Abstract
Chondrodysplastic dwarfism in Japanese brown cattle is an autosomal recessive disorder characterized by short limbs. Previously, we mapped the locus responsible for the disease on the distal end of bovine chromosome 6. Here, we narrowed the critical region to ≈2 cM by using linkage analysis, constructed a BAC and YAC contig covering this region, and identified a gene, LIMBIN (LBN), that possessed disease-specific mutations in the affected calves. One mutation was a single nucleotide substitution leading to an activation of a cryptic splicing donor site and the other was a one-base deletion resulting in a frameshift mutation. Strong expression of the Lbn gene was observed in limb buds of developing mouse embryos and in proliferating chondrocytes and bone-forming osteoblasts in long bones. These findings indicate that LBN is responsible for bovine chondrodysplastic dwarfism and has a critical role in a skeletal development.
Longitudinal growth of long bones arises from continuous proliferation and differentiation of chondrocytes followed by endochondrial ossification at the epiphysial growth plates located at both ends of long bone. Mutations of genes involved in the process cause several types of dwarfism in human and mouse; these findings have provided valuable insight into our understanding of skeletal development, a process we know very little about (1–15).
Bovine chondrodysplastic dwarfism (BCD) in Japanese brown cattle is an autosomal recessive disorder with the phenotype of short limbs, joint abnormality, and ateliosis (16, 17). Long bones of the affected animals have insufficient endochondrial ossification with irregularly arranged chondrocytes, abnormal formation of the cartilaginous matrix, and partial disappearance of the epiphysial growth plates. The axial skeletal structures and craniofacial skeleton are not significantly affected. Disproportionate dwarfism also has been reported in other cattle breeds including Dexter, Holstein, Aberdeen-Angus, Hereford, and Shorthorn breeds (18, 19), but their clinical features, associated with vertebral and craniofacial abnormalities, are different from those of BCD.
In a previous study, we mapped the locus (bcd) responsible for this disease in an 11-cM region between microsatellite markers BP7 and BM9257 on the distal end of bovine chromosome (BTA) 6 and demonstrated that the region corresponded to human chromosome (HSA) 4p (20). Various genes responsible for human hereditary diseases with skeletal abnormalities have been localized to this region. For example, achondroplasia (7, 8), hypochondroplasia (11), and thanatophoric dysplasia (15), characterized by short-limb dwarfism, are caused by mutations in fibroblast growth factor receptor 3 (MIM no. 134934). An association between rare alleles of the MSX1 locus and limb deficiency malformations has been suggested (MIM no. 142983; ref. 21). EVC was identified recently as the gene responsible for Ellis-van Creveld syndrome (MIM no. 225500), which is characterized by short limbs, short ribs, postaxial polydactyly, and dysplastic nails and teeth (22).
In the present study, we narrowed the bcd locus to a 2.4-cM region, constructed yeast and bacterial artificial chromosome (YAC, BAC) contigs covering this region, sequenced candidate genes located in the region, and finally identified causative mutations in a gene that is expressed in the epiphysial growth plates of long bone.
Materials and Methods
Construction of Physical Map.
A bovine YAC library was screened by using a PCR-based method (23). A bovine BAC library (RPCI-42) was screened by colony hybridization with high-density filters purchased from Children's Hospital (Oakland, CA). YAC end sequences were obtained by the end rescue method using the vectorette method, (24) and the BAC end sequence was identified by direct sequencing of BAC DNA by using T7 and SP6 primers with BigDye chemistry (Applied Biosystems). Microsatellite loci were isolated by using biotinylated (CA)12 probe, as described by Kijas et al. (25). The primer sequences and PCR conditions to amplify microsatellites are described in the GenBank database. Primer sequences of sequence-tagged site (STS) markers for bovine genes and ends of clones are described in the supporting information, which is published on the PNAS web site, www.pnas.org.
Pedigree Material and Haplotype Analysis.
We used two paternal half-sib pedigrees derived from sire A and sire B in a commercial herd of Japanese brown cattle. Sire B is the offspring of sire A. The pedigree of sire A was composed of sire A, 91 dams, 91 affected offspring, and 187 unaffected offspring. The pedigree of sire B was composed of sire B, 28 dams, and 28 affected offspring. We used 95 Japanese black and 106 Holstein cattle as normal controls. The dwarf symptoms of the affected calves were diagnosed by visual examination and by histologic examination of growth plate cartilage after necropsy. Microsatellite genotyping was performed according to standard procedures (26). We determined the most likely haplotype configurations as described by Coppieters et al. (27).
Reverse Transcriptase (RT)-PCR.
