Abstract
A specialized nucleotide excision repair pathway known as transcription-coupled repair (TCR) counteracts the toxic effects of DNA damage in transcriptionally active genes. The clustering of active genes into gene-rich chromosomal domains predicts that the sites of TCR are unevenly distributed through the genome. To elucidate the genomic organization and chromosomal localization of TCR, we isolated DNA fragments encompassing TCR-mediated repair sites from UV-C irradiated xeroderma pigmentosum group C cells, which can only repair the transcribed strand of active genes. This DNA was used as a molecular probe to visualize TCR in normal metaphase spreads by reverse fluorescence in situ hybridization. Whereas DNA repair sites in normal human cells are evenly distributed through the genome, TCR is highly localized at specific chromosomal domains. Particularly, clusters of TCR sites were identified at early-replicating gene-rich bands and telomeric regions of several chromosomes. High gene-density chromosomes such as chromosome 19 and the GC-rich domains of several chromosomes (T bands) are preferential locations of TCR. Our results demonstrate that the intragenomic localization of TCR resembles the uneven distribution of the human transcriptome, CpG islands, and hyperacetylated histones, enforcing the basic link between DNA repair, transcription, and nuclear organization in a complex genome.
Keywords: chromosome, DNA repair, xeroderma pigmentosum
In a continuous gene–environment interaction, the integrity of the genetic material in general and, in particular, the transcriptionally active regions is threatened by endogenous and exogenous factors such as chemical mutagens and radiation. These factors induce a wide variety of DNA lesions that exert their toxic effects by interference with transcription and replication. In nondividing cells, toxic effects are predominantly generated by DNA damage in transcriptionally active genes. This damage blocks transcription elongation and hence leads to stalling of the RNA polymerase at the site of a lesion (1, 2).
To reduce the harmful effects of exposure to DNA-damaging agents, the human genome has evolved more than 130 DNA repair genes (3). Within this group, over 30 proteins constitute the nucleotide excision repair (NER) pathway, a versatile system responsible for the repair of bulky DNA helix-distorting lesions. NER consists of two subpathways: the global genome repair pathway and a specific subpathway termed transcription-coupled repair (TCR) (recently reviewed in refs. 2 and 4). A wide variety of DNA lesions, including DNA damage induced by reactive oxygen species, seem to be targeted to TCR when located in transcriptionally active genes. Processing of DNA lesions by this pathway depends on ongoing transcription and leads to an accelerated removal of DNA lesions from the template strand of active genes and restoration of DNA damage-inhibited transcription (2, 5–7). The genomic regions subjected to TCR are larger than the actual transcription units, as TCR seems to extend beyond the polyadenylation sites of active genes (8). The situation is further complicated by the observations that preferential repair of transcriptionally active genes might occur without requirement for transcription (9) and, on the other hand, may be absent even when transcription is going on (10). In addition, recent evidences suggest that TCR is genomic context-dependent (11) and that there are TCR in RNA pol I-transcribed genes of yeast (12). All of these findings pose questions toward the nature of genomic sequences subjected to preferential repair and TCR.
In mammalian cells, only a small fraction of the genome codes for transcriptionally active genes. Recently, it has been shown that transcriptionally active genes are organized in clusters and that these gene-rich regions are separated by large intergenic DNA tracts (13). This clustering of transcriptionally active chromosomal regions is further exemplified by the uneven distribution of genes (14, 15), sites of hyperacetylated histones (16, 17), and CpG islands (18) in human chromosomes. The recently reported human transcriptome map also reflects the heterogeneous distribution of expressed genes in our genome (13). Because TCR is basically confined to RNA pol II-transcribed genes (2, 6, 7), the structural organization of genes within the human genome predicts that genomic regions subjected to TCR would also be heterogeneously distributed between and within chromosomes.
To assess the intragenomic organization and the distribution of TCR along the human chromosomes, we have used cells from patients with the hereditary UV-sensitive syndrome xeroderma pigmentosum (XP) known to be deficient in NER (reviewed in ref. 4). Genetic complementation experiments uncovered the presence of seven XP genes (XPA–XPG). All complementation groups including group A are defective in both NER subpathways with the exception of XP-C, in which only the global genome repair pathway is compromised; hence, XPC cells solely perform proficient TCR of active genes (19). In contrast, normal human fibroblasts exhibit efficient repair of UV-induced photolesions by both the global genome repair pathway and TCR. The unique repair phenotype of XPC cells provided us with a tool to isolate DNA fragments harboring TCR-mediated repair patches from UV-irradiated cells. This DNA was used as a probe to visualize TCR in normal metaphase spreads by reverse fluorescence in situ hybridization (FISH) and hence to visualize the intragenomic distribution of TCR sites at a single cell level.
