Abstract
Formation of oligosaccharides occurs both in the cytosol and in the lumen of the endoplasmic reticulum (ER). Luminal oligosaccharides are transported into the cytosol to ensure that they do not interfere with proper functioning of the glycan-dependent quality control machinery in the lumen of the ER for newly synthesized glycoproteins. Once in the cytosol, free oligosaccharides are catabolized, possibly to maximize the reutilization of the component sugars. An endo-β-N-acetylglucosaminidase (ENGase) is a key enzyme involved in the processing of free oligosaccharides in the cytosol. This enzyme activity has been widely described in animal cells, but the gene encoding this enzyme activity has not been reported. Here, we report the identification of the gene encoding human cytosolic ENGase. After 11 steps, the enzyme was purified 150,000-fold to homogeneity from hen oviduct, and several internal amino acid sequences were analyzed. Based on the internal sequence and examination of expressed sequence tag (EST) databases, we identified the human orthologue of the purified protein. The human protein consists of 743 aa and has no apparent signal sequence, supporting the idea that this enzyme is localized in the cytosol. By expressing the cDNA of the putative human ENGase in COS-7 cells, the enzyme activity in the soluble fraction was enhanced 100-fold over the basal level, confirming that the human gene identified indeed encodes for ENGase. Careful gene database surveys revealed the occurrence of ENGase homologues in Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana, indicating the broad occurrence of ENGase in higher eukaryotes. This gene was expressed in a variety of human tissues, suggesting that this enzyme is involved in basic biological processes in eukaryotic cells.
The N-glycosylation of proteins has been recognized as one of the most common co- and posttranslational modification reactions of proteins in eukaryotes (1). Accumulating evidence indicates that the N-glycosylation process plays critical roles in protein folding, intracellular transport, and degradation of proteins (2–4).
The processing events of N-glycan chains in the endoplasmic reticulum (ER) as well as in Golgi apparatus have been extensively studied (5, 6). In recent years, it has been found that, during the N-glycosylation of proteins in the ER, a significant number of unconjugated, free oligosaccharides (free OSs) are formed (7–9). The occurrence of free OSs has been reported in a wide variety of cells (10–14), and their sequential processing event of free OSs in mammalian cells has also been well characterized (15–19). When the free OSs are formed in the lumen of the ER, they are transported out of the ER to the cytosol (15). This export process is likely to be of importance for cells because of the potential competition between glycoproteins and free OSs in the glycan-dependent folding/degradation process that operates in the ER (2–4).
Endo-β-N-acetylglucosaminidase (ENGase, EC 3.2.1.96) is one of the key enzymes in the processing event of free OSs in the cytosol (8). This cytosolic enzyme activity has been described in a wide variety of animal cells (20–24). The combined actions of ENGase and a cytosolic α-mannosidase (18, 19) were found to be essential for free OSs to be transferred into lysosome, where they are further degraded (25). To determine the biological significance of this enzyme, as well as the cytosolic catabolic pathway for free OS, detailed information on the ENGase is imperative. The gene encoding the cytosolic ENGases, however, had not been identified in any eukaryotic system. Here, we report the complete purification of ENGase from hen oviduct, the organ from which purification and characterization of this enzyme were reported earlier (24, 26). Based on partial amino acid sequences of the purified protein and expressed sequence tag (EST) database analysis, the cDNA for a human homologue of the cytosolic ENGases has been identified. By expressing the candidate gene in a mammalian cell line, the expression of ENGase activity in the cell extract has been confirmed. Further database analysis revealed the wide occurrence of this enzyme activity in eukaryotes, which is consistent with the presumption that this enzyme plays an important role in basic catabolic processes in eukaryotes.
Materials and Methods
Assay for ENGase.
ENGase was assayed by using paper chromatography as described earlier (27) except that the solvent composition was modified to 1-butanol/ethanol/water = 2/1/1. The substrate used in this study was the glycoasparagine Asn(Man4GlcNAc4) (GP-IVB; ref. 28) and was 14C-labeded as described (27, 29). The specific radioactivity of 14C-labeled GP-IVB was 1.2 × 105 cpm/nmol. The assay reaction (10 μl) included 120 mM sodium citrate buffer (pH 5.5), 3 μl of enzyme fraction, and 1 μl of 14C-labeled GP-IVB (4,500 cpm; 3.75 μM). One unit was defined as the amount of enzyme that catalyzes the hydrolysis of 1 μmol of 14C-labeled GP-IVB at room temperature per min. For control reaction of ENGase, endo H (Glyko, Novato, CA) was used. Reaction was carried out by using 5 units of enzyme and buffer, provided by the manufacturer, at 37°C for 3 h.