Total RNA of fetal calf long bone was extracted with Trizol (Invitrogen) and reverse-transcribed with SuperScript II Reverse Transcriptase using oligo-d(T)12 and random hexamers, according to the manufacturer's instructions (Invitrogen). The cDNAs were amplified with an annealing temperature of 65°C using Avantage GC2 kit (CLONTECH), according to the manufacturer's instructions. To amplify bovine EVC and geneX, we used primer pairs as follows: EVC, 5′-GTCCACTCAGTGCATCCTGC-3′ and 5′-AGGGCTCTGAGCAGTTGCCA-3′; and geneX, 5′-CTGGAGTCCACTGATGAACTGACC-3′ and 5′-CAATGTGAGAACCGAGAGCCTTGC-3′.
Cloning of Bovine LBN cDNA.
A fetal calf long bone cDNA library (28) was screened by using standard methods with the RT-PCR product of LBN as a probe amplified by a primer pair: bov1, 5′-GAGAGAGGGTGATATTCTCTGG-3′ and 5′-GATAAAGAGCTTTTCACCCGTG-3′. In addition, RT-PCR using bovine fetal long bone total RNA was performed by using two sets of primer pairs as follows: bov2, 5′-GGCTTTCAGGAAGAAGTTTCTGC-3′ and 5′-CTCCAGGAGCTTCTGACCCTTGC-3′; and bov3, 5′-GGAAGTATCTGGCCCATTTTGCC-3′ and 5′-CTCCAGCGAGGCTGTGTAGCTGAC-3′. The RT-PCR products were directly sequenced by using internal primers. Sequencer software (Gene Codes, Ann Arbor, MI) was used for the sequence assembly.
Human and Mouse Ortholog Cloning.
The cDNA contig assembly of human LBN was produced by using the RT-PCR and rapid amplification of cDNA ends (RACE) methods (Invitrogen), according to the manufacturer's instructions using human kidney total RNA (CLONTECH). The primers were designed based on human genome draft sequences. The cDNA contig assembly of mouse Lbn was produced by using the mouse embryonic day (E)-11 limb buds λ-cDNA library (Stratagene) screening and 5′ RACE method with mouse embryo E14.5 total RNA. We created multiple alignments of amino acid sequences of LBN using GENETYX-MAC software (Software Development, Tokyo). The protein sequences were analyzed by using the programs SMART (available at http://smart.embl-heidelberg.de; ref. 29) for transmembrane and coiled coil domain searches, and PSORTII (available at http://psort.nibb.ac.jp; ref. 30) for protein localization prediction.
Mutation Analysis.
RT-PCR amplifications of bovine LBN cDNA segments covering the entire coding region were performed by using long bone total RNA derived from a 3-week old affected and normal fetal calf with two sets of primer pairs as follows: bov4, 5′-GCAAGGGTCAGAAGCTCCTGGAG-3′ and 5′-CAATGTGAGAACCGAGAGCCTTGC-3′; and bov5, 5′-CGTCATTGTGCCTTTGGACTTTCAG-3′ and 5′-GCAGCATCGTCAGCTTCGACAGG-3′. We used 38 PCR cycles with an annealing temperature of 65°C, and the RT-PCR products were sequenced by using internal primers. The genomic segments containing a region of C1356T and 2054–2055delCAinsG mutations were amplified with primer pairs as follows; bov6, 5′-TACAGCAGGAGGAGGACCTTGC-3′ and 5′-TTAGTTCACTGGAACCCAGCAC-3′; and bov7, 5′-GCCTGCAGAACTCAGGAATGAC-3′ and 5′-CGTGAAGATCAAGTGCTCCCAGTG-3′, respectively. We used 32 PCR cycles with an annealing temperature of 58°C, and the products were sequenced by using the same primers. For confirmation of the mutations, the PCR products also were cloned in a pGEM-T Easy vector (Promega) according to the manufacturer's instructions and sequenced.
Expression Analysis.
Northern blots contained 2 μg of mouse poly(A)+ RNA from adult tissues (MNT filter, CLONTECH), long bone and cranial bone from postnatal day (P)-14 and whole embryos at various stages. A mouse Lbn cDNA fragment (nucleotides 266-2086) was used as a probe for hybridization. We probed the same set of filter with a G3pdh probe as a loading control. Whole-mount in situ hybridization was performed according to the protocol described by Conlon and Rossant (31). The digoxigenin (DIG)-labeled riboprobes corresponding to the 5′ region of mouse Lbn cDNA (nucleotides 149-1558) was used. For in situ hybridization analysis, mouse tibias at E17 and P35 were fixed in 4% (vol/vol) paraformaldehyde in PBS overnight at 4°C and decalcified in 10% (wt/vol) EDTA for 1 day and 7 days, respectively. Specimens were embedded in paraffin and sectioned at 6 μm. DIG-11-UTP-labeled riboprobes corresponding to the 3′ region of mouse Lbn cDNA (nucleotides 2801–3044) were prepared by using a DIG RNA labeling kit (Roche Molecular Biochemicals), according to the manufacturer's instruction. Hybridization was performed overnight at 48°C with washes at 50°C as described by Ikegame et al. (32). Cell nuclei were stained with 1% (wt/vol) methyl green.
