Abstract
Imprinting is an epigenetic modification leading to monoallelic expression of some genes, and disrupted imprinting is believed to be a barrier to human stem cell transplantation, based on studies that suggest that epigenetic marks are unstable in mouse embryonic germ (EG) and embryonic stem (ES) cells. However, stem cell imprinting has not previously been examined directly in humans. We found that three imprinted genes, TSSC5, H19, and SNRPN, show monoallelic expression in in vitro differentiated human EG-derived cells, and a fourth gene, IGF2, shows partially relaxed imprinting at a ratio from 4:1 to 5:1, comparable to that found in normal somatic cells. In addition, we found normal methylation of an imprinting control region (ICR) that regulates H19 and IGF2 imprinting, suggesting that imprinting may not be a significant epigenetic barrier to human EG cell transplantation. Finally, we were able to construct an in vitro mouse model of genomic imprinting, by generating EG cells from 8.5-day embryos of an interspecific cross, in which undifferentiated cells show biallelic expression and acquire preferential parental allele expression after differentiation. This model should allow experimental manipulation of epigenetic modifications of cultured EG cells that may not be possible in human stem cell studies.
Genomic imprinting is defined as an epigenetic modification (i.e., DNA alteration other than sequence) in the germ line that leads to preferential expression of a specific parental allele in somatic cells of the offspring. Imprinting involves DNA methylation and possibly other as-yet-unidentified changes such as histone modification. Imprinting must be reprogrammed in the germ line, because a maternal allele in one generation may be a paternal allele in the next. Reprogramming involves erasure of epigenetic marks and establishment of new marks. The timing of this erasure is not known in humans. A recent study of embryos derived by pronuclear transplantation from primordial germ cells indicates that imprinting erasure begins between day 10.5 and 11.5, i.e., after colonization of the gonadal ridge, depending on the gene (1). Several studies of pluripotent cells cultured from mouse embryos suggest that imprinting is abnormal in them. For example, abnormal or variable imprinting was found in embryonic germ (EG) cells derived from as early as 8.5-day embryos (2). Tada et al. (3) found that most imprinted genes have lost parental origin-specific marks in day 11.5 and 12.5-derived EG cells, and they did not reacquire imprinted gene expression during differentiation. Dean et al. (4) found aberrant allele-specific expression and methylation in embryonic stem (ES) cells and embryos derived from an interspecific cross that did not become corrected during development, as well as marked developmental abnormalities in these mice. Humpherys et al. (5) found abnormal expression of four imprinted genes in mice derived from ES cells (5). These authors caution, “Because ES cells are a potential in vitro source of many cell types for transplantation medicine, it will be important to assess whether the epigenetic state of human ES cells is as unstable as that of murine ES cells” (5).
Because of these concerns, we sought to examine imprinted genes in cultured cells derived from human stem cells. The Johns Hopkins University Institutional Review Board explicitly banned any examination of the parental tissues of the human EG cells, so we could not determine parental origin per se; however, we could examine the ratio of expression of parental alleles as well as the methylation of binding sites for the insulator protein CTCF within an imprinting control region (ICR) upstream of the H19 gene. Furthermore, because the human EG cells were derived from the gonadal ridge and adjacent mesentery of 5- to 11-week embryos (6), at a time and location that mouse studies would predict complete erasure of imprints, we could determine whether there was biallelic expression of imprinted genes and loss of methylation, as seen in the mouse. Such imprinting erasure would represent a significant impediment to the clinical use of these cells. Finally, because of the inherent ethical, legal, and practical limitations of human stem cell research, we sought to complement these human studies by developing a mouse model system in which allele-specific expression of imprinted genes could arise in vitro.
Materials and Methods
Imprinting and Methylation Analysis of Human EG-Derived Cell Lines.
