Abstract
The maltose transport complex of Escherichia coli, a member of the ATP-binding cassette (ABC) superfamily, is made up of two nucleotide-binding subunits, MalK2, which hydrolyze ATP with positive cooperativity, and two transmembrane subunits, MalF and MalG. The ABC family is defined in part by the canonical signature motif LSGGQ whose exact function remains controversial. Taking advantage of the dual function of vanadate as a transition state analogue and as a photoactive chemical, we demonstrate that vanadate catalyzes the UV-dependent cleavage of the polypeptide backbone at both the LSGGQ motif and the nucleotide-binding, or Walker A, motif when it is trapped in the nucleotide-binding site of the bacterial maltose transporter. This highly specific cleavage pattern indicates that residues in both motifs are immediately adjacent to ATP during hydrolysis, and are therefore likely to participate directly in ATP-binding and/or hydrolysis. Because the LSGGQ motif is too distant from the nucleotide in the structure of an ABC monomer for cleavage to occur, these data support a model in which the LSGGQ motif contacts the nucleotide across the interface of a MalK dimer, as seen in the crystal structure of Rad50. This architecture provides a basis for the cooperativity observed in the nucleotide-binding domains of ABC transporters and a function for this highly conserved family signature motif.
ATP-binding cassette (ABC) transporters function in both prokaryotes and eukaryotes to transport a large variety of substrates into and out of cells. Medically important members of this family include the cystic fibrosis transmembrane regulator (1) and a multiple drug-resistance protein, P-glycoprotein, which extrudes drugs from cancer cells (2, 3). The maltose transporter traverses the cytoplasmic membrane of Escherichia coli and has proven to be a useful model system for the study of this family. The transporter (MalFGK2) is a complex made up of two transmembrane subunits, MalF and MalG, and two ATP-hydrolyzing subunits, MalK2, which are exposed to the cytoplasm (4, 5). The ATPase subunit of an ABC transporter contains a nucleotide-binding subdomain similar to that seen in F1-ATPase (6), and a helical subdomain unique to ABC transporters resulting from the insertion of approximately 70 aa between the canonical Walker A and Walker B motifs that bind MgATP (7, 8). This subdomain contains a family signature motif, LSGGQ, whose function remains controversial. In the maltose transport system, a soluble maltose-binding protein (MBP) stimulates the ATPase activity of the transporter complex by stabilizing the transition state conformation of the transporter during ATP hydrolysis (9).
In a reaction that requires ATP, vanadate acts as a transition state analogue to stabilize an inhibitory complex of MBP and the transporter with ADP and vanadate tightly bound in one of the two nucleotide-binding sites (9, 10). It presumably occupies the position of the γ phosphate adjacent to the ADP, as observed in the crystal structure of vanadate-trapped myosin (11). In myosin (12), adenylate kinase (13) and F1-ATPase (14), UV irradiation of the vanadate-trapped species induces a highly specific cleavage of the polypeptide backbone adjacent to the vanadate ion at the third residue of the Walker A motif (7). This residue is the point of closest contact between the γ phosphate and the polypeptide backbone of the Walker A motif (11, 15, 16). Because different residues occupy this position in these three proteins, proximity to vanadate rather than side-chain chemistry may dominate the photocleavage reaction.
Vanadate photocleavage can be used to identify residues in close contact to nucleotide as long as vanadate binding is restricted to the nucleotide-binding pocket. At high concentrations of vanadate, it binds and cleaves myosin at a second site remote from the Walker A (17). However, cleavage can be restricted to the nucleotide-binding site if nucleotide is present to trap vanadate in the nucleotide-binding site and excess vanadate is removed by gel filtration. Based on the nucleotide dependence of vanadate photocleavage of myosin light chain kinase, it has been suggested that the regulatory domain is in direct contact with ATP at the active site (18). In ABC transporters, ATP is bound on the surface of a nucleotide-binding subunit (8, 19–21), raising the possibility that residues from another subunit may complete the nucleotide-binding site. We have determined that both the Walker A and the LSGGQ motifs of MalK are cleaved by vanadate. These results are consistent with nucleotide binding at a dimer interface between the Walker A and LSGGQ, as seen in the crystal structure of Rad50 (22).
Materials and Methods
Protein Expression and Purification.