Materials and Methods
Cell Culturing.
Normal human (VH25), XPA (XP25RO), and XPC (XP21RO) primary fibroblasts were cultured as described (20, 21). A total of 12 90-mm Petri dishes were established per cell line, and the cells were allowed to grow until total confluence. The confluent state was maintained for at least 2 weeks before UV irradiation.
Irradiation and Labeling of Repair Sites.
Before UV irradiation, culture medium was supplemented with BrdUrd and fluorodeoxyuridine (final concentrations of 10−5 M and 10−6 M, respectively), and cells were incubated for 30 min at 37°C. Subsequently, the supplemented medium was collected, and the Petri dishes were rinsed with 2 ml of PBS and irradiated with 10 J/m2 of UV-C light at a dose rate of 0.2 J/m2 per sec. The conditioned medium was returned to the cultures, and the cells were incubated for an additional 24 h.
Purification of Parental DNA.
Cells were lysed in a buffer containing 150 mM NaCl, 10 mM EDTA, 10 mM Tris⋅HCl (pH 8.0), 0.5% SDS, 100 μg/ml proteinase K at 37°C for 16 h. High molecular weight DNA was extracted with saturated phenol and chloroform, ethanol precipitated, and dissolved in 3 ml of 1 mM EDTA/10 mM Tris⋅HCl, pH 8.0 (TE). The DNA was then digested with EcoRI (20 units/μl) overnight at 37°C in a water bath and density fractionated in CsCl gradients by centrifuging the DNA samples at 22°C for 72 h at 50,000 × g. In total, 32 fractions of 200 μl were collected. The fractions containing the parental DNA were collected, pooled, and dialyzed overnight at 4°C against TE. The dialyzed DNA was then N-buthanol concentrated and dissolved to obtain a final concentration of 1 μg/ml of parental DNA in TE.
Immunoextraction of DNA Fragments Containing BrdUrd-Labeled Repair Patches.
DNA fragments containing BrdUrd-labeled repair patches were immunoextracted using a mouse anti-BrdUrd antibody, a biotinylated goat anti-mouse antibody, and streptavidin-coated polymeric magnetic beads essentially as described (20, 22). The fractions containing the bound (i.e., immunoextracted) DNA fragments (repaired DNA) and the nonbound fragments (unrepaired DNA) were neutralized, and the concentration of human DNA in both fractions was measured for all cell lines by slot blot hybridization with radioactively labeled human DNA; the specificity of the immunoextraction was then tested by hybridization with strand-specific DNA probes of the human adenosine deaminase gene, as described (20).
Amplification and Labeling of the DNA Fractions.
Repaired (bound) and unrepaired (nonbound) DNA fractions were directly labeled with digoxigenin-11-dUTP by random primed reactions (DIG-High Prime kit, Boehringer Mannheim) or simultaneously amplified and labeled with biotin-16-dUTP by degenerate oligonucleotide primed (DOP)-PCR. In the random primed reactions, 300 ng of DNA were labeled overnight following the manufacturer's instructions. The DOP-PCR were carried out according to ref. 23 with some modifications. Briefly, each 25-μl reaction consisted of 50 ng of template DNA, 1.5 mM MgCl2, 0.2 mM dATP, dCTP, and dGTP, 0.1 mM dTTP, 0.1 mM biotin-16-dUTP, 1 μM DOP primer, and 1.25 units of Taq polymerase (GIBCO). The PCRs were heated at 93°C for 3 min, followed by nine cycles at 94°C for 1 min, 30°C for 1 min, and 72°C for 3 min each, 30 additional cycles at 94°C for 1 min, 56°C for 1 min and 72°C for 2 min each, and a final extension of 72°C for 10 min. Labeled DNA was used for in situ hybridization to normal human lymphocyte metaphase spreads to chromosomally localize the repaired and unrepaired DNA fractions.
