Abstract
Metabolic abnormalities underlying diabetes are primarily the result of the lack of adequate insulin action and the associated changes in protein phosphorylation and gene expression. To define the full set of alterations in gene expression in skeletal muscle caused by diabetes and the loss of insulin action, we have used Affymetrix oligonucleotide microarrays and streptozotocin-diabetic mice. Of the genes studied, 235 were identified as changed in diabetes, with 129 genes up-regulated and 106 down-regulated. Analysis revealed a coordinated regulation at key steps in glucose and lipid metabolism, mitochondrial electron transport, transcriptional regulation, and protein trafficking. mRNAs for all of the enzymes of the fatty acid β-oxidation pathway were increased, whereas those for GLUT4, hexokinase II, the E1 component of the pyruvate dehydrogenase complex, and subunits of all four complexes of the mitochondrial electron transport chain were all coordinately down-regulated. Only about half of the alterations in gene expression in diabetic mice could be corrected toward normal after 3 days of insulin treatment and euglycemia. These data point to as of yet undefined mechanisms for highly coordinated regulation of gene expression by insulin and potential new targets for therapy of diabetes mellitus.
Type 1 and type 2 diabetes are characterized by a loss of insulin action on peripheral tissues, caused by either insulin deficiency from autoimmune destruction of β cells or a combination of insulin resistance and altered insulin secretion, respectively (1). This loss of insulin action leads to profound changes in cellular function, including changes in glucose and lipid metabolism (2) as well as changes in gene expression and protein phosphorylation, which cumulatively contribute to the clinical manifestations and complications of the disease. Many studies have been done to assess whether specific genes involved in a particular pathway are changed in various tissues in animal models and humans with diabetes (3–6), and a large number of insulin-regulated genes have been identified, particularly in liver and adipose tissue (7). However, until recently there has been no way to define the full set of alterations in gene in expression that may contribute to the pathophysiology of these disorders. Furthermore, studies of the role of insulin on gene expression in skeletal muscle, the major site of glucose disposal in response to insulin and the major site of insulin resistance in type 2 diabetes, have been limited (3).
The development of cDNA and oligonucleotide arrays now allows characterization of global patterns of transcriptional changes, but these have received limited use in diabetes (8–10). As a first step toward determining the genome anatomy of insulin action, in the present study we have analyzed the pattern of gene expression in skeletal muscle of normal mice and mice with streptozotocin-diabetes by using Affymetrix (Santa Clara, CA) oligonucleotide arrays to address the following questions: (i) What are the changes in the transcriptional program of skeletal muscle in insulin-deficient diabetes on a global scale, and to what extent are they reversible by insulin treatment? (ii) Can these transcriptional changes be related to the pathophysiology and clinical manifestations of diabetes? (iii) Can patterns of alterations be identified that can lead to discovery of common pathways of regulation?
Materials and Methods
The methods are described in detail in the supporting information, which is published on the PNAS web site, www.pnas.org.
Sample Preparation.
Total RNA was isolated from skeletal muscle from 3 treatment groups of 9 mice each. The control group and the streptozotocin (STZ)-induced diabetic group had random-fed blood glucose levels of <200 mg/dl and >400 mg/dl for 4 weeks, respectively. The insulin-treated STZ-diabetic group had random-fed blood glucose levels of <200 mg/dl for 3 days before harvesting skeletal muscle. Equal quantities of total RNA were pooled together from 3 individual mice within the same treatment group. A hybridization mixture containing 15 μg of biotinylated c-RNA, generated as per the protocols provided by Affymetrix and adjusted for possible carryover of residual total RNA, was prepared and hybridized to mouse Affymetrix MG-U74A-v2 chips. The chips were washed, scanned, and analyzed with GENECHIP MAS V.4.0.
Data Analysis.
All chips were subjected to global scaling to a target intensity of 1,500 to take into account the inherent differences between the chips and their hybridization efficiencies. The background and the scaled noise of each of the chips were averaged. The next step was to calculate the means and SDs of the expression intensities of genes for the 3 samples in each treatment group.
After this calculation, 3 separate independent filters of significance were applied serially to obtain a list of those genes that had a significant change between the control and the diabetic groups. The first filter excluded all genes that had a mean expression value that was below the sum of the average background and the average standard difference threshold (SDT; equal to 4 times the scaled noise) in both the control and the diabetic groups. Those genes that passed this first filter were subjected to the second significance filter, which was to consider only those genes for which the difference between the means of the diabetic and control groups was ≥2 times the sum of the SDs of both groups. Finally, the third filter selected those genes that had an absolute difference between the means of the control and the diabetic groups that was greater than the average SDT. Genes that passed all three filters were then labeled as being significantly changed between the control and the diabetic groups.
Quantitative Reverse Transcription (RT)-PCR.