Results
Fine Genetic and Physical Mapping.
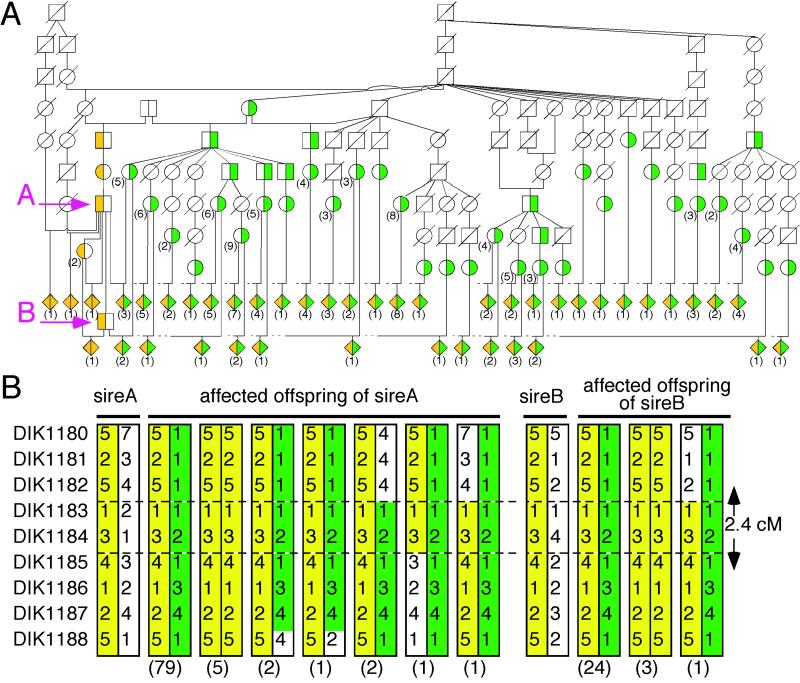
We constructed a bovine radiation hybrid (RH) map of BTA 6 by using a bovine-hamster whole-genome RH panel (33) and identified that the critical 11-cM region from BP7 to BM9257 corresponded to HSA 4p16 and 4q21 (M.T., unpublished data). To refine further the bcd locus, we constructed YAC and BAC clone contigs by using STS markers amplifying inserts of the clones as well as bovine orthologs of genes located in HSA 4p16 and 4q21 and developed 14 polymorphic microsatellite markers (DIK1178-DIK1190, and DIK1192) from the clones (Fig. 1A). We genotyped a paternal half-sibling pedigree of 119 affected calves using the markers to determine the precise localization of bcd and the disease-associated haplotypes. None of the affected calves had recombination with DIK1183 and DIK1184, narrowing the critical region to 2.4 cM between DIK1182 and DIK1185 (Fig. 2B). Interestingly, the haplotypes of the most affected calves (111/119) were heterozygous for two specific haplotypes (Fig. 2), suggesting that both haplotypes were responsible for BCD and that two distinct mutations associated with these haplotypes might be present in the population of Japanese brown cattle.
Fig 1.
Identification of bovine LBN gene. (A) Physical map consisting of YAC and BAC clones in the critical region on BTA 6. Informative and uninformative markers in our pedigree, STS markers for genes and ends of the clones are shown in blue, green, red, and black, respectively. Dotted lines show regions corresponding to HSA 4. (B) Transcription units on the human orthologous region on HSA 4p16. Genes and UniGene clusters localized by using the GRL database are shown in red and purple, respectively. C4000112, C4000113, and C4000114 are putative exons on the UCSC human genome database. The black boxes indicate a part of the human LBN gene. (C) Genomic structure of bovine LBN. M, translation initiation codon; *, stop codon.
Fig 2.
Pedigrees with BCD and haplotypes consisting of nine markers in the critical region on BTA 6. Green, haplotype associated with a C1356T mutation; yellow, haplotype associated with a 2054–2055delCAinsG mutation. The number of animals is shown in parentheses. (A) This pedigree includes grandsire A and sire B, which were identified as carriers of the disease, 90 affected offspring, 90 dams of the affected offspring, and some ancestors. The shaded diamonds represent affected offspring. White with diagonal line denotes animals not genotyped. (B) Haplotypes of nine microsatellites (Left) for grandsire A, sire B, and the 119 affected offspring are shown.
Candidate Gene Identification.
