde K. E. & Miller, K. I. (1995) Adv. Protein Chem. 47, 1-81. [DOI] [PubMed] [Google Scholar]
- 8.Burmester T. (2001) Mol. Biol. Evol. 18, 184-195. [DOI] [PubMed] [Google Scholar]
- 9.van Holde K. E., Miller, K. I. & Decker, H. (2001) J. Biol. Chem. 276, 15563-15566. [DOI] [PubMed] [Google Scholar]
- 10.Burmester T. (2002) J. Comp. Physiol. B 172, 95-117. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger N. B., Dangott, L. J. & Ryan, M. C. (1999) Proc. Natl. Acad. Sci. USA 96, 2013-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budd G. (1996) Lethaia 29, 1-14. [Google Scholar]
- 13.Hou X. G. & Bergström, J. (1995) Zool. J. Linn. Soc. London 114, 3-19. [Google Scholar]
- 14.Ellerton H. D., Ellerton, N. F. & Robinson, H. A. (1983) Prog. Biophys. Mol. Biol. 41, 143-248. [DOI] [PubMed] [Google Scholar]
- 15.Brusca R. C. & Brusca, G. J., (1990) Invertebrates (Sinauer, Sunderland, MA).
- 16.Bradford M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 17.Decker H., Ryan, M., Jaenicke, E. & Terwilliger, N. (2001) J. Biol. Chem. 276, 17796-17799. [DOI] [PubMed] [Google Scholar]
- 18.Chirgwin J. M., Przbyla, A. E., MacDonald, R. J. & Rutter, W. J. (1979) Biochemistry 18, 5294-5299. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J., (1993) phylip, Phylogeny Inference Package (Univ. of Washington, Seattle), Version 3.6a2.
- 20.Felsenstein J. (1985) Evolution (Lawrence, Kans.) 39, 783-791. [DOI] [PubMed] [Google Scholar]
- 21.Strimmer K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964-969. [Google Scholar]
- 22.Whelan S. & Goldman, N. (2001) Mol. Biol. Evol. 18, 691-699. [DOI] [PubMed] [Google Scholar]
- 23.Hazes B., Magnus, K. A., Bonaventura, C., Bonaventura, J., Dauter, Z., Kalk, K. H. & Hol, W. G. J. (1993) Protein Sci. 2, 597-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusche K. & Burmester, T. (2001) Mol. Biol. Evol. 18, 1566-1573. [DOI] [PubMed] [Google Scholar]
- 25.Edgecombe G. D., Wilson, G. D. F., Colgan, D. J., Gray, M. R. & Cassis, G. (2000) Cladistics 16, 155-203. [DOI] [PubMed] [Google Scholar]
- 26.Giribet G., Edgecombe, G. D. & Wheeler, W. C. (2001) Nature (London) 413, 157-161. [DOI] [PubMed] [Google Scholar]
- 27.Ballard J. W., Olsen, G. J., Faith, D. P., Odgers, W. A., Rowell, D. M. & Atkinson, P. W. (1992) Science 258, 1345-1348. [DOI] [PubMed] [Google Scholar]
- 28.Boore J. L., Collins, T. M., Stanton, D., Daehler, L. L. & Brown, W. M. (1995) Nature (London) 376, 163-165. [DOI] [PubMed] [Google Scholar]
- 29.Giribet G. & Ribera, C. (2000) Cladistics 16, 204-231. [DOI] [PubMed] [Google Scholar]
- 30.Friedrich M. & Tautz, D. (1995) Nature (London) 376, 165-167. [DOI] [PubMed] [Google Scholar]
- 31.Regier J. C. & Shultz, J. W. (2001) Mol. Phylogenet. Evol. 20, 136-148. [DOI] [PubMed] [Google Scholar]
- 32.Shultz J. W. & Regier, J. C. (2000) Proc. R. Soc. London Ser. B 267, 1011-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang U. W., Friedrich, M., Tautz, D., Park, C. J. & Kim, W. (2001) Nature (London) 413, 154-157. [DOI] [PubMed] [Google Scholar]
- 34.Mangum C. P., Scott, J. L., Black, R. E. L., Miller, K. I. & van Holde, K. E. (1985) Proc. Natl. Acad. Sci. USA 82, 3721-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaenicke E., Decker, H., Gebauer, W., Markl, J. & Burmester, T. (1999) J. Biol. Chem. 274, 29071-29074. [DOI] [PubMed] [Google Scholar]
- 36.Canfield D. E. & Teske, A. (1996) Nature (London) 382, 127-132. [DOI] [PubMed] [Google Scholar]
- 37.Budd G. E. & Jensen, S. (2000) Biol. Rev. 75, 253-295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.