Total RNA from each of the same 9 pooled RNA samples, 3 from each of the 3 treatment groups that were hybridized to the microarrays, was used as a template in a florescence-based, quantitative one-step RT-PCR, with specific primers for each of the selected genes (Tables 1 and 2) to confirm the results.
Table 1.
Genes up-regulated in STZ-diabetes
| Accession no. | Gene/protein | Relative change in diabetes | Insulin response |
|---|---|---|---|
| Lipid metabolism | |||
| Y14004 | Acyl-CoA thioesterase 1 | 3.71 | +++ |
| U69543 | Hormone-sensitive lipase | 2.38 | ++ |
| AI840013 | Δ3,Δ2-Enoyl-CoA isomerase | 1.93 | +++ |
| AF030343 | Enoyl-CoA hydratase | 1.92 | +++ |
| AV238359 | Carnitine acetyltransferase (similar to) | 1.65 | +++ |
| Z14050 | Dodecenoyl-CoA Δ-isomerase | 1.44 | +++ |
| X85983 | Carnitine acetyltransferase | 1.41 | +++ |
| U07159 | Acetyl-CoA dehydrogenase, medium chain | 1.38 | +++ |
| AW122615 | 47.6-kDa protein B0303.3-like (thiolase family) | 1.35 | +++ |
| AI849271 | 3-Ketoacyl-CoA thiolase (homolog-EST) | 1.33 | +++ |
| Glucose metabolism | |||
| D42083 | Fructose-bisphosphatase 2 | 4.48 | ++ |
| X13586 | 2,3-Bisphosphoglycerate mutase | 1.60 | +++ |
| AW123952 | Lactate dehydrogenase 1, A chain | 1.37 | +++ |
| Others-metabolism | |||
| U48896 | UDP-glucoronosyltransferase 8 | 40.98 | ++ |
| X89998 | 17β-Hydroxysteroid dehydrogenase 4 | 1.75 | ++ |
| AB005450 | Carbonic anhydrase 14 | 1.73 | +++ |
| M33934 | IMP dehydrogenase | 1.28 | + |
| AW046273 | Electron transfer flavoprotein-β-subunit homolog | 1.24 | +++ |
| Transport/trafficking | |||
| U15976 | Fatty acid transporter 1 | 28.18 | +++ |
| X14961 | Fatty acid binding protein 3 | 1.91 | +++ |
| AW125607 | SNAP-α homolog | 1.72 | +++ |
| AF035643 | VAMP-5 (myobrevin) | 1.71 | ++ |
| AF020185 | Dynein, cytoplasmic, light chain 1 | 1.58 | + |
| AI835359 | TRAP-γ signal sequence receptor γ | 1.29 | – |
| U27106 | Adaptor protein complex AP-2, μ 1 | 1.22 | ++ |
| Others | |||
| AV171056 | Ubiquitin-conjugating enzyme E2D2 | 8.26 | +++ |
| AF037454 | Ubiquitin–protein ligase | 1.90 | + |
| AW125420 | Plic-1 homolog | 1.36 | +++ |
| D87898 | ADP-ribosylation factor-1 (ARF1) | 1.27 | + |
Tables 1 and 2 list the up- and down-regulated genes in the STZ-diabetic mice as compared with the controls. The first column lists the GenBank accession no. for the respective gene or EST sequence. The second column lists the name of either the gene or the gene product. The third column lists the relative expression intensity values of the STZ-diabetic as compared to the control. The fourth column lists the insulin response (see Materials and Methods). +++ indicates a >75% response; ++ indicates 50–75% response; + indicates 25–50% response; and – indicates a response of <25%. EST, expressed sequence tag.
Genes that lost their significance when negative intensity values were used without transformation.
Genes that were confirmed with quantitative RT-PCR.
Table 2.