The maltose transport complex MalFGK2 was overexpressed in E. coli as described (10). Modification of MalK with a polyhistidine tag allows purification of the entire MalFGK2 complex on cobalt affinity resin (CLONTECH) to greater than 90% homogeneity (10). Briefly, membrane proteins (3 mg/ml) were solubilized with 1% n-dodecyl β-d-maltoside in 20 mM Hepes, pH 8.0/5 mM MgCl2/20% glycerol. The affinity resin was equilibrated and loaded in buffer A [50 mM Hepes (pH 8.0)/300 mM NaCl/10% glycerol/0.01% n-dodecyl β-d-maltoside] and eluted with buffer A containing 100 mM imidazole. Purified transport complexes were dialyzed against buffer B [20 mM Hepes (pH 8.0)/5 mM MgCl2/10% glycerol/0.01% n-dodecyl β-d-maltoside]. Three different variants of MalK were used in these experiments. N-His6-MalK is MalK with an N-terminal 23-aa extension containing a 6-histidine repeat and has been described (23). N-VRRAS-His6-MalK is a modification of N-His6-MalK containing an insertion of 6 aa, VRRASV, immediately following the N-terminal methionine. MalK-His6-C has a 10-aa extension, ASASHHHHHH, added to the C terminus of MalK. Oligonucleotides encoding these N- or C-terminal extensions were added to the ends of the malK gene by using PCR.
Phosphorylation of N-VRRAS-His6-MalK by Protein Kinase A.
Two hundred and fifty units of protein kinase A (Sigma) was resuspended in 250 μl of buffer containing 20 mM Tris⋅HCl (pH 8.0), 1 mM DTT, and 100 mM NaCl. Six micromolar purified MalFGK2 containing the N-VRRAS-His6-MalK protein was incubated with 1 unit of protein kinase A per 25 μl of final volume, 0.2 μM [γ-32P]ATP, 20 mM Tris⋅HCl (pH 8.0)/10% glycerol/10 mM DTT/100 mM NaCl/20 mM MgCl2/0.1% n-dodecyl β-d-maltoside at 37°C for 1 h (24). The sample was desalted into buffer B before treatment with vanadate.
Vanadate Trapping and Photocleavage.
To trap vanadate and ADP in the nucleotide-binding site of MalFGK2, 6 μM purified MalFGK2 and 6 μM MBP were typically incubated with 15 mM MgCl2, 15 mM ATP, 0.1 mM maltose, and 0.5 mM vanadate in 20 mM Hepes (pH 8)/10% glycerol/0.01% n-dodecyl β-d-maltoside for 20 min at 37°C, as described (9, 10). Vanadate was boiled for 2 min before use to break down higher-order complexes (25). For some experiments, excess vanadate was removed with a desalting column. Vanadate-trapped complexes were then treated with UV light for 2 h using a Blak-Ray Longwave UV lamp (model B-100, UVP, San Gabriel, CA).
Electrophoresis.
SDS/PAGE was performed according to Laemmli (26) with either 11% or 15% polyacrylamide. Proteins were visualized by silver staining according to the method of Heukeshoven and Dernick (27). For Western blot analysis, proteins were transferred to nitrocellulose as described by Towbin (28) and detected with polyclonal anti-MalK antiserum (29) and SuperSignal West Dura Extended (Pierce). For autoradiography, gels were soaked in water, dried under a vacuum, and exposed to Hyperfilm MP (Amersham Pharmacia). Quantitation of radioactive bands was performed on a PhosphorImager (Molecular Dynamics, Storm 860).
Isolation of Cleavage Fragments for Mass Spectrometry.
After vanadate-induced photocleavage of MalFGK2, cleavage fragments were isolated from SDS/PAGE gels following the procedure recommended by Galvani et al. (31) for analysis by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS). Briefly, gels were stained for 15 min in 0.1% Coomassie blue/50% methanol/10% acetic acid, then destained overnight in water/methanol/acetic acid (4:1:1, vol/vol). The desired bands were cut from the gel and sonicated for 10 min in 50% methanol to remove the dye. Bands were then sliced very thinly with a razor blade and placed in 100 μl of a solution of formic acid/acetonitrile/isopropyl alcohol/water (50:25:15:10, vol/vol) and sonicated for 1 h. Recovery of the band was monitored by loading one-tenth of the total sample on an SDS/PAGE gel and visualizing by Coomassie blue stain or by transferring to nitrocellulose for Western blot analysis. MS was done at the Harvard Microchemistry and Proteomics Analysis Facility.