As shown in Fig. 1 A and B, the gene order from PPP2R2C to MSX1 covering the critical region is conserved between cattle and human. In this region, the human genome database GRL (http://grl.gi.k.u-tokyo.ac.jp; ref. 34) disclosed four genes (CRMP1, EVC, C4orf6, and HSA250839) and six human UniGene clusters (Hs.256259, Hs.257022, Hs.87306, Hs.192146, Hs.133062, and Hs.122053; Fig. 1B). We examined the expression of these possible candidate genes in long bones of fetal calves with RT-PCR and/or Northern hybridization. Among them, EVC and bovine expressed sequence tag AW345074 (designated geneX), an ortholog of Hs.87306, were expressed in long bones of fetal calves (data not shown), suggesting that these two genes were the most potent candidate genes for BCD. No causative mutation was found, however, in the bovine EVC gene by sequencing the entire coding region of the gene of affected animals (data not shown).
To characterize the complete coding sequence of geneX, we isolated two clones from a bovine fetal long bone cDNA library with AW345074 as a probe, covering a 1,775-bp region of geneX containing a poly(A)+ tail but not the 5′ region of the coding sequence. As of April 1, 2001, the University of California Santa Cruz (UCSC) human genome database (http://genome.ucsc.edu) located three predicted exons C4000112, C4000113, and C4000114 upstream of Hs.87306, a human ortholog of geneX. We confirmed that the three predicted exons were included in human geneX transcript with RT-PCR by using human kidney RNA (data not shown). Bovine sequences corresponding to C4000112, C4000113, and C4000114 were determined by shotgun sequencing of bovine BAC 162M16 and 232M6. Primers for amplifying the 5′ region of the cDNA were designed from these sequences. The nucleotide sequences of the RT-PCR products using these primers allowed reconstruction of a 5,759-base transcript containing a putative 3630-base ORF (position 1686–5315). geneX encoded a protein consisting of 1209 amino acids (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). To determine the genomic structure of the gene, we sequenced bovine BAC 232M6 and 162M16 using primers designed from the cDNA sequence and identified that the gene consists of 22 exons (Fig. 1C). The human and mouse orthologs also were isolated by a combination of cDNA cloning, RT-PCR, and RACE. The human and mouse geneX encode 1,228 amino acids with 78.7% homology, and 1,220 amino acids with 68.4% homology with bovine geneX, respectively (Fig. 6). The SMART (29) and PSORTII (30) program searches suggested that a putative transmembrane domain, two coiled-coil domains, and three nuclear localization signals were conserved between cattle, human, and mouse genes (Fig. 6).
Identification of Two Mutations in the Gene.
We examined the expression of geneX in the long bones of normal and affected calves using RT-PCR; there was no qualitative difference observed (data not shown). Expression of geneX was not detected by Northern hybridization, probably because of the low level of expression (data not shown).
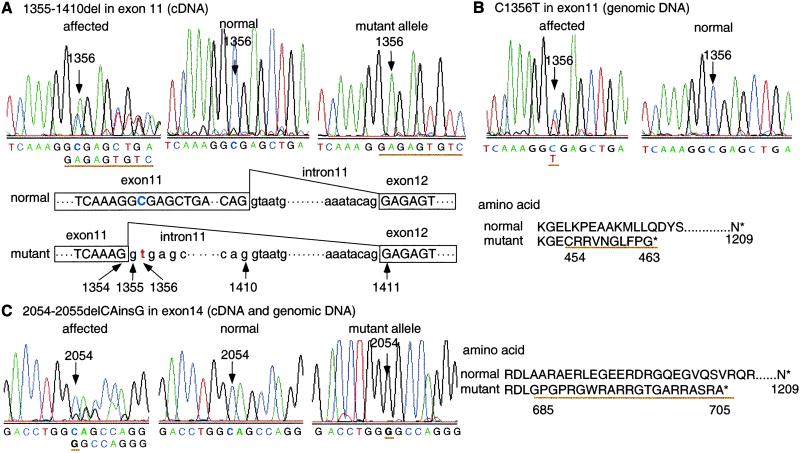
To determine whether mutations in geneX cause BCD, we compared the nucleotide sequences of geneX between normal and affected calves, and identified two distinct mutations in the affected calves. One mutation was a 56-base deletion at position 1355–1410 in exon 11 (Fig. 3A). The deletion caused a frameshift and a premature termination at codon 464, resulting in a 62% shortened protein. Comparison of the genomic sequence of the region between the affected and normal calves revealed no differences in the nucleotide sequence except for a C to T transition at position 1356 (C1356T; Fig. 3B). Remarkably, the C1356T mutation created a cryptic splice donor site in exon 11 (AAGGT1356GAGC) that substituted for the authentic splice donor site and led to improper splicing at position 1355, resulting in the 56-base RNA deletion between 1355 and 1410. The second mutation was a CA to G substitution at position 2054–2055 (2054–2055delCAinsG; Fig. 3C). The substitution also caused a frameshift and a premature termination at codon 706, resulting in a 42% shortened protein. We confirmed the CA to G substitution in the genomic DNA of the affected calves.