Purification of ENGase from Hen Oviduct.
Step 1: toyopearl butyl-650 M chromatography.
All of the purification steps were carried out at 4°C or on ice. Hen oviduct was provided by Saitama Experimental Animals Supply (Kitakatsushika, Saitama, Japan) and stored at −80°C until use. A total of 4 kg (from 75 hens) of oviduct was used for this purification. For one extraction, an average 200 g of sample was used. Oviduct was homogenized in a Waring blender in 2 vol of 50 mM Tris⋅HCl (pH 7.5) with 2 mM DTT including various protease inhibitors (1.0 μg/ml leupeptin (Peptide Institute, Osaka), 1.0 μg/ml aprotinin (Sigma), 100 μM PMSF (Wako Pure Chemical, Osaka), and 0.5 mg/ml soybean trypsin inhibitor (Sigma)). After clearing extract by centrifugation (9,900 × g, 12 min), solid NaCl was added to the sample to make the final concentration to 2.0 M. After removing any precipitate formed, the supernatant was divided into eight aliquots, and each (≈1.2 liters) was applied to a Toyopearl Butyl-650M column (Tosoh, Tokyo; 5.5 × 10 cm). The column was washed with 300 ml of 0.4 M ammonium sulfate/50 mM Tris⋅HCl containing 2 mM DTT. ENGase was recovered from the flow-through/wash fractions.
Step 2: toyopearl phenyl-650M chromatography.
The combined flow-through fraction (≈1.5 liters for each column operation) was dialyzed against 10 mM Tris⋅HCl (pH 7.5) for 2 days, and the precipitate was removed by centrifugation (9,900 × g, 30 min). One-fifth volume of saturated ammonium sulfate was then added to the supernatant obtained. After clearing the sample by centrifugation, the supernatant was applied to a Toyopearl Phenyl-650M column (Tosoh, 6.8 × 3 cm). After washing the column with 0.8 M ammonium sulfate/50 mM Tris⋅HCl (pH 7.5), ENGase-active fractions were eluted with 200 ml of 0.3 M ammonium sulfate/50 mM Tris⋅HCl (pH 7.5).
Step 3: toyopearl DEAE-650M chromatography.
The combined ENGase-active fraction was dialyzed against 50 mM Tris⋅HCl (pH 7.5) for 2 days, and cleared by centrifugation. A one-eighth fraction was applied to a Toyopearl DEAE-650 M column (Tosoh, 2.2 × 5.5 cm). The column was washed with 60 ml of 50 mM Tris⋅HCl (pH 7.5), and ENGase-active fraction was eluted with 60 ml of 90 mM NaCl/50 mM Tris⋅HCl (pH 7.5).
Step 4: Sephacryl S-200 chromatography (pH 6.5).
To the combined fraction, an equal amount of saturated ammonium sulfate (pH 7.1) was added, and the mixture was left on ice for 30 min. ENGase-active fraction was collected by centrifugation (17,000 × g, 90 min) at 4°C. The pellet was dissolved with 40 ml of 50 mM Tris⋅HCl (pH 7.5), and an aliquot 2.5 ml was applied to a Sephacryl S-200 column (Amersham Pharmacia Biosciences, 2.0 × 115 cm) equilibrated with 50 mM Mes-NaOH (pH 6.5). ENGase was eluted at fractions 63–71 (3.5 ml per fraction).
Step 5: POROS HS chromatography.
The combined ENGase-active fraction (total 320 ml) was applied to a POROS HS column (Waters, 1.6 × 2.5 cm), and elution was carried out by linear gradient of 0–0.6 M NaCl in 10 mM Mes-NaOH buffer (pH 6.5). ENGase-active fractions were eluted with 260–360 mM NaCl.
Step 6: Sephacryl S-200 chromatography (pH 7.5).
ENGase-active fractions were concentrated to 1 ml by using YM-10 (Amicon), and then applied to a Sephacryl S-200 column (1.2 × 120 cm) equilibrated with 50 mM Tris⋅HCl (pH 7.5). fractions 59–65 (1.2 ml per fraction) were ENGase-active.
Step 7: TALON immobilized metal affinity chromatography.
The ENGase-active fraction was applied to a TALON metal affinity-column (CLONTECH, 0.4 × 0.8 cm). After washing the column with 50 mM Tris⋅HCl (pH 7.5), the ENGase-active fraction was eluted with 1 ml of 10 mM imidazole/50 mM Tris⋅HCl (pH 7.5) fraction.