MALDI-TOF MS Analysis.
Samples were purified before MALDI-TOF MS analysis by using a C4 ZipTip (Millipore), eluted with 1 μl of 50% acetonitrile/0.1% trifluoroacetic acid (TFA) and mixed with 1 μl of saturated sinapinic acid in 1:3:2, vol/vol, formic acid/water/isopropyl alcohol (FWI) on a 96 × 2 Teflon sample target and allowed to air dry. Positive-ion MALDI spectra were acquired in linear mode using a 25-kV acceleration voltage and a grid voltage of 91 V with an Applied Biosystems Voyager DE-STR mass spectrometer.
Liquid Chromatography/Electrospray Ionization (ESI) MS Analysis.
Sample 17C from the MALDI-TOF MS analysis was loaded onto a 75 μm × 10 cm Poros microcapillary column (New Objective, Woburn, MA). The column was equilibrated with 5% acetonitrile/0.1% acetic acid (buffer A) and a reverse-phase liquid chromatography gradient of 2% B per minute (B: 0.1% acetic acid/acetonitrile) was run. Data were acquired in positive-ion mode by using a Micromass QTof-1 (Atlas Park, Simonway, Manchester, UK) mass spectrometer operated in MS mode using a capillary spray voltage of 3.35 kV and a cone voltage of 40 V. The highly charged spectrum was deconvoluted by using the MaxEnt 1 algorithm.
Results
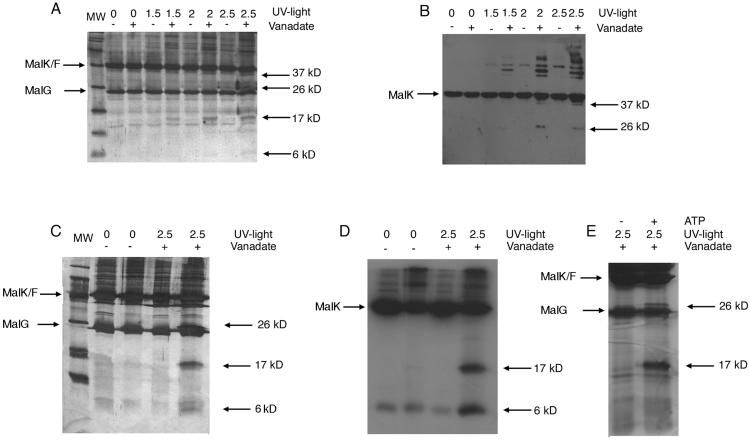
The conditions for stable inhibition of the maltose transport complex by vanadate have been described (9, 10). ATP hydrolysis is required to see stable inhibition of ATPase activity and trapping of ADP along with vanadate in one of the two nucleotide-binding sites of MalFGK2. Because ADP cannot replace ATP in the trapping reaction, it is assumed that vanadate binds the transporter immediately after ATP hydrolysis and after phosphate dissociation, but before the release of ADP. MBP is also required for vanadate inhibition because it stimulates ATP hydrolysis in a wild-type transporter. However, a binding-protein-independent mutant can hydrolyze ATP and trap vanadate in the absence of MBP. When a vanadate-trapped binding-protein-independent MalF500GK2 transporter was irradiated with UV light, four cleavage fragments of 37, 26, 17 and 6 kDa are visible in a silver-stained gel (Fig. 1A). Essentially the same cleavage pattern is observed in the wild-type MalFGK2 transporter (Fig. 1C) although the 37-kDa band is likely to be obscured by the presence of MBP on the gel. The 37-kDa and 26-kDa fragments were identified as originating from MalK based on their interaction with antibody specific to MalK (Fig. 1B). A protein kinase A recognition site with the sequence VRRAS was placed at the N terminus of MalK to aid in the identification of the other two fragments. When the transporter complex was phosphorylated by protein kinase A using [γ-32P]ATP before vanadate trapping, the 17-kDa and 6-kDa cleavage fragments were radiolabeled, demonstrating that they both contain the N terminus of MalK (Fig. 1D). Quantitation of the radioactivity associated with each band in Fig. 1D indicated that ≈5% of the MalK protein was cleaved in each event. Thus all four fragments have been positively identified as originating from MalK and the sizes of the fragments were consistent with two separate cleavage events having occurred in the MalK protein (Fig. 2). If the protein were cut in the Walker A motif, fragments of 6 kDa and 37 kDa would result. Cleavage at the Walker A motif, which contacts the γ phosphate of ATP, is seen in many other ATPases, including P-glycoprotein, an ABC protein (12–14, 18, 30). If the protein were cut at the LSGGQ motif, fragments of 17 kDa and 26 kDa would result, and this would be a previously undocumented finding. When ATP was omitted from the sample to prevent trapping of vanadate in the nucleotide-binding site (10), this second cleavage event did not occur, even if the sample was not desalted before UV light exposure (Fig. 1E). This result indicates that vanadate trapped at the nucleotide-binding site is responsible for the cleavage event and that this region of the protein is located very close to the γ phosphate of ATP in the catalytic transition state.