Metaphase Preparations and in Situ Hybridization.
Peripheral blood was obtained from healthy donors, and human lymphocytes were cultured as described (24, 25). Some of the cultures received 0.1 μg/ml BrdUrd 6–7 h before harvesting to label the late-replicating chromosomal bands as described (26). Harvesting and slide preparations were performed by following standard cytogenetic procedures. FISH with digoxigenin-labeled DNA and antidigoxigenin detection with tetramethylrhodamine β-isothiocyanate-conjugated antibodies were performed as described in detail in ref. 27, and BrdUrd was simultaneously detected with mouse anti-BrdUrd and FITC-labeled goat anti-mouse antibodies as explained elsewhere (26). FISH with biotin-labeled DNA was performed as described above, and the immunodetection was done in green fluorescence with Alexa 488-conjugated streptavidin. BrdUrd detection in FISH experiments using DOP-PCR-labeled biotin probes was performed with a mouse anti-BrdUrd antibody, a digoxigenin-conjugated sheep anti-mouse antibody, and a rhodamine-conjugated antidigoxigenin sheep antibody as described in ref. 27.
Image Analysis and Editing.
Fluorescence images were captured either with a Zeiss Axioplan fluorescent microscope equipped with a cooled charge-coupled device camera (Photometrics, Tucson, AZ) and ip-lab software (Signal Analytics, Vienna, VA) or with a laser confocal microscope (Leica TCS 4D). Images were finally edited using the program PHOTODELUXE 1.0 (Adobe Systems, San Jose, CA).
Results and Discussion
Repair patches were labeled with BrdUrd for 24 h, and antibodies against BrdUrd-labeled DNA were used to immunoextract the fraction of the genome subjected to DNA repair by using magnetic beads (see Materials and Methods). The DNA bound to the beads was then used as a molecular probe to microscopically visualize the sites of repair at the chromosomal level by reverse FISH on normal chromosome spreads.
As expected, the concentration of DNA recovered from the magnetic beads as measured by slot blot hybridization with radioactive labeled human DNA was minimal in UV-irradiated XPA cells (data not shown), and it accounts for unspecific binding. This is consistent with the fact that XP25RO cells are completely deficient in NER and exhibit undetectable levels of unscheduled DNA synthesis after UV irradiation. Consistent with our previous data (20), the concentration of repaired fraction of XPC cells was intermediate between normal and XPA cells. The repaired fraction of XPC cells contained the transcribed strand only when assayed with strand-specific probes recognizing the 5′ part of the human adenosine deaminase gene (data not shown). This confirms the suitability and specificity of the methodology used to isolate the repaired fraction of the genome following UV irradiation and indicates that the contamination of the bound fraction with replicated DNA must be rather small and negligible.
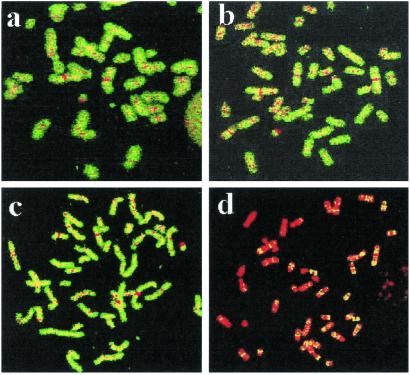
The repaired and unrepaired fractions isolated from normal human and XPC cells were labeled by random primed or DOP-PCR and used as a probe to assess their genomic distribution by FISH on normal metaphase spreads. Repair-labeled DNA from normal cells is evenly distributed along the human genome, without clearly discernible hotspots. In contrast, hybridization with repair-labeled DNA from XPC cells results in a highly localized pattern decorating specific chromosomal domains (Fig. 1). The observed pattern is not the result of an experimental artifact, as homogenous staining of all chromosomes was observed when unrepaired (nonbound) DNA from either normal cells (Fig. 1a) or XPC cells (Fig. 1c) were used as a probe. Normal human cells display rapid and efficient repair of 6–4 photoproducts and cyclobutane pyrimidine dimers throughout the genome; in cells exposed to 10 J/m2 UV-C, virtually all lesions are repaired during a 24-h repair period. At this level of resolution, NER occurs homogeneously throughout the genome over a period of 24 h, and poorly repaired regions are not manifested by reverse FISH despite the fact that UV-induced cyclobutane pyrimidine dimers are inefficiently repaired in some X chromosomal-located regions in human cells (7). At the level of resolution of this molecular cytogenetic approach, it is not possible to distinguish differences in transcription rates in different genes. Similarly, whole genome painting was obtained with the nonbound fraction isolated from UV-irradiated XPA cells, but, as predicted, we were unable to generate FISH signals when employing the bound DNA from XPA cells as a probe (data not shown).