Step 8: heparin-Sepharose chromatography.
The ENGase-active fraction was diluted with 5 vol of 10 mM sodium phosphate buffer (pH 7.0), and applied to a heparin-Sepharose column (HiTrap; Amersham Pharmacia; 0.7 × 2.5 cm) and eluted with gradient of NaCl (0–0.6 M) in 10 mM sodium phosphate buffer (pH 7.0). Major ENGase activity was detected in the flow-through fraction (fraction I) but some activity was also eluted in the fractions of 0.15–0.25 M NaCl (fraction II). Fraction II was concentrated by precipitation with 10% trichloroacetic acid (TCA), and used for amino acid analysis. Fraction I was divided into two parts. One (fraction Ia) was concentrated by TCA precipitation for amino acid sequence analysis, and the other was used for further purification.
Step 9: POROS HS rechromatography.
The rest of fraction I was applied to a POROS HS column (0.7 × 2.5 cm). The column was washed with 10 ml of 10 mM Mes-NaOH buffer (pH 6.5). The column was eluted with a linear gradient of NaCl (0–0.5 M) in 10 mM Mes-NaOH buffer (pH 6.5). The ENGase-active fraction eluted with 250–350 mM NaCl was collected.
Step 10: hydroxyapatite chromatography.
The obtained fraction was diluted with 5 vol of 50 mM Tris⋅HCl buffer (pH 7.5), and applied to a Gigapite hydroxyapatite column (Seikagaku Kogyo, Tokyo; 0.4 × 0.8 cm). After washing the column with 4 ml of 50 mM Tris⋅HCl (pH 7.5), the ENGase-active fraction was eluted with 4 ml of 50 mM sodium phosphate buffer (pH 7.0).
Step 11: Q-Sepharose chromatography.
The fraction obtained was diluted with 5 vol of 50 mM Tris⋅HCl buffer (pH 7.5), and applied on a Q-Sepharose column (HiTrap; 0.7 × 2.5 cm). ENGase-active fraction was eluted with 1 ml of 50 mM Tris⋅HCl containing 0.2 M NaCl. This fraction (fraction Ib) was further concentrated by 10% TCA precipitation for amino acid sequence analysis.
Chemical Analyses.
Protein concentration was determined by the bicinchoninic acid (BCA) method (Pierce) according to the manufacturer's instructions with BSA as a standard. N-terminal amino acid sequence analysis was carried out after 11% SDS/PAGE followed by blotting on a Problott membrane (Applied Biosystems). Proteins were visualized by staining with 0.1% Coomassie brilliant blue. The 55 kDA protein band was excised and analyzed with a PPSQ-10 protein sequenator (Shimadzu).
Internal amino acid sequences of ENGase were determined as follows: the samples were subjected to SDS/PAGE, and proteins were visualized by staining with 0.1% Coomassie brilliant blue. The 55-kDa protein was excised and digested with either trypsin (sequencing grade, Roche Diagnostics) or lysyl endopeptidase (Wako Pure Chemical, Osaka). Trypsin (0.2 μg) was added to one gel piece and incubated in 100 mM Tris⋅HCl (pH 8.5), 1 mM CaCl2 at 37°C for 16 h. Another gel piece was digested with 0.3 μg lysyl endopeptidase in 100 mM Tris⋅HCl (pH 9.0) at 37°C for 16 h. The resulting peptides were isolated by reversed phase chromatography on a μRPC C2/C18 column (2.1 × 100 mm, Amersham Pharmacia Biosciences). Automated Edman degradation was performed with either Procise 492cLC protein sequenator (Applied Biosystems) or PPSQ-10 as mentioned above.
Sequence Database Analyses.
Using the five internal amino acid sequences obtained from hen oviduct ENGase, the National Center for Biotechnology Information (NCBI) sequence database was surveyed to identify hen ENGase homologues. After obtaining the sequence of human ENGase from Image clone No. 4654169 (gene accession number BG491299), more specific analysis was carried out for Saccharomyces cerevisiae (SGD, http://genome-www.stanford.edu/Saccharomyces/), Caenorhabditis elegans (http://www.sanger.ac.uk/Projects/C elegans/), Drosophila melanogaster (http://www.fruitfly.org/), and Arabidopsis thaliana (http://www.arabidopsis.org), to examine the occurrence of ENGase homologues in these organisms.
Protein Expression of Human ENGase Homologue in COS-7 Cells.