Figure 1.
Detection of cleavage fragments generated by vanadate trapping and UV exposure. Purified MalFGK2 was vanadate trapped as described in Materials and Methods, and samples were then treated with UV light over a time course of 1.5, 2, and 2.5 h. (A) Cleavage fragments from the binding protein-independent MalF500GK2 transporter, containing N-His6-MalK, are visualized by silver staining after SDS/PAGE on a 15% gel in the presence or absence of vanadate and UV light, as indicated. MW, molecular weight standards. (B) Samples from A were separated by SDS/PAGE for Western blot analysis using a polyclonal antibody raised against MalK on both 11% and 15% gels to visualize the larger and smaller cleavage fragments, respectively. Only the 11% gel is shown, as the two smaller fragments did not react with the antibody. (C) Cleavage fragments from a wild-type transporter containing 32P-labeled N-VRRAS-His6-MalK protein are visualized by silver staining in the presence or absence of vanadate and UV light as indicated. (D) Autoradiogram of gel in C. (E) Silver-stained gel showing cleavage fragments from the wild-type transporter incubated with vanadate in the presence or absence of ATP and treated with UV light.
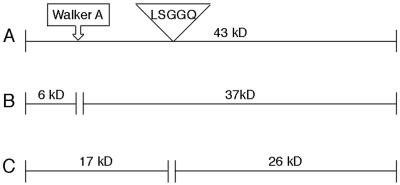
Figure 2.
Schematic representation of the potential cleavage sites in MalK. (A) Full-length N-His6-MalK (43 kDa) showing the location of Walker A and LSGGQ motifs. (B) Fragments (6 kDa and 37 kDa) produced from cleavage at the Walker A motif. (C) Fragments (17 kDa and 26 kDa) produced from cleavage at the LSGGQ motif.
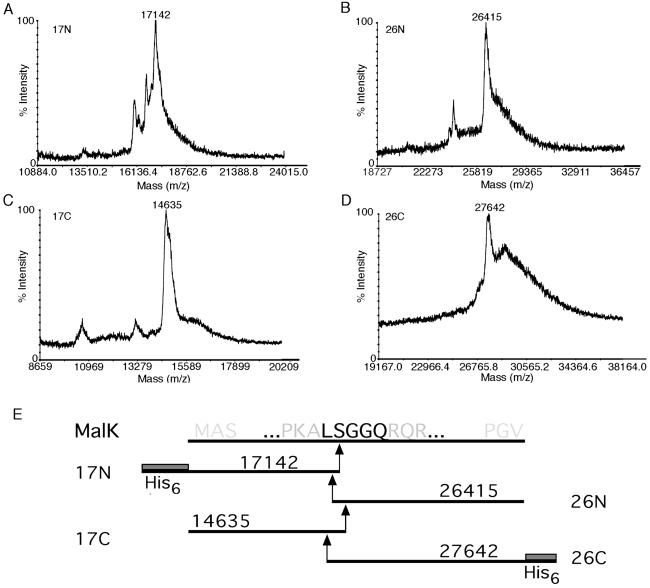
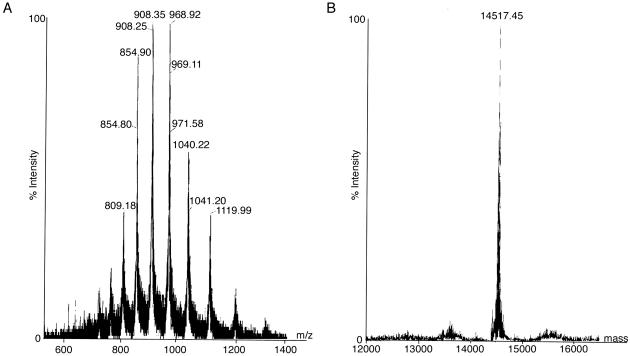
To identify the exact site of cleavage near the LSGGQ motif, we attempted to sequence the 26-kDa C-terminal cleavage fragment by Edman degradation, but it appeared to be blocked. Vanadate will cleave the polypeptide backbone at regions other than the peptide bond, leaving modifications that can prevent N-terminal sequencing at the site of cleavage (12, 13, 18). MS was used as an alternative approach for localizing the cleavage site in MalK. Two different MalK constructs with different modifications at the N terminus and C terminus were used to help identify the fragments in the spectrum. In the first experiment, MalK containing an N-terminal histidine-tagged extension (N-His6-MalK, 43,485 Da) was incorporated into the transporter complex. The N- and C-terminal fragments from this construct (17N and 26N) were isolated from the gel as described in Materials and Methods, and analyzed by using MALDI-TOF MS (Fig. 3 A and B). We estimate that ≈1 μg of cleavage fragment protein was recovered from 1 mg of total protein loaded on the SDS/PAGE gel. In the second experiment, MalK