Fig 1.
Chromosomal distribution of nucleotide excision repair. Chromosomal distribution of unrepaired (a and c) and repaired (b and d) fractions isolated from wild-type (a and b) and XPC (c and d) cells 24 h after UV irradiation. Both fractions were isolated as described in Materials and Methods; DOP-PCR was labeled with biotin and detected with Alexa 488-conjugated streptavidin as green fluorescence. Chromosomes are counterstained in red color with propidium iodine. Unrepaired and repaired fractions extracted from wild-type cells colored all chromosomes in a similar fashion (a and b). A similar pattern was obtained with the unrepaired fraction in XPC (c). The repaired fraction from XPC cells produced a specific banding pattern indicating clustering of TCR.
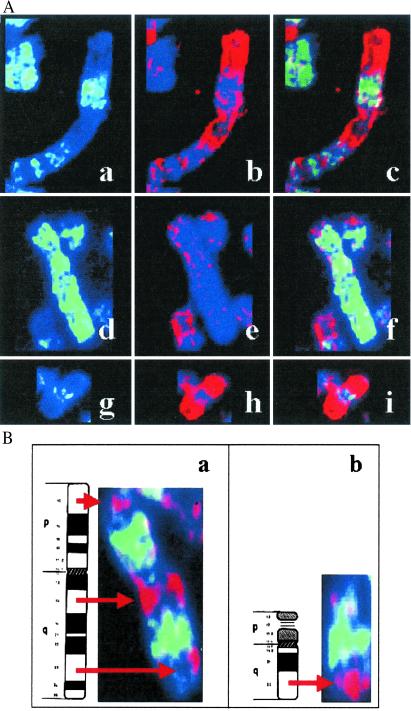
A detailed observation of the pattern obtained with the repaired fraction in XPC using random primed labeled probes uncovered a very heterogeneous picture: some chromosomes were completely painted by the TCR repair probe, whereas other chromosomes appeared banded or completely devoid of any labeling. Because the painting pattern observed was reproducible in repeated experiments and resembled an R-banding pattern, we applied a painting approach to combine hybridization with the XPC repair probe and immunodetection with antibodies against BrdUrd to stain the late-replicating DNA (26). This procedure allowed us to simultaneously visualize the sites of TCR (presumed to be the sites of transcription) along the chromosomes and the gene-poor late-replicating chromosomal bands (inverse of the R banding) (Fig. 2A). These coimmunodetection experiments clearly revealed clustering of sites of TCR at most of the early-replicating bands. Signal was strongest at subtelomeric regions of some chromosomes mostly representing a subset of R bands called T bands. T banding is obtained by R banding at elevated temperatures and identifies a subset of extremely GC-rich R bands, of which about half are at telomeric locations in the human karyotype (28). As shown in Fig. 2A, the XPC repair probe hybridizes poorly to the gene-poor chromosome 4 with the exception of its very terminal short arm. The very terminal part of chromosome 4p is known to be of high gene density, whereas the rest of chromosome 4 is gene-poor and overall late-replicating. A similar pattern was observed in chromosome 1, displaying strong hybridization of the XPC probe with gene-rich early-replicating bands. Also, the early replicating chromosome 19, known to have the highest gene density in the human genome, was found to be completely painted by the XPC probe.
Fig 2.
Clusters of sites of TCR at early-replicating chromosomal domains and T bands. (A) Clusters of sites of TCR at early-replicating chromosomal domains. Simultaneous detection of late-replicating DNA with FITC antibodies against BrdUrd incorporated at late S-phase (green) and of the repaired fraction from UV-irradiated XPC cells. The repaired DNA was visualized by FISH after random primed labeling of the probe and immunodetection with tetramethylrhodamine β-isothiocyanate-conjugated antibodies (red). Chromosome 1 (a–c), 4 (d–f), and 19 (g–i) are shown for late-replication banding in green (a, d, and g), repaired DNA in red (b, e, and h), and the merged image (c, f, and i). (B) Clustering of TCR at T bands. Clustering of TCR in red at T bands 11p15, 11q13, 11q23 (a), and 21q22 (b). As in a, green fluorescence indicates late-replicating bands, and the sites of TCR are shown in red fluorescence.