Standard molecular biological techniques were used (30). An EST clone encoding human ENGases (Image clone no. 4654169; gene accession no. BG491299) was obtained from Incyte Pharmaceuticals (Palo Alto, CA), and the sequence of the clone was confirmed by direct DNA sequencing. The ENGase ORF was then amplified by using the following primers: 5′-AAAAATCTAGAGCTAGCGCCACCATGGAGGCCGCGGCGGTGACGGTCAC-3′ and 5′-AAAAAGAATTCTCATGCAGGGGCTGAATAAAGCAT-3′. The amplified ≈2.2-kb fragment was digested with XbaI/EcoRI and was cloned into NheI/EcoRI sites of pcDNA3.1+ vector (pcDNA3.1-hENGase). The expression of human ENGase protein in COS-7 was carried out by transfection of pcDNA3.1-hENGase into COS-7 cells in six-well plates by using FUGENE 6 (Roche) according to the manufacturer's procedure. A protein extract was prepared 48 h after transfection, by suspending cells in 50 μl of 20 mM Hepes-KOH buffer (pH 7.0), containing 5 mM MgCl2, 150 mM potassium acetate, 250 mM sorbitol, and 10 mM DTT. After being frozen in liquid N2, samples were thawed on ice, and solutions were cleared by centrifugation at 20,000 × g for 10 min at 4°C. Enzyme activity in soluble fraction was determined as described above except that 10 μl of assay reaction was carried out in 120 mM Mes-NaOH buffer (pH 6.0). For protein determination, protein was determined by using a protein assay kit (Bio-Rad) with BSA as a standard.
Northern Blot Analysis.
Tissue distribution of ENGase homologues in human was examined by Northern blot analysis by using FirstChoice Northern Blots (Ambion, Austin, TX). A ≈0.9-kb fragment obtained after SspI/XhoI digestion of hENGase cDNA was used for a probe of Northern analysis. Hybridization was carried out according to the manufacturer's procedure and visualized by a PhosphorImager (Molecular Dynamics).
Results
Purification of ENGase from Hen Oviduct.
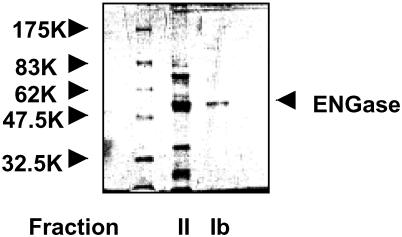
The ENGase was purified from hen oviduct through 11 steps as described in Materials and Methods. Fraction Ib exhibited a single band at ≈55 kDa by SDS/PAGE (Fig. 1). The specific activity of this fraction was estimated to be 3,000 milliunits/mg protein, which represented a 150,000-fold over the crude extract. The 55-kDa band was found as a dark band in all three ENGase-active fractions obtained during final purification steps, fractions Ia, Ib, and II. Moreover, the 55-kDa band comigrated with the ENGase activity by various column fractionations (data not shown), suggesting that this protein band represented the ENGase protein. Hence, this band from each of these fractions was subjected to N-terminal amino acid analysis. From the results shown below, we can safely conclude that all three fractions contained essentially the same enzyme protein.
Figure 1.
Analysis of purified ENGase fractions from hen oviduct by SDS/PAGE. The purified ENGase samples (fractions II and Ib, 10 μl each after precipitation with 10% TCA) were analyzed by 10% SDS/PAGE gel. Protein bands were visualized by Coomassie brilliant blue staining. The protein band for ENGase (≈55 kDa) from each fraction was subjected to amino acid sequence analysis.
Fraction Ia AEPLGTXVLXAAV
Fraction Ib AEPLG
Fraction II AEPLGTTVLHAAVDTRPQPAXYFDTG
(X is an unidentified residue.)
Identification of Human Homologue of Hen ENGase Through Sequence Database Analysis.
Having confirmed that fraction Ia contained the same 55-kDa protein, this sample was used for internal protein sequence analysis. After in-gel enzymatic digestion of the 55-kDa band, the resulting peptides were fractionated with a reversed phase column, and the sequence of purified peptides was determined. The five sequences obtained are shown in Table 1. By using these sequences, the NCBI sequence database was surveyed to identify proteins containing homologous sequences. It was found that a human hypothetical protein (FLJ21865; 377 aa according to protein accession no. NP_073596) showed sequence similarity to 4 of 5 (fractions 1, 2, 4, and 5) internal sequences obtained from the isolated hen oviduct protein (Table 1), strongly suggesting that this hypothetical protein was a human orthologue of the purified protein. Moreover, an EST sequence (AJ397822) from hen containing 2-aa fragments (fractions 1 and 2) was also identified. When the deduced amino acid sequence from the hen EST AJ397822 was compared with that of the C-terminal region of human FLJ21865 (amino acid 310–363), a high degree of homology was observed [identities = 39 (72%); similarities = 4 (7%)]. After sequencing Image clone no. 4654169, it was found that this protein actually consisted of 743 aa, extended toward the C terminus by ≈370 residues compared with the predicted sequence of hypothetical protein FLJ21865 (see below).