Genes down-regulated in STZ-diabetes
| Accession no. | Gene/protein | Relative change in diabetes | Insulin response |
|---|---|---|---|
| Lipid metabolism | |||
| AI843232 | Succinyl-CoA:3-ketoacid-CoA transferase homolog | 0.12 | +++ |
| AV327760 | Stearoyl-CoA desaturase 2 | 0.74 | + |
| AI850017 | Cytochrome b5 homolog (fatty acid synthesis) | 0.64 | + |
| AI839690 | NADH:cytochrome b5 reductase isoform 1 homolog | 0.70 | – |
| AW106745 | NAD(P)-dependent steroid dehydrogenase-like | 0.73 | – |
| AW124555 | Methylmalonyl-CoA epimerase homolog | 0.83 | +++ |
| Glucose metabolism | |||
| AW210370 | α1-4 to 1-6 glucan branching enzyme homolog | 0.54 | ++ |
| Y11666 | Hexokinase II | 0.59 | ++ |
| X51905 | Lactate dehydrogenase 2, B chain | 0.64 | ++ |
| AW125336 | Pyruvate dehydrogenase E1 component β-subunit | 0.80 | +++ |
| Electron transport chain | |||
| AA590675 | NADH dehydrogenase (ubiquinone) Fe-S protein 4 | 0.69 | + |
| AV260484 | Cytochrome c oxidase subunit VIII b | 0.73 | +++ |
| AI845556 | NADH dehydrogenase (ubiquinone) 1β-B22 subunit | 0.84 | +++ |
| AF037371 | Cytochrome c oxidase, subunit VIIa 3 | 0.85 | +++ |
| AI842835 | Ubiquinol cytochrome c reductase core protein 1 | 0.86 | +++ |
| AW121091 | Succinate dehydrogenase cytochrome b subunit | 0.89 | + |
| AI842835 | Ubiquinol cytochrome c reductase core protein 2 | 0.92 | +++ |
| Amino acid metabolism | |||
| AW047743 | Isovaleryl dehydrogenase precursor | 0.65 | +++ |
| L47335 | Branched chain ketoacid dehydrogenase E1, α | 0.66 | ++ |
| Others-metabolism | |||
| M29395 | UMP synthetase (pyrimidine synthesis) | 0.12 | + |
| AV250974 | Creatine kinase-mitochondrial sarcomeric homolog | 0.51 | +++ |
| AI181132 | Creatine kinase precursor (sarcomeric) | 0.71 | +++ |
| X04591 | Creatine kinase, brain isoform | 0.84 | – |
| Transcription, transcriptional regulation and translation | |||
| M28845 | Early growth response 1 | 0.32 | ++ |
| X57638 | Peroxisome proliferator activated receptor α | 0.38 | ++ |
| Signaling | |||
| AV367375 | Protein kinase C ζ | 0.51 | – |
| Transport/trafficking | |||
| M23383 | GLUT4 | 0.67 | +++ |
| AI836322 | GDP dissociation inhibitor 1 | 0.77 | +++ |
| U76832 | Syntaxin 4 | 0.85 | ++ |
| Structural | |||
| L22545 | Procollagen type XVIII, α 1 | 0.31 | +++ |
| M60474 | Myristoylated alanine-rich protein kinase C substrate | 0.51 | +++ |
See Table 1 legend for details.
Genes that lost their significance when negative intensity values were used without transformation.
Genes that were confirmed with quantitative RT-PCR.
Results and Discussion
Of the 14,288 genes and expressed sequence tags (all henceforth referred to as genes) represented on the Affymetrix mouse MG-U74A v.2 oligonucleotide array, 7,303 genes passed the first significance filter (see Materials and Methods and Fig. 5A, which is published as supporting information on the PNAS web site), i.e., had a minimum expression value that was greater than the sum of the average background plus the average standard difference threshold (4 times the noise). With the second significance filter, i.e., a difference between control and diabetic of greater than 2 times the sum of the SDs, 262 genes demonstrated significance in the diabetic state. Of these, the difference was also greater than 4 times the noise for 235 genes. These 235 genes were then deemed to be significantly changed between the control and the STZ-induced diabetic groups, because they survived application of all three significance filters (Fig. 5B).
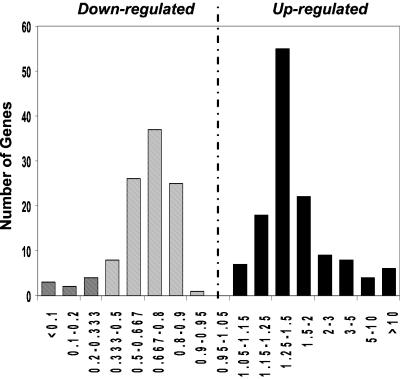
Of these 235 genes that were significantly changed in the STZ-diabetic group as compared with the controls, 129 were up-regulated and 106 down-regulated. When plotted in terms of fold-change between the two groups, there was a skewed bell-shaped distribution for these gene expression differences (Fig. 1). Interestingly, many of the changes were relatively modest (25–50% increase or decrease), but based on the pathway analysis (see below), the confirmation by reverse transcription–PCR (Fig. 7, which is published as supporting information on the PNAS web site; Tables 1 and 2), and the stringent statistical analysis used for defining these differences and published reports, these seem to be part of the overall program of changes.
Fig 1.
Relative expression of STZ to control. The number of genes significantly up- and down-regulated in STZ-diabetes are plotted to show the distribution by the magnitude of change (STZ/control). A skewed distribution is seen in both groups with the maximum number of affected genes showing a 25–50% up-regulation or a 20–33% down-regulation. Most of the genes that show a higher relative fold change are those that have a low expression intensity.