Fig 3.
Mutation analysis in bovine LBN gene. Sequence chromatograms of affected calves having heterozygous haplotypes (Left), normal calves (Center), and subcloned mutant alleles (Right). Amino acid sequence changes caused by mutations. (A) cDNA sequence in exon 11. A 56-bp deletion is observed at position 1355–1410 in the mutant allele, which is schematically illustrated. The deletion causes a frameshift, producing a premature termination at codon 464 with an extension of 10 aberrant amino acids. (B) Genomic sequence in exon 11. C to T substitution is observed at position 1356 in the affected calf (C1356T), which creates a cryptic splice donor site in exon 11 and leads to the improper splicing identified in the affected cDNA sequence. (C) cDNA and genomic sequences in exon 14. CA to G substitution is observed at positions 2054 and 2055 in the mutant allele (2054–2055delCAinsG), which causes a frameshift producing a premature termination at codon 706 with an extension of 21 aberrant amino acids.
To confirm whether the mutations cosegregate with the phenotype and whether these two mutations are distinct alleles of the gene, we performed a direct sequencing analysis of all affected and unaffected animals of the pedigree as well as animals of unrelated populations (see Table 1, which is published as supporting information on the PNAS web site). All 119 affected calves were either homozygous for 2054–2055delCAinsG or compound heterozygous for C1356T and 2054–2055delCAinsG. The two sires of the affected offspring were carriers of 2054–2055delCAinsG, and the dams were carriers of either of the two mutations. The result was consistent with that of the haplotype analysis (Fig. 2). All 187 unaffected relatives had one or two chromosomes carrying the wild-type allele. Furthermore, no mutation was observed in 201 animals of the unrelated populations (106 calves of Holstein and 95 calves of Japanese black cattle), confirming that these mutations are specific to the disease. These data provide strong evidence that both mutations are distinct alleles of geneX and are responsible for BCD. We designated this gene LIMBIN (LBN), because the BCD disorder was characterized as the formation of short limbs.
Expression of Mouse Lbn.
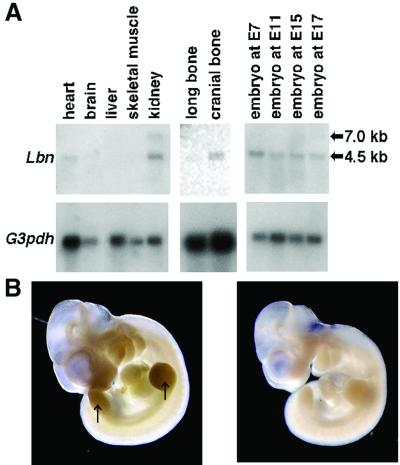
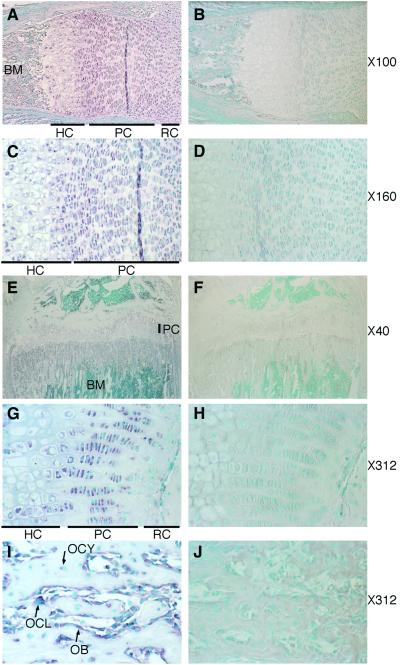
We examined the expression of the Lbn gene in mouse tissues to evaluate the potential roles of the gene in the development of long bone. Northern blot analysis revealed that Lbn was expressed as an ≈4.5-kb mRNA in long bone, cranial bone, kidney, heart, and embryos at E7, E11, E15, and E17 (Fig. 4A). Whole-mount in situ hybridization showed that Lbn was expressed in fore- and hindlimb buds, branchial arches, and facial primordia at the limb bud formation stage, E11 (Fig. 4B), suggesting that Lbn has a role in early embryonic morphogenesis as well. In situ hybridization analysis using mouse tibia at E17 revealed that mouse Lbn was strongly expressed in proliferating chondrocytes, and weakly in other resting and hypertrophic chondrocytes in the epiphysial growth plates (Fig. 5 A and C). At P35, Lbn also was expressed in proliferating chondrocytes, but not in resting and hypertrophic chondrocytes (Fig. 5 E and G). In the metaphysis, the expression was observed strongly in osteoblasts and osteoclasts on the bone surface and weakly in osteocytes (Fig. 5I). These data implicate involvement of the Lbn gene in the formation and growth of long bones.