Table 1.
Amino acid sequence analysis of internal fragments of purified hen ENGase and a comparison with deduced human hypothetical protein FLJ21865
| Fragments (protease used) | Sequence obtained | Deduced sequence (FLJ21865) |
|---|---|---|
| Fraction 1 (Lys-C) | SLSLIRK | SLELIRK (347–353)* |
| Fraction 2 (Lys-C) | HGLSAAIFA | HGFSVALFA (354–362) |
| Fraction 3 (trypsin) | AVSHQLAR | Not found |
| Fraction 4 (trypsin) | GGYLEDR | GGYLDDR (133–139) |
| Fraction 5 (trypsin) | WQNELNEQNR | WQDELNQHNR (281–290) |
Numbers in parentheses represent corresponding amino acid residues on complete ENGase sequence.
The Human Orthologue of the Candidate Protein Displays Enzyme Activity When Expressed in COS-7 Cells.
To confirm that this protein is indeed ENGase, the protein was expressed in COS-7 cells, and the extract was assayed for ENGase activity. The ORF of the candidate protein from human was cloned into pcDNA3.1+ vector for expression of the protein COS-7. As shown in Fig. 2, strong ENGase activity in COS-7 was observed in 60 min incubation only after transfection with the cDNA for human candidate protein (Table 2). This result convincingly shows that the human sequence homologue identified through sequence analysis is indeed ENGase.
Figure 2.
Identification of ENGase activity in COS-7 cells transfected with human ENGase cDNA. Analysis of the reaction products produced by incubation of 14C-labeled substrate (14C-GP IVB) with the extract of COS-7 cells transfected with or without prior transfection with the human ENGase (hENGase) was carried out by paper chromatography. Lane 1, COS-7 cell extract with vector alone. Lane 2, COS-7 cells with hENGase-expression vector. Lane 3, Authentic reaction product produced by incubation of the substrate with Endo H.
Table 2.
ENGase activity in COS-7 cells transfected with human ENGase cDNA
| ENGase activity (μunits/mg protein) | |
|---|---|
| pcDNA3.1 (vector alone) | 21 |
| pcDNA3.1-hENGase | 2.3 × 103 |
Human ENGase Is Distributed Widely in Human Tissues.
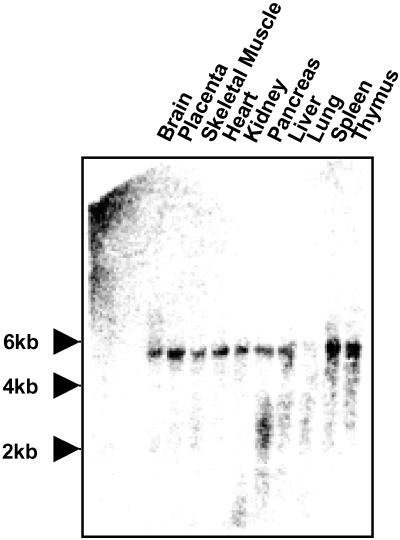
To study the tissue distribution of ENGase in human, Northern analysis was carried out. As shown in Fig. 3, the ENGase gene exhibited a broad tissue distribution, with highest expression in thymus and spleen.
Figure 3.
Distribution in various tissues of human ENGase mRNA. FirstChoice Northern Blots (Ambion) was used for Northern blot analysis, and positions for RNA sizing marker were indicated.
Detailed Sequence Analysis Revealed Wide Occurrence of ENGase Orthologues in Eukaryotes.