To assess further the effect of STZ-diabetes on gene expression, these 235 genes were identified specifically and assigned functional categories. Genes associated with substrate and energy metabolism constituted the single largest functional group, comprising about 20% of all of the genes with significant change in the diabetic group. Genes involved in transcription regulation (13%) and in transport/trafficking (9.5%) were the other two major functional categories showing significant change.
A similar analysis was performed on the STZ-diabetic mice treated with s.c. insulin pellets after 3 days of near euglycemia (see Materials and Methods). To assess reversibility of these changes, if one defined a positive insulin response as a change in gene expression of the diabetic toward the level found in controls of 50% or more, 62% of up-regulated genes and 50% of down-regulated genes were insulin-responsive with treatment to achieve 3 days of euglycemia (Fig. 3). The reversible set included 77% of the genes involved in metabolism, 53% of the genes involved in transcriptional regulation, and 73% of the genes involved in protein transport/trafficking. These are summarized in Tables 1 and 2 and discussed in detail below.
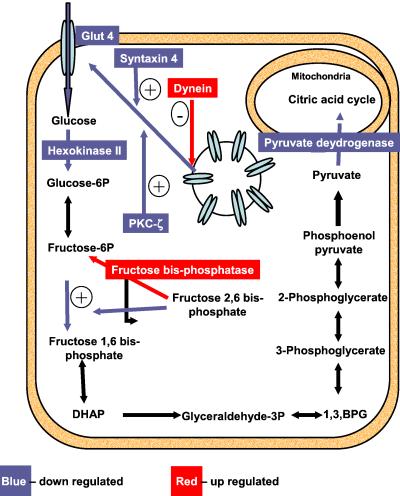
Fig 3.
Schematic representation of glucose uptake and the glycolytic pathway in skeletal muscle. The steps that exhibit up-regulation at the mRNA level are shown in red and those that are down-regulated are shown in blue.
Lipid Metabolism.
The predominant source of energy for resting skeletal muscle is fatty acid oxidation. Glucose and ketone body metabolism also contribute to varying degrees, depending on the activity and metabolic state of the muscle. However, in diabetes there is a diminished contribution of glucose and ketone bodies to skeletal muscle energy metabolism. Hence, there is an increased demand on the muscle for fatty acid oxidation for its energy needs. Thus, it is not surprising that there is a significant increase in the mRNAs for the various proteins involved in fatty acid oxidation in diabetes (Fig. 2). These include a 28-fold increase in the fatty acid transporter type 1 that regulates the entry of long chain fatty acids into the muscle across the plasma membrane; a 2.4-fold increase in hormone-sensitive lipase, further contributing to the increased pool of nonesterified fatty acids in the cytosol; and a 1.9-fold increase in the mRNA of fatty acid binding protein 3, a transport protein involved in the intracellular transport of long chain fatty acids and their CoA esters.
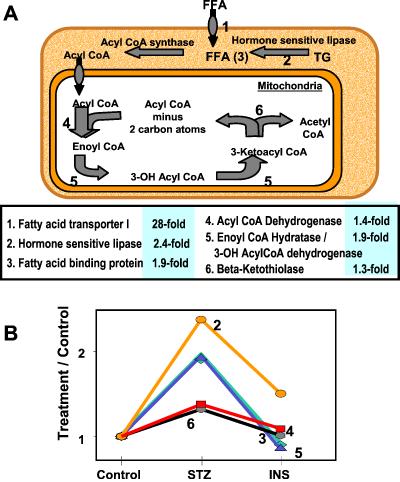
Fig 2.
Changes in the β-oxidation pathway in diabetes. (A) Schematic representation of the β-oxidation pathway of fatty acids. The numbers in the center panel are the relative values for the genes in the diabetic group as compared with the control. (B) The graph shows the fold up-regulation in expression for many of these genes in STZ-diabetes and its correction with insulin treatment.
Moreover, the transcripts of all enzymes involved in β-oxidation were increased in diabetes (Fig. 2). β-Oxidation includes multiple cycles leading to the formation of acetyl-CoA, with the first step of each cycle consisting of dehydrogenation, followed by hydration, a second dehydrogenation, and finally thiolysis. The mRNA for acetyl-CoA dehydrogenase, the rate-limiting enzyme catalyzing the first dehydrogenation of fatty acids with a carbon chain length of 4–16, was increased 3.7-fold, as was the mRNA for the enzyme that catalyzes the next step of hydration—enoyl-CoA hydratase. The mRNA for β-ketoacyl thiolase, which catalyzes the last step, was also increased. The expression of electron transfer flavoprotein-β (ETF), a necessary electron acceptor for many of the dehydrogenases in the mitochondria including acetyl-CoA dehydrogenase, was increased in the STZ-diabetic. The mRNAs for two enzymes specifically required for β-oxidation of unsaturated fatty acids, namely Δ2,Δ3-enoyl-CoA isomerase and dodecenoyl-CoA Δ- isomerase, were also increased. In keeping with increased fatty acid oxidation, the mRNAs for the rate-limiting enzyme in unsaturated fatty acid synthesis, steroyl-CoA desaturase and the required cytochrome b5 dehydrogenase were decreased. Succinyl- CoA:3-oxoacid-CoA transferase, the rate-limiting first step in extrahepatic metabolism of ketone bodies, was down-regulated in the diabetic group. Taken together, all of these changes in gene expression would be expected to lead to a coordinated increase in free fatty acids and fatty acid oxidation, decreased fatty acid synthesis, and ketone body utilization in muscle of STZ-diabetes.