An example to further assign TCR at T bands is chromosome 11. On chromosome 11, 70% of mapped genes of known function are located at three T bands of this chromosome (p15, q13, and q23; refs. 14 and 29). This is also reflected in the localization of clusters of CpG islands in these chromosomal bands (18). The same is true for chromosome 21q22. Detailed analysis of chromosomes 11 and 21 revealed that TCR is specifically clustered in these T bands, as shown in Fig. 2B. This chromosomal distribution clearly resembles the known distribution of hyperacetylated histones in gene-rich chromosomal domains (16). Thus, a high correlation between CpG islands, hyperacetylated histones, and TCR becomes evident from this study. This observation is in line with the notion that UV-induced repair in XPC cells is tightly associated with the nuclear skeleton (30, 31) and that DNA fractions attached to the nuclear matrix preferentially hybridize to gene-rich R bands and sites of high levels of transcription (32).
Here, we show that TCR is organized in clusters encompassing predominantly early-replicating gene-rich bands within the human genome. The close coupling of locations of TCR with gene-rich bands at the chromosomal level is consistent with biochemical studies with a few number of genes indicating that TCR is confined to transcribed regions of the genome. Although there may be other activities resembling the stalled RNA pol II recruiting the repair complex to the site of damage, our data strongly support the notion that a large amount of repair in XPC is actually associated with transcription. Moreover, the chromosomal distribution of TCR target sequences observed in the current study resembles the uneven distribution of the transcriptome, enforcing the basic link between DNA repair, transcription, and nuclear organization in a complex genome. The repaired fraction isolated from UV-irradiated XPC cells can be used as an instrument to visualize, at a single cell level, large-scale movements of transcriptionally active chromatin and the behavior of the whole genome in response to DNA damage in cell lines with known genetic defects in the processing of DNA damage.
Acknowledgments
We thank Mercè Martí and the rest of the staff of the Servei de Microscopia Electrònica of the Universitat Autònoma de Barcelona for their excellent technical help. This work was partially funded by the Spanish Ministry of Science and Technology (Project No. CICYT, PM98-0179) and the Commission of the European Union (Project Nos. F1S5-1999-00071 and QLG1-CT-1999-00181). J.S. is supported by a “Ramón y Cajal” project entitled “Genome stability and DNA repair,” awarded by the Spanish Ministry of Science and Technology and cofinanced by the Universitat Autònoma de Barcelona.
Abbreviations
TCR, transcription-coupled repair
FISH, fluorescence in situ hybridization
NER, nucleotide excision repair
XP, xeroderma pigmentosum
DOP, degenerate oligonucleotide primed
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Donahue B. A., Yin, S., Taylor, J. S., Reines, D. & Hanawalt, P. C. (1994) Proc. Natl. Acad. Sci. USA 91, 8502-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Svejstrup J. Q. (2002) Nat. Rev. Mol. Cell Biol. 3, 21-29. [DOI] [PubMed] [Google Scholar]
- 3.Wood R. D. (2001) Science 291, 1284-1289. [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers J. H. J. (2001) Nature (London) 411, 366-374. [DOI] [PubMed] [Google Scholar]
- 5.Mellon I., Spivak, G. & Hanawalt, P. C. (1987) Cell 51, 241-249. [DOI] [PubMed] [Google Scholar]
- 6.Leadon S. A. & Lawrence, D. A. (1991) Mutat. Res. 255, 67-78. [DOI] [PubMed] [Google Scholar]