Using the human ENGase sequence as an inquiry, further detailed analysis was carried out to determine the distribution of this enzyme in nature. First, the sequence analysis was carried out using the human ENGase sequence as an inquiry in the NCBI database. In EST databases, apparent orthologues (E value < 5e-07) for the human ENGase protein were found in mammals (Sus scrofa, Bos taurus, Mus musculus, and Rattus norvegicus), birds (Gallus gallus), fish (Danio rerio), insects (D. melanogaster and Anopheles gambiae), nematode (C. elegans), and various plants (Pinus taeda, Solanum tuberosum, Hordeum vulgare, Gossypium hirsutum, Glycine max, Triticum aestivum, and Arabidopsis thaliana). Moreover, an apparent orthologue was detected in the Trypanosoma brucei genomic sequence (data not shown). To more rigorously examine the sequences of ENGase orthologues in eukaryotes, the databases for eukaryotes for which the genome sequence has been completed (S. cerevisiae, C. elegans, D. melanogaster, and A. thaliana) were more closely examined. Apparent orthologues were found in all but in S. cerevisiae. Alignment of the predicted amino acid sequences from these organisms is shown in Fig. 4. They share sequence homology in a long stretch of amino acids (amino acids 122–434 in human ENGases) although they have relatively diverse N- and C-terminal sequences. Taken together, these results suggested the wide distribution of this enzyme in eukaryotes. Interestingly, when a nonredundant sequence database was surveyed, previously characterized bacterial/microbial ENGases, such as Endo-M from Mucor hiemalis (31, 32) or Endo-A from Arthrobacter protophormiae (33, 34), were also found to have less, but significant, homology with human ENGase (data not shown).
Figure 4.
Comparison of sequences of human ENGase and its homologues in other eukaryotes. hENGase, human ENGase (GenBank accession no. AF512564); DmENGase, D. melanogaster ENGase (AE003505; protein no. CG5613 in http://www.fruitfly.org); CeENGase, C. elegans ENGase (AB079783); AtENGase1/2, two ENGase orthologues found in A. thaliana genome (AB010692 and AC009991, protein nos. At5g05460 and At3g11040, respectively, in http://www.arabidopsis.org).
Discussion
The objective of this study was the identification of the gene encoding a cytosolic ENGase in animal cells. To this purpose, the complete purification of the enzyme was achieved from hen oviduct, where the purification of the enzyme was reported earlier (26). Based on the amino acid sequence of the purified enzyme, the human orthologue of ENGase was identified through DNA sequence database surveys. Expressing the protein in mammalian cells and detection of the activity in the extract confirmed that the human candidate protein is indeed an ENGase. Although the cytosolic ENGase had previously been detected and biochemically characterized (20–24), the gene encoding this cytosolic enzyme has not been reported.
There are two types of cytosolic deglycosylating enzymes that can generate free OSs from glycoproteins (7, 8). One is ENGase described in this study, and the other is PNGase (peptide: N-glycanase), which releases a free glycan from glycoprotein/glycopeptide by cleaving the amide bond between the proximal GlcNAc residue and the side chain of an asparagine residue (35). Because the gene encoding PNGase (PNG1) has been identified (36), rapid progress regarding functional studies of cytoplasmic PNGase has been made (35). PNGase-mediated deglycosylation of glycoproteins has been observed in various organisms (35). On the other hand, whether the cytosolic ENGase can work on glycoproteins in nature remains to be determined. Although the cytosolic ENGase activity was believed to act on free OSs in vivo, further studies have to be performed to determine whether it can also act in vivo on substrates other than free OSs, i.e., glycopeptides/glycoproteins or dolicholpyrophosphoryl oligosaccharides. In this connection, it should be noted that the cytosolic ENGase was reported to act on not only glycopeptides but also dolicholpyrophosphoryl oligosaccharides in vitro (37).
Although the sequential processing events of free OSs have been well characterized (15–19), the molecular nature of the enzyme/transporters involved in this process has not been identified, with the exception of cytosolic PNGase (35). Because free OSs in the lumen of the ER can potentially interfere with the glycan-dependent protein quality control system in the ER (2–4), the rapid clearance of them from the lumen of the ER may be of great importance. Once in the cytosol, action of ENGase on free OSs with a di-N-acetylchitobiose at their reducing termini [GN2 species (15)] to give rise to those bearing a single GlcNAc at their reducing termini (GN1 species) was shown to be required for the subsequent further degradation by a cytosolic α-mannosidase (18, 19). Both ENGase and the α-mannosidase are required to form a free OS with the structure Man5GlcNAc, which is then transported into the lysosomes by an unknown mechanism (25). These observations therefore raised the possibility that a defect of ENGase activity should lead to the accumulation of free OSs in the cytosol because of the lack of their processing. Now that the gene encoding ENGase has been identified, rapid clarification of the functional importance of this enzyme, as well as a further understanding of the processing of free OSs in the cytosol, is likely.