Peroxisome proliferator activated receptor α (PPARα) is known to up-regulate many of the fatty acid oxidation enzymes (11) and hence is a strong candidate for mediating these effects of diabetes. Surprisingly, however, PPARα mRNA was down-regulated in the STZ-diabetic group. Previous reports have shown that the expression of PPARα (12) is increased in the livers of diabetic rats, whereas it is decreased in pancreatic β-cells by high glucose (13). There is also evidence that PPARα may have differential effects on gene expression depending on the target tissue (11). It is likely that the decrease in PPARα observed in this study is either the result of a tissue-specific change induced by diabetes or opposing effects of hyperglycemia (causing a decrease) and a loss of insulin signaling (causing an increase) on the expression of PPARα, with the glucose effect predominating in skeletal muscle.
Carbohydrate Metabolism.
Impaired glucose disposal by muscle is one of the key features of diabetes. In the muscle of STZ-diabetic mice, this impairment seems to be caused, at least in part, by coordinated changes in the mRNAs for the transporters and enzymes involved in glucose uptake and metabolism (Fig. 3). GLUT4 is clearly the dominant insulin-responsive glucose transporter in muscle (14), and a decrease in GLUT4 expression as seen in this study (Table 2) would contribute to impaired muscle glucose uptake (15–19). The evidence of GLUT4 mRNA regulation in human diabetes is, however, not as conclusive (5, 20, 21). Protein kinase C (PKC)-ζ has recently been linked as a potential mediator of GLUT4 translocation (22), and thus a decrease in PKC-ζ expression and PKC-ζ signaling might impair GLUT4 translocation even further. However, recent data indicate that ablation of PKC-ζ in mice (23) does not impair whole-body glucose tolerance (J. Moscat, personal communication).
Once glucose enters the muscle cell, it is phosphorylated by hexokinase II before entry into any metabolic pathway. Hexokinase II catalyzes the first of the three irreversible reactions of glycolysis, and hence a change in the level or activity of this enzyme would affect the glycolytic flux. A decrease in hexokinase II mRNA, as seen in the diabetic group, would be expected to decrease intracellular glucose 6-phosphate. Although there are no data in humans with type 1 diabetes, studies done in obese and type 2 diabetic patients have shown similar findings (5, 24, 25). Mice with a heterozygous knockout of hexokinase II have a 50% decrease in hexokinase II mRNA and a corresponding 50% decrease in muscle glucose uptake, suggesting that hexokinase II may serve as a rate-limiting step for glucose uptake in the muscle (26). The second irreversible and rate-limiting step in glycolysis is the conversion of fructose 6-phosphate to fructose 1,6-bisphosphate and is catalyzed by phosphofructokinase-1 (PFK-1). One of the key allosteric activators of PFK-1 is fructose 2,6-bisphosphate, whose level is regulated by the enzyme fructose-bisphosphatase 2. In states of relative insulin deficiency and glucagon excess, as occur in diabetes, fructose-bisphosphatase 2 acts as a phosphatase leading to a decrease in fructose 2,6-bisphosphate. In the muscle of the STZ-diabetic mice, we found that the expression of this enzyme was increased. This increase would cause an inhibition of the PFK-1-catalyzed step by decreasing the level of fructose 2,6-bisphosphate.
The committed step for glucose oxidation is the conversion of pyruvate into acetyl CoA in the mitochondria by the pyruvate dehydrogenase complex. In the diabetic group, there was a down-regulation of the mRNA for E1 (dehydrogenase) component of this complex. When this decrease is coupled with further inhibition of the enzyme by the increased amounts of acetyl-CoA present in the mitochondria (generated by the increased fatty acid oxidation and decreased acetyl-CoA utilization as a result of decreased functioning of the citric acid cycle), there would be a major decrease in glucose oxidation in the muscle of the diabetic mice.
The α-1,4- to 1,6-glucan-branching enzyme was also decreased in expression in diabetic mice. This decrease could lead to glycogen with longer but fewer chains, and when coupled with the decreased signaling to glycogen synthase, less readily usable glycogen in the muscle of the diabetic mice.