containing a C-terminal histidine-tagged extension (MalK-His6-C, 42,087 Da) was used (Fig. 3 C and D). The results of these experiments are summarized in Fig. 3E, where the mass of each of the four fragments is shown to be consistent with cleavage within the LSGGQ motif. More specifically, the data cluster around the first and second residues of the motif, within a range of 170 Da. The range most likely reflects the error associated with the measurements after extraction of fragments from an SDS/PAGE gel. The resolution in this experiment might be adversely affected by residual salts and detergents, adduct formation due to exposure to acrylamide and formic acid, or heterogeneity of the sample (31). To obtain a more accurate mass measurement, the sample containing fragment 17C, which displayed the best signal-to-noise ratio in the MALDI-TOF MS analysis was further purified by reverse-phase liquid chromatography and analyzed by ESI MS (Fig. 4). The deconvoluted mass of the fragment was 14,517 Da. The mass accuracy in this determination was estimated to be within 0.02%, based on an external calibration with myoglobin (16,952.6 Da). If the N-terminal methionine is still present on the fragment, then the cleavage has occurred before the serine in the LSGGQ motif, leaving at least part of the leucine in the N-terminal cleavage fragment. However, if methionine aminopeptidase has removed the N-terminal methionine during synthesis (32), then the size is more consistent with cleavage after the serine residue (14,510 Da). Only MalK fragment lacking the N-terminal methionine was found by Edman degradation. However, formylation could have masked a population of MalK with N-terminal methionine intact.
Figure 3.
MALDI-TOF mass spectrometric analysis of cleavage fragments. After SDS/PAGE, the bands containing the cleavage fragments were excised and protein was extracted as described in Materials and Methods for analysis by using MALDI-TOF MS. (A) Spectrum of N-terminal fragment, 17N, from N-His6-MalK. (B) Spectrum of C-terminal fragment, 26N, from N-His6-MalK. (C) Spectrum of N-terminal fragment, 17C, from MalK-His6-C. (D) Spectrum of C-terminal fragment, 26C, from MalK-His6-C. (E) Schematic illustrating the predicted site of cleavage in MalK, based on masses of fragments determined in A–D. Mass of the N-His6 extension is 2,521 Da, mass of MalK is 40,964 Da, and mass of the C-His6 extension is 1,138 Da.
Figure 4.
ESI MS of the 17C fragment. After extraction from an SDS/PAGE gel, sample 17C was further purified by reverse-phase liquid chromatography on a C4 column, then analyzed by ESI. (A) Mass spectrum with charge state distribution from approximately +11 to +20. (B) Deconvoluted spectrum showing mass of 17C fragment.