With respect to the distribution of the cytosolic ENGase orthologue in nature, it was found that this protein occurs in a wide variety of multicellular organisms, but not in S. cerevisiae. The absence of an orthologue in S. cerevisiae is consistent with the failure to detect the enzyme activity biochemically in this yeast extract (38). It will be interesting to learn about the difference in the sequence of events in cytosolic oligosaccharide processing between yeast and mammalian cells. Studies aimed at clarifying this issue are needed.
It should also be noted that the plant A. thaliana has two distinct ENGase orthologues. Both of these proteins lack an apparent signal sequence, and are predicted to be located in the cytosol (based on psort analysis; http://psort.nibb.ac.jp/). Whether these predicted proteins have overlapping functions or play distinct roles in plants should be experimentally evaluated.
One interesting structural feature of the cytosolic ENGase is that this protein shares reduced, but significant, homology with fungal/bacterial ENGases such as Endo-M (protein accession no. AB060586; refs. 31 and 32) or Endo-A (U59168; ref. 34). These fungal/bacterial ENGases are secreted enzymes (31, 33) and are apparently different from the cytosolic ENGases in terms of physiological functions. These ENGases (Endo-M, Endo-A, and cytosolic ENGases, which we propose should be called class II ENGases) are structurally distant from class I ENGase (Endo H and its structurally related enzymes), for which extensive structural studies have been carried out (39–41). To our knowledge, however, structural studies on the class II ENGases have not been carried out in detail. Because the cytosolic ENGases were found to have a certain similarity in primary sequence with bacterial/fungal secreted ENGases, rapid progress on clarification of the catalytic mechanism of the class II ENGases is likely.
Acknowledgments
We thank Dr. Guangtao Li, Ms. Luxuan Guo (State Univ. of New York, Stony Brook), Ms. Keiko Hiraishi (Kyowa Hakko Kogyo Co.), and Mr. Yong-Chen Lu (Academia Sinica) for experimental help on this project. We also thank members of the Inoue lab/Emori lab (University of Tokyo) and the Lennarz lab (State Univ. of New York, Stony Brook) for useful discussion. This study was supported by a research grant for young researchers from the Japan Society for the Promotion of Science (to T.S.). The research was also supported in part by National Institutes of Health Grant GM33184 (to W.J.L.), a Grant-in-Aid for Scientific Research on Priority Area (No. 14011205) from the Ministry of Education, Science, Sports and Culture, Japan (to Y.E.), and a research grant from Mizutani Foundation for Glycosciences (to T.S.).
Abbreviations
- ER
endoplasmic reticulum
- OS
oligosaccharide
- ENGase
endo-β-N-acetylglucosaminidase
- EST
expressed sequence tag
- TCA
trichloroacetic acid
References
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frigerio L, Lord J M. Curr Biol. 2000;10:R674–R677. doi: 10.1016/s0960-9822(00)00680-1. [DOI] [PubMed] [Google Scholar]
- 3.Parodi A J. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 5.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 6.Herscovics A, Orlean P. FASEB J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 7.Cacan R, Verbert A. Trends Glycosci Glycotechnol. 1997;9:365–377. [Google Scholar]
- 8.Suzuki T, Yan Q, Lennarz W J. J Biol Chem. 1998;273:10083–10086. doi: 10.1074/jbc.273.17.10083. [DOI] [PubMed] [Google Scholar]
- 9.Moore S E H. Trends Cell Biol. 1999;9:441–446. doi: 10.1016/s0962-8924(99)01648-7. [DOI] [PubMed] [Google Scholar]
- 10.Oliver G J A, Harrison J, Hemming F W. Eur J Biochem. 1975;58:223–229. doi: 10.1111/j.1432-1033.1975.tb02367.x. [DOI] [PubMed] [Google Scholar]
- 11.Cacan R, Hoflack B, Verbert A. Eur J Biochem. 1980;106:473–479. doi: 10.1111/j.1432-1033.1980.tb04594.x. [DOI] [PubMed] [Google Scholar]