Human EG cell lines were cultured and maintained under conditions previously described (6). DNA was extracted as described (7). To identify polymorphisms, we amplified by PCR and sequenced genomic DNA from individual EG-derived clones. Total RNA was isolated by using the RNeasy Minikit (Qiagen, Valencia, CA). To eliminate DNA contamination from RNA preparations, samples were treated with preamplification-grade DNase I (Invitrogen, Carlsbad, CA) according to the supplied protocol. Reverse transcriptase (RT)-PCR was done with the Superscript II preamplification system (Invitrogen) and was performed for each sample in the presence and absence (negative controls) of RT. Samples were sequenced only when no bands were obtained with the negative controls. IGF2 imprinting analysis using the ApaI polymorphism was done as previously described (8). For the analysis of the CpA dinucleotide repeat polymorphism in exon 9 of IGF2, we used the primers IGF2CAF, 5′-TCCCATCCTAAAAAGCACTCA-3′, and IGF2CAR, 5′-GGACTTTGGCCTGATCCATA-3′. H19 was analyzed by using primers H1, H2, and H3 (9). TSSC5 genomic PCR was performed with primers TSSC5GF, 5′-CTTCAGCAGGGACAGCAGTCAGG-3′, and TSSC5GR, 5′-GAGGAGGCTGCTCCACTCGCTGG-3′. RT-PCR of TSSC5 was done with the primers TSSC5RTF, 5′-GCTCTTCATGGTCATGTTCTCCA-3′, and TSSC5RTR, 5′-GGAGCAGTGGTTGTACAGAGG-3′. SNRPN was analyzed with primers SNRPN-F250, 5′-CTTAGCTGAGACACCAAGAGG-3′, and SNRPN-R496, 5′-GCAGCATCTTGCTACTCTTGC-3′. NAP2 was analyzed by using primers NAP2-F1589, 5′-GCCTGTAGCTCTGGACTTCC-3′, and NAP2-R2314, 5′-CCAGGACCTTCAGACAGGATG-3′. DNA and cDNA sequencing was performed on an ABI-377 automated sequencer, following protocols recommended by the manufacturer (Perkin–Elmer). Methylation analysis was carried out by treatment with sodium metabisulfite before PCR, and cloning and sequencing of individual PCR products, as described (10). PCR reagents were purchased from Invitrogen. The final PCR mixture contained 1× PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 0.5 μM each primer, and 1.25 units of Taq DNA polymerase, in a 25-μl reaction volume. Amplification was performed as follows: denaturation at 94°C for 2 min; 40 cycles of 94°C for 1 min, 60°C for 30 s, and 72°C for 1 min; and a final extension step at 72°C for 9 min.
Derivation and Differentiation of Mouse EG Cells.
Day 8.5 embryos were derived from crosses between 7- to 8-week male CAST/Ei (The Jackson Laboratory) and female 129/SvEv (Taconic Farms) mice, and were dissected according to Buehr and McLaren (11). Primary cultures were performed in EG culture medium (DMEM with 4.5 g/liter glucose, 15% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, 2 mM l-glutamine, 0.01 mM each nonessential amino acid, and 0.1 mM 2-mercaptoethanol) supplemented with leukemia inhibitory factor (LIF, 1,000 units/ml), basic fibroblast growth factor (bFGF, 1 ng/ml), and murine stem cell factor (SCF, 60 ng/ml). Cultures were trypsinized after 9 days and replated in EG culture medium without bFGF and SCF supplementation. Colonies were picked, and individual EG cell lines were propagated on irradiated STO feeder layers in EG medium with LIF (1,000 units/ml). All four EG lines characterized in detail had a normal male karyotype with no chromosomal abnormalities, and they all formed teratocarcinomas in nude mice, as expected for EG cell lines. Spontaneous differentiation of EG cells on plastic (12), retinoic acid, dimethyl sulfoxide, and culture in methylcellulose medium (12, 13) was performed as described. For green fluorescent protein (GFP) labeling of EG cells, the pEGFP-N3 vector (CLONTECH) was transfected into SJEG-1 cells by electroporation (250 μF, 0.2 kV). Clones with stable vector integration were obtained by G418 selection (500 μg/ml). The αmMHCneo vector was kindly provided by Lauren Field (14). SJEG-1 cells were transfected by electroporation (250 μF, 0.2 kV). Stable transfected lines were obtained by hygromycin selection (200 μg/ml). Transfected EG cells were differentiated on plastic and then on tissue culture surfaces. Upon the appearance of spontaneously contracting cells, G418 (400 μg/ml) was added until the culture fully consisted of rhythmically contracting muscle bundles. For generating chimeric mice, 8–12 EG cells were injected into C57BL/6 blastocysts. The injected embryos were transferred to pseudopregnant CD-1/VAF female mice.