Discussion
In this report, we have used vanadate-induced photooxidative cleavage to identify amino acids at the ATP-binding site of an ABC transporter. In addition to an anticipated cleavage event that maps to the Walker A motif of MalK, a second cleavage event occurred at the serine residue in the conserved family signature sequence LSGGQ of MalK. The nucleotide dependence of this photocleavage event indicated that vanadate trapped in the nucleotide-binding site of MalK was responsible for the cleavage. Vanadate-induced photocleavage is a highly specific event. In myosin, the side chain of serine in the Walker A motif is oxidized by vanadate (33), leading to cleavage of the peptide at that site. Consistent with these data, the side chain of this serine forms a hydrogen bond with vanadate oxygen in the structure of myosin in complex with MgADP and vanadate (11). Specific cleavage by vanadate at the serine in the LSGGQ of MalK suggests that this serine makes a similar contact to vanadate in the trapped species and, by analogy, to the γ phosphate of ATP in the catalytic transition state. These data strongly support the idea, suggested by Manavalan et al. (34), that the LSGGQ motif might contact the γ phosphate of ATP. Mutations in the LSGGQ motif inhibit ATP hydrolysis (35, 36). However, this hypothesis was not supported by the first structure of an ABC, HisP, the nucleotide-binding subunit from the histidine permease (8). The LSGGQ motif was over 25 Å from the nucleotide in the structure of the HisP monomer. HisP formed a dimer in the crystal, but the LSGGQ motif faced away from the dimer interface and did not contact nucleotide. Jones and George (21) proposed an alternative model for dimerization of HisP that aligned the LSGGQ motif of one subunit with the Walker A motif of the second subunit, thereby creating two nucleotide-binding sites between these two sets of motifs along the dimer interface. The concept of nucleotide-binding sites shared between two subunits is appealing because of its ability to explain the requirement for two nucleotide-binding domains or subunits in all ABC transporters. This dimer architecture was later observed in the structure of the Rad50 catalytic domain (22). Although classified as an ABC protein, Rad50 belongs to a subfamily that functions in DNA repair and maintenance rather than transport (37). The nucleotide-binding subdomain of Rad50 shares essentially the same fold as that in ABC transporters (22). Although the helical subdomain differs, the location of the family signature LSGGQ is conserved. In both the theoretical model for HisP (21) and the structure of Rad50 (22), the LSGGQ motif is located at the end of an α-helix whose positive dipole points to the γ phosphate of ATP, helping to balance the negative charge on the phosphate. Vanadate-induced photocleavage of the LSGGQ motif in MalK provides biochemical evidence that the Rad50 structure is an appropriate model for MalK–MalK interaction in the context of the intact MalFGK2 complex as it hydrolyzes ATP. Our data are not consistent with the crystallographic dimer of MalK from Thermococcus litoralis (19) or the crystallographic dimers of HisP (8) and MsbA (20). Although the structure of the monomer is conserved in each, the dimer interface varies between structures and the LSGGQ motif is distant from the nucleotide-binding site in all cases.
In the LSGGQ motif of the Rad50 dimer, both the serine, with the side-chain oxygen, and the second glycine, with the main-chain nitrogen, contact the γ phosphate of ATP (22). The serine residue in the LSGGQ motif of MalK is precisely where we have shown that vanadate, which occupies the position of the γ phosphate in the trapped intermediate, cleaves the polypeptide chain after UV irradiation. Thus, it is likely that in the catalytic transition state, the LSGGQ motif of MalK lies in essentially the same position relative to nucleotide as in the crystal structure of the Rad50 catalytic domain. Because vanadate is trapped in just one of the two nucleotide-binding sites of MalK, cleavage of the LSGGQ motif would be expected to occur in one subunit of the dimer, whereas cleavage of the Walker A motif would occur in the other subunit. The fact that this highly conserved signature motif has now been shown to lie in the nucleotide-binding pocket of MalK and Rad50, two distantly related members of the ABC superfamily, raises the possibility that all ABC proteins adopt the same dimeric configuration. In the very recent structure of the ABC transporter for vitamin B12 (38), the nucleotide-binding subunits of this transporter dimerize in a similar conformation, placing the LSGGQ motif close to the Walker A motif and providing further support for the universality of this dimeric interface.