- 7.Venema J., Bartosova, Z., Natarajan, A. T., Vanz Eeland, A. A. & Mullenders, L. H. F. (1992) J. Biol. Chem. 267, 8852-8856. [PubMed] [Google Scholar]
- 8.Tolbert D. M. & Kantor, G. J. (1996) Cancer Res. 56, 3324-3330. [PubMed] [Google Scholar]
- 9.Nouspikel T. & Hanawalt, P. C. (2000) Mol. Cell. Biol. 20, 1562-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beecham E. J., Mushinski, J. F., Shacter, E., Potter, M. & Bohr, V. A. (1991) Mol. Cell. Biol. 11, 3095-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Z., Hu, W., Komissarova, E., Pao, A., Hung, M.-C., Adair, G. M. & Tang, M.-S. (2002) J. Biol. Chem. 277, 12777-12783. [DOI] [PubMed] [Google Scholar]
- 12.Conconi A., Bespalov, V. A. & Smerdon, M. J. (2002) Proc. Natl. Acad. Sci. USA 99, 649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caron H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.-C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 14.Schuler G. D., Boguski, M. S. & Stewart, G. A. (1996) Science 274, 540-546. [PubMed] [Google Scholar]
- 15.Deloukas P., Schuler, G. D., Gyapay, G., Beasley, E. M., Soderlund, C., Rodriguez-Tome, P., Hui, L., Matise, T. C., McKusick, K. B., Beckmann, J. S., et al. (1998) Science 282, 744-746. [DOI] [PubMed] [Google Scholar]
- 16.Jeppesen P. (1997) BioEssays 19, 67-74. [DOI] [PubMed] [Google Scholar]
- 17.Surrallés J., Jeppesen, P., Morrison, H. & Natarajan, A. T. (1996) Am. J. Hum. Genet. 59, 1091-1096. [PMC free article] [PubMed] [Google Scholar]
- 18.Craig J. M. & Bickmore, W. A. (1994) Nat. Genet. 7, 376-382. [DOI] [PubMed] [Google Scholar]
- 19.Venema J., Van Hoffen, A., Karcagi, V., Natarajan, A. T., van Zeeland, A. A. & Mullenders, L. H. F. (1991) Mol. Cell. Biol. 11, 4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoffen A., Kalle, W. H., de Jong-Versteeg, A., Lehmann, A. R., van Zeeland, A. A. & Mullenders, L. H. F. (1999) Nucleic Acids Res. 27, 2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volker M., Mone, M. J., Karmakar, P., van Hoffen, A., Schul, W., Vermeulen, W., Hoeijmakers, J. H., van Driel, R., van Zeeland, A. A. & Mullenders, L. H. F. (2001) Mol. Cell 8, 213-224. [DOI] [PubMed] [Google Scholar]
- 22.Kalle W. H., Hazekamp-van Dokkum, A. M., Lohman, P. H., Natarajan, A. T., van Zeeland, A. A. & Mullenders, L. H. F. (1993) Anal. Biochem. 208, 228-236. [DOI] [PubMed] [Google Scholar]
- 23.Verma R. S. & Babu, A., (1995) Human Chromosomes: Principles and Techniques (McGraw–Hill, New York).
- 24.Surrallés J., Hande, P. H., Marcos, R. & Lansdorp, P. (1999) Am. J. Hum. Genet. 65, 1616-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callén E., Samper, E., Ramírez, M. J., Creus, A., Marcos, R., Ortega, J. J., Olivé, T., Badell, I., Blasco, M. A. & Surrallés, J. (2002) Hum. Mol. Genet. 11, 439-444. [DOI] [PubMed] [Google Scholar]
- 26.Surrallés J. & Natarajan, AT. (1998) Mutat. Res. 414, 117-124. [DOI] [PubMed] [Google Scholar]
- 27.Surrallés J., Sebastian, S. & Natarajan, A. T. (1997) Mutagenesis 12, 437-442. [DOI] [PubMed] [Google Scholar]
- 28.Holmquist G. P. (1992) Am. J. Hum. Genet. 51, 17-37. [PMC free article] [PubMed] [Google Scholar]
- 29.van Heyningen V. & Little, P. F. R. (1994) Cytogenet. Cell Genet. 69, 128-158. [DOI] [PubMed] [Google Scholar]
- 30.Mullenders L. H. F., van Kesteren van Leeuwen, A. C., van Zeeland, A. A. & Natarajan, A. T. (1988) Nucleic Acids Res. 16, 10607-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karmakar P. & Natarajan, A. T. (2000) Mutagenesis 15, 115-120. [DOI] [PubMed] [Google Scholar]
- 32.Craig J. M., Boyle, S., Perry, P. & Bickmore, W. A. (1997) J. Cell Sci. 110, 2673-2682. [DOI] [PubMed] [Google Scholar]