Fig 4.
Lbn expression in mouse tissues. (A) Northern blot analysis. An ≈4.5-kb Lbn mRNA can be seen in heart and kidney in adult, long bone, and cranial bone at P14 and whole embryos at all stages tested. An additional ≈7.0-kb band is faintly visible in kidney. (Lower) Loading controls with a G3pdh probe. (B) Whole-mount in situ hybridization with Lbn antisense probe (Left) and sense probe as a control (Right). A definite signal is observed in fore- and hindlimb buds (arrow), branchial arches, and facial primordia. The magnification is ×12.5.
Fig 5.
Lbn expression in mouse long bones. In situ hybridization with Lbn antisense probe (Left) and sense probe as a control (Right). (A–D) Mouse tibia at E17. Lbn is expressed strongly in proliferating chondrocytes and weakly in other resting and hypertrophic chondrocytes in the epiphysial growth plates. (E–J) Mouse tibia at P35. Lbn is expressed strongly in proliferating chondrocytes in the epiphysial growth plates, osteoblasts, and osteoclasts and weakly in osteocytes in the metaphysis. RC, resting chondrocytes; PC, proliferating chondrocytes; HC, hypertrophic chondrocytes; OB, osteoblasts; OCL, osteoclasts; OCY, osteocytes; BM, bone marrow. The magnification is shown on the right.
Discussion
In the present study, we identified the gene LBN as a causative gene for BCD. This conclusion is supported by the following evidence: (i) the position of the LBN gene on cattle chromosome, (ii) two frameshift mutations causing 62% and 42% deletion of the LBN protein, (iii) the perfect cosegregation of the mutations with the phenotype, and (iv) expression of LBN in the limb buds and growth plate chondrocytes. Although the function of LBN remains unknown, LBN should have essential roles for a skeletal development.
Mouse Lbn was strongly expressed in the proliferating chondrocytes in mouse tibia (Fig. 5 C and G). Several ligands and their receptors, including Indian hedgehog, PTH-related peptide, PTH/PTH-related peptide receptor, fibroblast growth factor receptor 3, and others are expressed in proliferating and/or prehypertrophic chondrocytes and regulate chondrocytic proliferation and differentiation that are critical to bone morphogenesis (35–38). In addition, extracellular matrix proteins such as proteoglycan and collagen have important roles in normal bone development (1–3, 39). As long bones of the affected animals show a disturbance in chondrocytic differentiation and abnormal formation of cartilaginous matrix (17), the loss of function of LBN might have a direct or indirect effect on proliferating chondrocytes through interactions with these factors and disturb a proper differentiation to hypertrophic chondrocytes to form a proper cartilaginous matrix. Moreover, the expression of mouse Lbn was observed in cranial bone, osteoblasts, osteoclasts, osteocytes, and kidney (Figs. 4A and 5I), suggesting that Lbn has a role in osteogenesis and/or Ca2+ homeostasis as well.
LBN might have a positional relation to EVC. LBN and EVC genes are both responsible for short-limb disorder (22) and are arranged in a head-to-head configuration in cattle, human, and mouse, with transcription-start sites separated by 1,869 and 1,754-bp in human and mouse, respectively. In addition, LBN and EVC genes have similar expression patterns. Ruiz-Perez et al. (22) reported that EVC is expressed in human fetal tissues including developing bone, heart, kidney, and lung at Carnegie stages 19 and 21. We also demonstrated that mouse Lbn is expressed in developing bone and adult heart and kidney (Fig. 4A). Because of the contiguous genomic organization and similarity of expression patterns, both genes might be coexpressed and coregulated by common sequences within the intervening region. Kunte et al. (40) reported that UFD1L and CDC45L genes expressed in limb buds and pharyngeal arches are arranged in a head-to-head configuration, and an 884-bp intervening region could direct bidirectional transcription activation of both genes in a neural crest-derived cell line.
LBN is a gene involved in bone formation and shows no structural homology with any other known gene. Future studies of LBN, both in vivo and in vitro, and further pathological examination of BCD will provide new insight into the complex biologic processes of mammalian skeletal development.
Supplementary Material
Acknowledgments
We thank the Kumamoto Federation of Agricultural Mutual Aid Association and the Japan Registry Association of Akaushi for collecting samples and pedigree data; H. Tsukazawa, K. Maruyama, and Y. Nishijima for technical assistance; S. Miura for assistance in whole-mount in situ hybridization; N. E. Cockett for fgfr3 primers; and K. Hara, M. Agaba, A. Takasuga, T. Watanabe, and S. Hirotsune for insightful suggestions. This work was supported in part by grants from the Japan Racing and Livestock Promotion Foundation (to Y.S.), the Agriculture and Livestock Industries Corporation (to Y.S.), the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.K.), and the Livestock Improvement Association of Japan, Inc. (to T.K.).