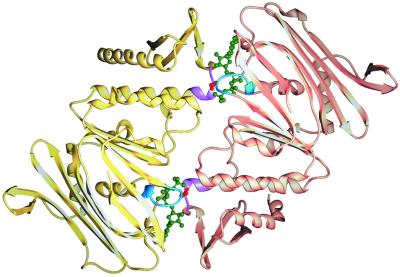
Fig. 5 illustrates how the LSGGQ motif of Rad50 completes the nucleotide-binding site of the opposing subunit, burying two nucleotides along the dimer interface. Binding of ATP actually promotes the dimerization of two Rad50 monomers (22). Residues from both subunits likely position the nucleotide and water for attack, thereby contributing to catalysis (22, 39). It has been suggested that regulation of ATPase activity in the intact transporter might be controlled by the degree of association of the two nucleotide-binding subunits (9, 40, 41). Interaction of the periplasmic binding protein with the transporter may control the ATPase activity of the transporter by bringing the LSGGQ motif into the nucleotide-binding site when ATP is bound, generating the relatively closed nucleotide-binding pocket observed in the structure (Fig. 5). In the structure of the B12 transporter, which lacks ATP, the LSGGQ motif is farther from the Walker A motif than in Rad50, 9.8 Å vs. 6.6 Å, perhaps reflecting this conformational change (38). The requirement that ATP must bind to both nucleotide-binding sites before this conformational change can take place could explain the positive cooperativity in the hydrolysis of ATP by the maltose and histidine transporters (42, 43), and the negative effect that mutations in one binding site can have on hydrolysis in the second binding site (23, 44–46).
Figure 5.
Structure of Rad50cd dimer (Protein Data Bank ID code 1F2U). The polypeptide chains of Rad50 are differentially colored to reflect the two catalytic domains that associate to form a dimer. The location of the Walker A (blue) and LSGGQ (magenta) motifs can be viewed in the context of the dimer. ATP is shown in green. The side chain of the serine at the site of cleavage in the LSGGQ motif, shown in red, is within hydrogen-bonding distance of the γ phosphate of ATP. The figure was prepared by using spock software (47).
The use of MALDI-TOF MS to measure the mass of the cleavage fragments has allowed a reasonably precise determination of the site of cleavage in the LSGGQ motif. Determining the mass for both the N- and C-terminal fragments increased the degree of confidence in the determination and provided a rough measure of the error associated with the measurement. The predicted cleavages from each of the four fragments fell within one residue of the leucine–serine peptide bond. Because vanadate does not cleave precisely at the peptide bond, undefined modifications may occur on both the N-terminal and C-terminal residues at the cleavage site. Hence, even with a more exact determination of mass, the nature of the modification is not likely to be determined by this method, without additional chemical analysis (12). The finding that the LSGGQ motif interacts with the γ phosphate of ATP clarifies the nature of the nucleotide-binding site and provides a key to understanding how the structure of an ABC transporter relates to its function.
Acknowledgments
We thank members of the Harvard Microchemistry and Proteomics Analysis Facility for helpful discussions, D. Fetsch and F. Gimble for preparation of figures and J. Chen for the MalK-His6-C construct. This work is supported by grants from the National Institutes of Health (GM49261) and the Welch Foundation (Q-1391).
Abbreviations
- ABC
ATP-binding cassette
- ESI
electrospray ionization
- MALDI-TOF
matrix-assisted laser desorption ionization–time of flight
- MBP
maltose-binding protein
- MS
mass spectroscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 9609.
References
- 1.Riordan J R, Rommens J M, Kerem B-S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Chen C-J, Chin J E, Ueda K, Clark D P, Pastan I, Gottesman M M, Roninson I G. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Hamon Y, Chimini G. J Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 4.Davidson A L, Nikaido H. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 5.Ehrmann M, Ehrle R, Hofmann E, Boos W, Schlosser A. Mol Microbiol. 1998;29:685–694. doi: 10.1046/j.1365-2958.1998.00915.x. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 7.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung L-W, Wang I X, Nikaido K, Liu P-Q, Ames G F-L, Kim S-H. Nature (London) 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Sharma S, Quiocho F A, Davidson A L. Proc Natl Acad Sci USA. 2001;98:1525–1530. doi: 10.1073/pnas.041542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S, Davidson A L. J Bacteriol. 2000;182:6570–6576. doi: 10.1128/jb.182.23.6570-6576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith C A, Rayment I. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 12.Grammer J C, Loo J A, Edmonds C G, Cremo C R, Yount R G. Biochemistry. 1996;35:15582–15592. doi: 10.1021/bi961901g. [DOI] [PubMed] [Google Scholar]