Imprinting Analysis of Mouse EG Cells.
Preparation of RNA and cDNA was as described for human EG cells. Snrpn was analyzed by single-stranded conformational polymorphism (SSCP) or single-nucleotide primer extension (SNuPE) using an A/G polymorphism we identified at position 918. Primers were Snrpn-U/L, 5′-CACCAAGACCTAAGATACA-3′ and 5′-GCTTGCAGGTACACAATTTC-3′ for PCR, and Snrpn-I2, 5′-GCAGGTACACAATTTCACAAGAAGCATT-3′ for the SNuPE assay. Kvlqt1 was analyzed by SSCP-PCR, using a C/A polymorphism we identified at position 1826. Primers were mLQT1–108/208, 5′-CCACCAATCAAGGTCATCAGGCGCATGC-3′ and 5′-GAGCTCCTTCAGGAACCCTCATCAGGG-3′ for the first PCR, and mLQT1-U/L2, 5′-TTTGTTCATCCCCATCTCAG-3′ and 5′-TTGTTCGATGGTGGGCAGG-3′ for nested PCR. Igf2 was analyzed by SSCP-PCR using an A/G polymorphism we identified at position 780. Primers were Igf2-U/L, 5′-TTGTTTAGAGCCAATCAAAT-3′ and 5′-GATCTCTCTGCTCCACTTCC-3′. H19 was analyzed by SNuPE (15) and by DNA sequencing using a G/T polymorphism we identified at position 1596. Primers were H19-U/L2, 5′-CCACTACACTACCTGCCTCAGAATCTGC-3′ and 5′-GGAACTGCTTCCAGACTAGG-3′, for PCR, and H19-L1, 5′-ACGGAGATGGACGACAGGTG-3′, for sequencing. L23mrp was analyzed by SSCP-PCR using a C/T polymorphism we identified at position 410. Primers were L23mrp-101/201, 5′-GTCGCTCCTTCCTGACCAGGAGGTGC-3′ and 5′-CTTGTCAGAACCACTGTGCTCACTAG-3′. In each case, the experiments were performed redundantly by at least two of the following three methods: SSCP, SNuPE, and direct sequencing. The final PCR mixture contained 1× PCR buffer, 0.1 mM dNTPs, 1 mM MgCl2, 0.5 μM each primer, and 1 unit of Taq DNA polymerase, in a 25-μl reaction volume. Amplification was performed as follows: denaturation at 94°C for 1 min; 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and a final extension step at 72°C for 5 min. For SSCP, 2 μl of the PCR products was used for subsequent SSCP carried out in a 20-μl volume containing 1× PCR buffer (BRL), 1 mM MgCl2, 0.2 mM dNTP, 0.5 mM unlabeled primer, 0.1 mM end-labeled primer, and 0.5 unit of Taq DNA polymerase.
Results
Monoallelic Expression of Imprinted Genes in Human EG Cell-Derived Lineages.
We examined five separate pluripotent human EG cell cultures derived from primordial germ cells obtained from the gonadal ridges and attached mesenteries of 5- to 11-week postfertilization female embryos. A detailed description of these lines and confirmation of their genuine EG cell properties has been reported previously (6). It is extremely difficult to obtain sufficient quantities of fully undifferentiated human EG cells, as a substantial percentage spontaneously differentiate in culture, and markers useful for isolating undifferentiated mouse stem cells, such as OCT3/4, are not specific for undifferentiated stem cells in humans (6). Nevertheless, differentiated cells derived from human stem cells are the intended substrate for eventual human tissue transplantation. We therefore performed these experiments on differentiated cells derived from the EG cells, as described (6). We examined genomic imprinting in five differentiated monolayer cultures of lineage-restricted cell types (16). All of the lines, LV.EB, SL.RC, EU.EE, SD.EP, and SD.EC, were mesenchymal fibroblast-like cultures (ref. 16 and data not shown).