Abbreviations
BCD, bovine chondrodysplastic dwarfism
YAC, yeast artificial chromosome
BAC, bacterial artificial chromosome
STS, sequence-tagged site
RT, reverse transcriptase
En, embryonic day n
Pn, postnatal day n
DIG, digoxigenin
References
- 1.Lee B., Vissing, H., Ramirez, F., Rogers, D. & Rimoin, D. (1989) Science 244, 978-980. [DOI] [PubMed] [Google Scholar]
- 2.Vissing H., D'Alessio, M., Lee, B., Ramirez, F., Godfrey, M. & Hollister, D. W. (1989) J. Biol. Chem. 264, 18265-18267. [PubMed] [Google Scholar]
- 3.Warman M. L., Abbott, M., Apte, S. S., Hefferon, T., McIntosh, I., Cohn, D. H., Hecht, J. T., Olsen, B. R. & Francomano, C. A. (1993) Nat. Genet. 5, 79-82. [DOI] [PubMed] [Google Scholar]
- 4.Foster J. W., Dominguez-Steglich, M. A., Guioli, S., Kowk, G., Weller, P. A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I. D., Goodfellow, P. N., et al. (1994) Nature (London) 372, 525-530. [DOI] [PubMed] [Google Scholar]
- 5.Hastbacka J., de la Chapelle, A., Mahtani, M. M., Clines, G., Reeve-Daly, M. P., Daly, M., Hamilton, B. A., Kusumi, K., Trivedi, B., Weaver, A., et al. (1994) Cell 78, 1073-1087. [DOI] [PubMed] [Google Scholar]
- 6.Karaplis A. C., Luz, A., Glowacki, J., Bronson, R. T., Tybulewicz, V. L., Kronenberg, H. M. & Mulligan, R. C. (1994) Genes Dev. 8, 277-289. [DOI] [PubMed] [Google Scholar]
- 7.Rousseau F., Bonaventure, J., Legeai-Mallet, L., Pelet, A., Rozet, J. M., Maroteaux, P., Le Merrer, M. & Munnich, A. (1994) Nature (London) 371, 252-254. [DOI] [PubMed] [Google Scholar]
- 8.Shiang R., Thompson, L. M., Zhu, Y. Z., Church, D. M., Fielder, T. J., Bocian, M., Winokur, S. T. & Wasmuth, J. J. (1994) Cell 78, 335-342. [DOI] [PubMed] [Google Scholar]
- 9.Wagner T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Bricarelli, F. D., Keutel, J., Hustert, E., et al. (1994) Cell 79, 1111-1120. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe H., Kimata, K., Line, S., Strong, D., Gao, L. Y., Kozak, C. A. & Yamada, Y. (1994) Nat. Genet. 7, 154-157. [DOI] [PubMed] [Google Scholar]
- 11.Bellus G. A., McIntosh, I., Smith, E. A., Aylsworth, A. S., Kaitila, I., Horton, W. A., Greenhaw, G. A., Hecht, J. T. & Francomano, C. A. (1995) Nat. Genet. 10, 357-359. [DOI] [PubMed] [Google Scholar]
- 12.Briggs M. D., Hoffman, S. M., King, L. M., Olsen, A. S., Mohrenweiser, H., Leroy, J. G., Mortier, G. R., Rimoin, D. L., Lachman, R. S., Gaines, E. S., et al. (1995) Nat. Genet. 10, 330-336. [DOI] [PubMed] [Google Scholar]
- 13.Hecht J. T., Nelson, L. D., Crowder, E., Wang, Y., Elder, F. F., Harrison, W. R., Francomano, C. A., Prange, C. K., Lennon, G. G., Deere, M., et al. (1995) Nat. Genet. 10, 325-329. [DOI] [PubMed] [Google Scholar]
- 14.Schipani E., Kruse, K. & Juppner, H. (1995) Science 268, 98-100. [DOI] [PubMed] [Google Scholar]
- 15.Tavormina P. L., Shiang, R., Thompson, L. M., Zhu, Y. Z., Wilkin, D. J., Lachman, R. S., Wilcox, W. R., Rimoin, D. L., Cohn, D. H. & Wasmuth, J. J. (1995) Nat. Genet. 9, 321-328. [DOI] [PubMed] [Google Scholar]
- 16.Moritomo Y., Ishibashi, T., Ashizawa, H. & Shibata, T. (1989) J. Jpn. Vet. Med. Assoc. 42, 173-177. [Google Scholar]
- 17.Moritomo Y., Ishibashi, T. & Miyamoto, H. (1992) J. Vet. Med. Sci. 54, 453-459. [DOI] [PubMed] [Google Scholar]