Electron Transport Chain and Energy Metabolism.
The mitochondrial electron transport chain consists of five large enzyme complexes (Complexes I–V) situated in the inner mitochondrial membrane that couple oxidation to phosphorylation and provide most of the ATP that is necessary for cell survival. In the diabetic group, mRNAs for two of the polypeptides in Complex I (NADH-ubiquinone dehydrogenase) and two of the subunits each of Complexes III (cytochrome c oxidoreductase) and IV (cytochrome c oxidase) were decreased (Fig. 4). The mRNA for the cytochrome b subunit of succinate dehydrogenase (Complex II) of the electron transport chain was also decreased in the diabetic group. All this would lead us to predict that in diabetic muscle there is a decreased ability of the mitochondria to generate ATP as compared with the controls. These changes would be accompanied by an increase in reactive oxygen species, as occurs whenever there is a decrease in the function of the mitochondrial electron transport chain (27). Further, there was a decrease in the mRNAs for creatine kinase homologs in diabetic mice that would decrease the level of available creatine phosphate, a critical energy reservoir for the active muscle. Similar results have been reported in skeletal muscle (28) and myocardium (29) of STZ-diabetic rats. Cumulatively, these changes could lead to some manifestations of diabetes, and if similar changes are present in other tissues, such as the nervous system and cardiovascular system, contribute to complications of the disease.
Fig 4.
(A) Schematic representation of the electron transport chain. (B) Relative expression of the genes for polypeptides of the indicated electron transport chain complexes are plotted to reflect the down-regulation of these in diabetes and their subsequent correction with insulin treatment.
Proteasome Pathway.
Three ubiquination enzymes were increased in expression in the diabetic muscle. These include ubiquitin-conjugating enzyme E2D2 and ubiquitin protein ligase, both important in coupling ubiquitin to proteins that are earmarked for degradation via the proteasome. PLIC, a protein that is involved in the attachment of ubiquitin to the proteasome, a necessary step in protein degradation, is also increased at the mRNA level. All of the increases seen in the ubiquination pathway are consistent with published reports (30–32) and might explain the increased proteolysis in skeletal muscle that could contribute to the muscle atrophy and myopathy seen with uncontrolled diabetes. One proteasome subunit, b5, was decreased in the diabetic group.
Transport and Trafficking.
Many of the proteins involved in protein trafficking and transport were differentially regulated in diabetes. The transcripts for α-SNAP, syntaxin-4 (a t-SNARE), and VAMP-5 (a v-SNARE) were all increased in diabetes. α-SNAP has been implicated in the regeneration of the SNARE complexes after the fusion of GLUT4 vesicles to the plasma membrane (33). Syntaxin-4 is present at the plasma membrane and is critical in the docking of the GLUT4 vesicles and insulin-stimulated glucose transport in skeletal muscle (34). VAMP-5 is normally increased 8–10-fold in myogenesis (35) but has thus far not been associated with insulin action and diabetes. ADP-ribosylation factor-1 (ARF-1), a member of the small GTPase family of proteins involved in vesicle transport, was increased. Although other members of the ARF family, such as ARF-6, have been associated with GLUT4 translocation (36), ARF-1 has yet to be directly implicated in glucose transport. In addition, the mRNA for GTP dissociation inhibitor-1 (RhoGDI-1) was decreased. RhoGDI-1 interacts with Rab4 and has a potential role in insulin-mediated glucose transport. The expression of dynein, a microtubule-based motor protein, which has recently been reported to be involved in internalization of GLUT4 transporters (37), thus attenuating glucose transport into the cell, was increased in its mRNA expression in the diabetic group.
Others.
Procollagen-8α mRNA was decreased in the diabetic group, as was reported in a study of a diabetes-induced model of impaired wound healing (38). The mRNA for MARCKS (myristoylated alanine-rich protein kinase C substrate) protein was also decreased in diabetes. Excess phosphorylation by protein kinase C or a decrease in the protein level of MARCKS could impair myoblast fusion and thus muscle differentiation and repair (39, 40). These both could contribute to the poor muscle and connective tissue-reparative process seen in patients with uncontrolled diabetes. Finally, early growth response-1 (EGR-1) mRNA was decreased in diabetes. This zinc finger protein is necessary for the postinjury repair associated with the smooth muscle and vascular endothelium (41), and although the changes seen in this study in skeletal muscle can be extrapolated to vascular smooth muscle, this could contribute to the pathophysiology of wound healing in diabetes.