- 13.Cremo C R, Loo J A, Edmonds C G, Hatlelid K M. Biochemistry. 1992;31:491–497. doi: 10.1021/bi00117a027. [DOI] [PubMed] [Google Scholar]
- 14.Ko Y H, Bianchet M, Amzel L M, Pedersen P L. J Biol Chem. 1997;272:18875–18881. doi: 10.1074/jbc.272.30.18875. [DOI] [PubMed] [Google Scholar]
- 15.Berry M B, Phillips G N., Jr Proteins. 1998;32:276–288. doi: 10.1002/(sici)1097-0134(19980815)32:3<276::aid-prot3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 16.Braig K, Menz R I, Montgomery M G, Leslie A G, Walker J E. Structure Fold Des. 2000;8:567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 17.Cremo C R, Long G T, Grammer J C. Biochemistry. 1990;29:7982–7990. doi: 10.1021/bi00486a029. [DOI] [PubMed] [Google Scholar]
- 18.Maruta S, Mitsuhashi S, Yamada M, Ikebe M. J Biochem. 1998;124:557–564. doi: 10.1093/oxfordjournals.jbchem.a022148. [DOI] [PubMed] [Google Scholar]
- 19.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang G, Roth C B. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 21.Jones P M, George A M. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner K P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 23.Davidson A L, Sharma S. J Bacteriol. 1997;179:5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baichoo N, Heyduk T. Biochemistry. 1997;36:10830–10836. doi: 10.1021/bi970714v. [DOI] [PubMed] [Google Scholar]
- 25.Goodno C C. Methods Enzymol. 1982;85:116–123. doi: 10.1016/0076-6879(82)85014-3. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Heukeshoven J, Dernick R. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson A L, Nikaido H. J Biol Chem. 1990;265:4254–4260. [PubMed] [Google Scholar]
- 30.Hrycyna C A, Ramachandra M, Ambudkar S V, Ko Y H, Pedersen P L, Pastan I, Gottesman M M. J Biol Chem. 1998;273:16631–16634. doi: 10.1074/jbc.273.27.16631. [DOI] [PubMed] [Google Scholar]
- 31.Galvani M, Bordini E, Piubelli C, Hamdan M. Rapid Commun Mass Spectrom. 2000;14:18–25. doi: 10.1002/(SICI)1097-0231(20000115)14:1<18::AID-RCM826>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Creighton T E. Proteins, Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 33.Cremo C R, Grammer J C, Yount R G. Biochemistry. 1988;27:8415–8420. doi: 10.1021/bi00422a018. [DOI] [PubMed] [Google Scholar]
- 34.Manavalan P, Dearborn D G, McPherson J M, Smith A E. FEBS Lett. 1995;366:87–91. doi: 10.1016/0014-5793(95)00463-j. [DOI] [PubMed] [Google Scholar]
- 35.Schmees G, Stein A, Hunke S, Landmesser H, Schneider E. Eur J Biochem. 1999;266:420–430. doi: 10.1046/j.1432-1327.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- 36.Szakacs G, Ozvegy C, Bakos E, Sarkadi B, Varadi A. Biochem J. 2001;356:71–75. doi: 10.1042/0264-6021:3560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudet R, Wiley D C. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locher K P, Lee A T, Rees D C. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Y R, Blecker S, Martsinkevich O, Millen L, Thomas P J, Hunt J F. J Biol Chem. 2001;276:32313–32321. doi: 10.1074/jbc.M100758200. [DOI] [PubMed] [Google Scholar]
- 40.Mannering D E, Sharma S, Davidson A L. J Biol Chem. 2001;376:12362–12368. doi: 10.1074/jbc.M011686200. [DOI] [PubMed] [Google Scholar]
- 41.Davidson A L. J Bacteriol. 2002;184:1225–1233. doi: 10.1128/JB.184.5.1225-1233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson A L, Laghaeian S S, Mannering D E. J Biol Chem. 1996;271:4858–4863. [PubMed] [Google Scholar]
- 43.Liu C E, Liu P Q, Ames G F L. J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- 44.Azzaria M, Schurr E, Gros P. Mol Cell Biol. 1989;9:5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkower C, Michaelis S. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loo T W, Clarke D M. J Biol Chem. 1996;270:21449–21452. doi: 10.1074/jbc.270.37.21449. [DOI] [PubMed] [Google Scholar]
- 47.Christopher J A. spock (Texas A & M Univ., College Station, TX), Version 1.0b135. 1998. [Google Scholar]