- 18.Julian L. M., Tyler, W. S. & Gregory, P. W. (1959) J. Am. Vet. Med. Assoc. 135, 104-109. [PubMed] [Google Scholar]
- 19.Weaver A. D. (1975) Vet. Ann. 15, 7-9. [Google Scholar]
- 20.Yoneda K., Moritomo, Y., Takami, M., Hirata, S., Kikukawa, Y. & Kunieda, T. (1999) Mamm. Genome 10, 597-600. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S. J., Beaty, T. H., McIntosh, I., Hefferon, T. & Panny, S. R. (1998) Am. J. Med. Genet. 75, 419-423. [PubMed] [Google Scholar]
- 22.Ruiz-Perez V. L., Ide, S. E., Strom, T. M., Lorenz, B., Wilson, D., Woods, K., King, L., Francomano, C., Freisinger, P., Spranger, S., et al. (2000) Nat. Genet. 24, 283-286. [DOI] [PubMed] [Google Scholar]
- 23.Takeda H., Yamakuchi, H., Ihara, N., Hara, K., Watanabe, T., Sugimoto, Y., Oshiro, T., Kishine, H., Kano, Y. & Kohno, K. (1998) Anim. Genet. 29, 216-219. [DOI] [PubMed] [Google Scholar]
- 24.Ogilvie D. J. & James, L. A. (1995) in YAC Protocols, ed. Markie, D. (Humana, Totowa, NJ), pp. 131–138.
- 25.Kijas J. M., Fowler, J. C., Garbett, C. A. & Thomas, M. R. (1994) Biotechniques 16, 656-662. [PubMed] [Google Scholar]
- 26.Hirano T., Nakane, S., Mizoshita, K., Yamakuchi, H., Inoue-Murayama, M., Watanabe, T., Barendse, W. & Sugimoto, Y. (1996) Anim. Genet. 27, 365-368. [DOI] [PubMed] [Google Scholar]
- 27.Coppieters W., Kvasz, A., Farnir, F., Arranz, J. J., Grisart, B., Mackinnon, M. & Georges, M. (1998) Genetics 149, 1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takasuga A., Hirotsune, S., Itoh, R., Jitohzono, A., Suzuki, H., Aso, H. & Sugimoto, Y. (2001) Nucleic Acids Res. 29, E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz J., Milpetz, F., Bork, P. & Ponting, C. P. (1998) Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai K. & Horton, P. (1999) Trends Biochem. Sci. 24, 34-36. [DOI] [PubMed] [Google Scholar]
- 31.Conlon R. A. & Rossant, J. (1992) Development (Cambridge, U.K.) 116, 357-368. [DOI] [PubMed] [Google Scholar]
- 32.Ikegame M., Ishibashi, O., Yoshizawa, T., Shimomura, J., Komori, T., Ozawa, H. & Kawashima, H. (2001) J. Bone Miner. Res. 16, 24-32. [DOI] [PubMed] [Google Scholar]
- 33.Womack J. E., Johnson, J. S., Owens, E. K., Rexroad, C. E., 3rd, Schlapfer, J. & Yang, Y. P. (1997) Mamm. Genome 8, 854-856. [DOI] [PubMed] [Google Scholar]
- 34.Honkura T., Ogasawara, J., Yamada, T. & Morishita, S. (2002) Nucleic Acids Res. 30, 221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amizuka N., Warshawsky, H., Henderson, J. E., Goltzman, D. & Karaplis, A. C. (1994) J. Cell Biol. 126, 1611-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng C., Wynshaw-Boris, A., Zhou, F., Kuo, A. & Leder, P. (1996) Cell 84, 911-921. [DOI] [PubMed] [Google Scholar]
- 37.Lanske B., Karaplis, A. C., Lee, K., Luz, A., Vortkamp, A., Pirro, A., Karperien, M., Defize, L. H., Ho, C., Mulligan, R. C., et al. (1996) Science 273, 663-666. [DOI] [PubMed] [Google Scholar]
- 38.Vortkamp A., Lee, K., Lanske, B., Segre, G. V., Kronenberg, H. M. & Tabin, C. J. (1996) Science 273, 613-622. [DOI] [PubMed] [Google Scholar]
- 39.Chintala S. K., Miller, R. R. & McDevitt, C. A. (1994) Arch. Biochem. Biophys. 310, 180-186. [DOI] [PubMed] [Google Scholar]
- 40.Kunte A., Ivey, K., Yamagishi, C., Garg, V., Yamagishi, H. & Srivastava, D. (2001) Mech. Dev. 108, 81-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.