Summary and Conclusion
In summary, we have shown that insulin-deficient diabetes results in profound changes in gene expression in skeletal muscle and that many, but not all, of these changes are reversed with 3 days of euglycemia. Most importantly, we have demonstrated patterns of regulation for genes involved in substrate and energy metabolism, protein degradation and protein trafficking in the diabetic that can be linked to the pathophysiology of diabetes and suggest mechanisms of coordinated regulation. Possible candidates for such coordinated regulation over such a wide variety of targets would include networks of insulin dependent, interacting transcription factors, or possible co-activators and/or co-repressors that could link multiple transcription pathways simultaneously. These represent potential novel therapeutic targets for diabetes and other states of altered insulin action.
Supplementary Material
Acknowledgments
We thank Mary Loeken and Melissa Horal for their help with insulin pellet administration, Isaac Kohare and Atul Butto for discussion about data analysis, Mousumi Moulik and Mandira Moulik for their help in animal studies, and Julie Marr and Martha Strom for their secretarial assistance. This study was supported by the National Institutes of Health Grants DK36836-15, DK33201, and DK45935 (to C.R.K.).
Abbreviations
STZ, streptozotocin
References
- 1.Kahn C. R. (1994) Diabetes 43, 1066-1084. [DOI] [PubMed] [Google Scholar]
- 2.Saltiel A. R. & Kahn, C. R. (2001) Nature (London) 414, 799-806. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien R. M., Lucas, P. C., Yamasaki, T. & Granner, D. K. (1993) Adv. Second Messenger Phosphoprotein Res. 28, 245-253. [PubMed] [Google Scholar]
- 4.Suzuki A., Yasuno, T., Kojo, H., Hirosumi, J., Mutoh, S. & Notsu, Y. (2000) Jpn. J. Pharmacol. 84, 113-123. [DOI] [PubMed] [Google Scholar]
- 5.Ducluzeau P. H., Perretti, N., Laville, M., Andreelli, F., Vega, N., Riou, J. P. & Vidal, H. (2001) Diabetes 50, 1134-1142. [DOI] [PubMed] [Google Scholar]
- 6.Vestergaard H., Bjorbaek, C., Hansen, T., Larsen, F. S., Granner, D. K. & Pedersen, O. (1995) J. Clin. Invest. 96, 2639-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Brien R. M. & Granner, D. K. (1996) Physiol. Rev. 76, 1109-1161. [DOI] [PubMed] [Google Scholar]
- 8.Joussen A. M., Huang, S., Poulaki, V., Camphausen, K., Beecken, W. D., Kirchhof, B. & Adamis, A. P. (2001) Invest. Ophthalmol. Visual Sci. 42, 3047-3057. [PubMed] [Google Scholar]
- 9.Wada J., Zhang, H., Tsuchiyama, Y., Hiragushi, K., Hida, K., Shikata, K., Kanwar, Y. S. & Makino, H. (2001) Kidney Int. 59, 1363-1373. [DOI] [PubMed] [Google Scholar]
- 10.Nadler S. T., Stoehr, J. P., Schueler, K. L., Tanimoto, G., Yandell, B. S. & Attie, A. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11371-11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook W. S., Yeldandi, A. V., Rao, M. S., Hashimoto, T. & Reddy, J. K. (2000) Biochem. Biophys. Res. Commun. 278, 250-257. [DOI] [PubMed] [Google Scholar]
- 12.Asayama K., Sandhir, R., Sheikh, F. G., Hayashibe, H., Nakane, T. & Singh, I. (1999) Mol. Cell. Biochem. 194, 227-234. [DOI] [PubMed] [Google Scholar]
- 13.Roduit R., Morin, J., Masse, F., Segall, L., Roche, E., Newgard, C. B., Assimacopoulos-Jeannet, F. & Prentki, M. (2000) J. Biol. Chem. 275, 35799-35806. [DOI] [PubMed] [Google Scholar]
- 14.Shepherd P. R. & Kahn, B. B. (1999) N. Engl. J. Med. 341, 248-257. [DOI] [PubMed] [Google Scholar]
- 15.Kainulainen H., Breiner, M., Schurmann, A., Marttinen, A., Virjo, A. & Joost, H. G. (1994) Biochim. Biophys. Acta 1225, 275-282. [DOI] [PubMed] [Google Scholar]
- 16.Dombrowski L. & Marette, A. (1995) FEBS Lett. 374, 43-47. [DOI] [PubMed] [Google Scholar]
- 17.Charron M. J. & Katz, E. B. (1998) Mol. Cell. Biochem. 182, 143-152. [PubMed] [Google Scholar]