- 12.Hanover J A, Lennarz W J. J Biol Chem. 1982;257:2787–2794. [PubMed] [Google Scholar]
- 13.Anumula K R, Spiro R G. J Biol Chem. 1983;258:15274–15282. [PubMed] [Google Scholar]
- 14.Belárd M, Cacan R, Verbert A. Biochem J. 1988;255:235–242. [PMC free article] [PubMed] [Google Scholar]
- 15.Moore S E H, Spiro R G. J Biol Chem. 1994;269:12715–12721. [PubMed] [Google Scholar]
- 16.Moore S E H, Bauvy C, Codogno P. EMBO J. 1995;14:6034–6042. doi: 10.1002/j.1460-2075.1995.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmiécik D, Herman V, Stroop C J M, Michalski J-C, Mir A M, Labiau O, Verbert A, Cacan R. Glycobiology. 1995;5:483–494. doi: 10.1093/glycob/5.5.483. [DOI] [PubMed] [Google Scholar]
- 18.Kumano M, Omichi K, Hase S. J Biochem. 1996;119:991–997. doi: 10.1093/oxfordjournals.jbchem.a021340. [DOI] [PubMed] [Google Scholar]
- 19.Grand T, Saint-Pol A, Hauew J F, Alonso C, Wieruszeski J-M, Strecker G, Michalski J-C. Eur J Biochem. 1994;223:99–106. doi: 10.1111/j.1432-1033.1994.tb18970.x. [DOI] [PubMed] [Google Scholar]
- 20.Nishigaki M, Muramatsu T, Kobata A. Biochem Biophys Res Commun. 1977;59:638–645. doi: 10.1016/s0006-291x(74)80027-6. [DOI] [PubMed] [Google Scholar]
- 21.Pierce R J, Spik G, Montreuil J. Biochem J. 1980;185:261–264. doi: 10.1042/bj1850261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overdijk B, Koref M J V D, Lisman J J W, Pierce R J, Montreuil J, Spik G. FEBS Lett. 1981;128:364–366. doi: 10.1016/0014-5793(81)80118-4. [DOI] [PubMed] [Google Scholar]
- 23.Delmonte F, Kieda C, Monsigny M. C R Acad Sci. 1976;283:1121–1124. [PubMed] [Google Scholar]
- 24.Tarentino A L, Maley F. J Biol Chem. 1976;251:6537–6543. [PubMed] [Google Scholar]
- 25.Saint-Pol A, Bauvy C, Codogno P, Moore S E H. J Cell Biol. 1997;136:45–59. doi: 10.1083/jcb.136.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato T, Hatanaka K, Mega T, Hase S. J Biochem. 1997;122:1167–1173. doi: 10.1093/oxfordjournals.jbchem.a021877. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1994;269:17611–17618. [PubMed] [Google Scholar]
- 28.Nomoto H, Inoue Y. Eur J Biochem. 1983;135:243–250. doi: 10.1111/j.1432-1033.1983.tb07644.x. [DOI] [PubMed] [Google Scholar]
- 29.Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1991;266:22110–22114. [PubMed] [Google Scholar]
- 30.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 31.Kadowaki S, Yamamoto K, Fujisaki M, Izumi K, Tochikura T, Yokoyama T. Agric Biol Chem. 1990;54:97–106. [PubMed] [Google Scholar]
- 32.Kadowaki S, Yamamoto K, Fujisaki M, Tochikura T. J Biochem. 1991;110:17–21. doi: 10.1093/oxfordjournals.jbchem.a123536. [DOI] [PubMed] [Google Scholar]
- 33.Takegawa K, Nakoshi M, Iwahara S, Yamamoto K, Tochikura T. Appl Environ Microbiol. 1989;55:3107–3112. doi: 10.1128/aem.55.12.3107-3112.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takegawa K, Yamabe K, Fujita K, Tabuchi M, Mita M, Izu H, Watanabe A, Asada Y, Sano M, Kondo A, et al. Arch Biochem Biophys. 1997;338:22–28. doi: 10.1006/abbi.1996.9803. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki T, Park H, Lennarz W J. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, Park H, Hollingsworth N M, Sternglanz R, Lennarz W J. J Cell Biol. 2000;149:1039–1051. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauew J-F, Michalski J-C, Strecker G, Spik G, Montreuil J. Glycobiology. 1991;1:487–492. doi: 10.1093/glycob/1.5.487. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Park H, Kitajima K, Lennarz W J. J Biol Chem. 1998;273:21526–21530. doi: 10.1074/jbc.273.34.21526. [DOI] [PubMed] [Google Scholar]
- 39.Waddling C A, Plummer T H, Jr, Tarentino A L, Van Roey P. Biochemistry. 2000;39:7878–7885. doi: 10.1021/bi0001731. [DOI] [PubMed] [Google Scholar]
- 40.Rao V, Guan C, Van Roey P. Structure (London) 1995;3:449–457. doi: 10.1016/s0969-2126(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 41.Rao V, Cui T, Guan C, Van Roey P. Protein Sci. 1999;8:2338–2346. doi: 10.1110/ps.8.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]