- 18.Zisman A., Peroni, O. D., Abel, E. D., Michael, M. D., Mauvais-Jarvis, F., Lowell, B. B., Wojtaszewski, J. F., Hirshman, M. F., Virkamaki, A., Goodyear, L. J., et al. (2000) Nat. Med. 6, 924-928. [DOI] [PubMed] [Google Scholar]
- 19.Kim J. K., Zisman, A., Fillmore, J. J., Peroni, O. D., Kotani, K., Perret, P., Zong, H., Dong, J., Kahn, C. R., Kahn, B. B., et al. (2001) J. Clin. Invest. 108, 153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen P. H., Vestergaard, H., Lund, S., Vedel, P., Junker, S., Kahn, B. B. & Pedersen, O. (1993) Diabet. Med. 10, 699-706. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen O., Bak, J. F., Andersen, P. H., Lund, S., Moller, D. E., Flier, J. S. & Kahn, B. B. (1990) Diabetes 39, 865-870. [DOI] [PubMed] [Google Scholar]
- 22.Braiman L., Alt, A., Kuroki, T., Ohba, M., Bak, A., Tennenbaum, T. & Sampson, S. R. (2001) Mol. Cell. Biol. 21, 7852-7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitges M., Sanz, L., Martin, P., Duran, A., Braun, U., Garcia, J. F., Camacho, F., Diaz-Meco, M. T., Rennert, P. D. & Moscat, J. (2001) Mol. Cell 8, 771-780. [DOI] [PubMed] [Google Scholar]
- 24.Vestergaard H. (1999) Dan. Med. Bull. 46, 13-34. [PubMed] [Google Scholar]
- 25.Pendergrass M., Koval, J., Vogt, C., Yki-Jarvinen, H., Iozzo, P., Pipek, R., Ardehali, H., Printz, R., Granner, D., Defronzo, R. A., et al. (1998) Diabetes 47, 387-394. [DOI] [PubMed] [Google Scholar]
- 26.Heikkinen S., Pietila, M., Halmekyto, M., Suppola, S., Pirinen, E., Deeb, S. S., Janne, J. & Laakso, M. (1999) J. Biol. Chem. 274, 22517-22523. [DOI] [PubMed] [Google Scholar]
- 27.Wallace D. C. (1999) Science 283, 1482-1488. [DOI] [PubMed] [Google Scholar]
- 28.Su C. Y., Payne, M., Strauss, A. W. & Dillmann, W. H. (1992) Am. J. Physiol. 263, E310-E316. [DOI] [PubMed] [Google Scholar]
- 29.Depre C., Young, M. E., Ying, J., Ahuja, H. S., Han, Q., Garza, N., Davies, P. J. & Taegtmeyer, H. (2000) J. Mol. Cell. Cardiol. 32, 985-996. [DOI] [PubMed] [Google Scholar]
- 30.Lecker S. H., Solomon, V., Price, S. R., Kwon, Y. T., Mitch, W. E. & Goldberg, A. L. (1999) J. Clin. Invest. 104, 1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merforth S., Osmers, A. & Dahlmann, B. (1999) Mol. Biol. Rep. 26, 83-87. [DOI] [PubMed] [Google Scholar]
- 32.Mitch W. E., Bailey, J. L., Wang, X., Jurkovitz, C., Newby, D. & Price, S. R. (1999) Am. J. Physiol. 276, C1132-C1138. [DOI] [PubMed] [Google Scholar]
- 33.Cheatham B. (2000) Trends Endocrinol. Metab. 11, 356-361. [DOI] [PubMed] [Google Scholar]
- 34.Yang C., Coker, K. J., Kim, J. K., Mora, S., Thurmond, D. C., Davis, A. C., Yang, B., Williamson, R. A., Shulman, G. I. & Pessin, J. E. (2001) J. Clin. Invest. 107, 1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Q., Subramaniam, V. N., Wong, S. H., Tang, B. L., Parton, R. G., Rea, S., James, D. E. & Hong, W. (1998) Mol. Biol. Cell 9, 2423-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langille S. E., Patki, V., Klarlund, J. K., Buxton, J. M., Holik, J. J., Chawla, A., Corvera, S. & Czech, M. P. (1999) J. Biol. Chem. 274, 27099-27104. [DOI] [PubMed] [Google Scholar]
- 37.Huang J., Imamura, T. & Olefsky, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 13084-13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darby I. A., Bisucci, T., Hewitson, T. D. & MacLellan, D. G. (1997) Int. J. Biochem. Cell Biol. 29, 191-200. [DOI] [PubMed] [Google Scholar]
- 39.Inoguchi T., Xia, P., Kunisaki, M., Higashi, S., Feener, E. P. & King, G. L. (1994) Am. J. Physiol. 267, E369-E379. [DOI] [PubMed] [Google Scholar]
- 40.Kim S. S., Kim, J. H., Kim, H. S., Park, D. E. & Chung, C. H. (2000) Biochem. J. 347, 139-146. [PMC free article] [PubMed] [Google Scholar]
- 41.Gousseva N., Kugathasan, K., Chesterman, C. N. & Khachigian, L. M. (2001) J. Cell. Biochem. 81, 523-534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.