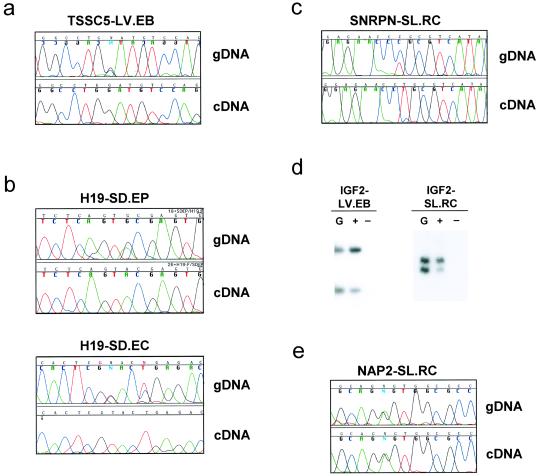
We performed RT-PCR analysis of eight genes, of which five were heterozygous for polymorphisms in the transcribed region in at least one EG line, and thus suitable for analysis of allele-specific expression. Four of these genes, TSSC5, H19, SNRPN, and IGF2, normally show differential expression of the two alleles in somatic cells, and the fifth, NAP2, is not imprinted and thus is normally expressed equally from both alleles. TSSC5 was examined by RT-PCR using a G/C polymorphism we identified at nucleotide 1166, which is within exon 11, and for which line LV.EB was heterozygous. Sequencing of the cDNA product of this gene showed that only the G allele was expressed (Fig. 1a). Imprinting of H19 was then examined by using a G/A polymorphism at nucleotide 1924 in exon 5, as described (9), for which lines SD.EP and SD.EC were heterozygous. Analysis of the cDNA showed a monoallelic expression pattern, with expression of the A allele in both lines (Fig. 1b), indicating normal imprinting of H19 in both lines. The SNRPN gene was then examined by RT-PCR using a C/T polymorphism we identified at nucleotide 375, which is within the 5′ untranslated region (UTR), for which line SL.RC was heterozygous. Sequencing of the cDNA product showed that only the T allele was expressed (Fig. 1c).
Fig 1.
Expression analysis of four imprinted genes and one nonimprinted gene in differentiated human EG cell derivatives. (a) Chromatogram showing the polymorphism in genomic DNA (gDNA) and monoallelic expression of the imprinted gene TSSC5 as shown by the cDNA from line LV.EB. (b) Chromatogram showing monoallelic expression of H19 in lines SD.EC and SD.EP. The reverse complement is displayed for line SD.EC. (c) Chromatogram showing monoallelic expression of SNRPN in line SL.RC. (d) Preferential allele expression of IGF2 in the EG lines LV.EB and SL.RC. Genomic DNA lane is denoted by the letter G, The lane marked + indicates that the RT reaction was performed in the presence of reverse transcriptase, and the lane marked − represents reactions performed in the absence of the enzyme. Analysis of IGF2 in line LV.EB was performed with the ApaI polymorphism, and in line SL.RC with the dinucleotide repeat polymorphism. Both polymorphisms are found in exon 9 (ref. 17). Allele ratio quantitation was performed with a PhosphorImager. (e) Chromatogram showing biallelic expression of the nonimprinted gene NAP2 in line SL.RC.
Finally, IGF2 was examined by using either of two transcribed polymorphisms, an ApaI polymorphism and a dinucleotide repeat polymorphism, both of which are in exon 9 (17). Two cell lines were heterozygous for at least one of the polymorphisms, LV.EB and SL.RC. Both showed partial relaxation of imprinting, with an allelic ratio 4:1 in line LV.EB and 5:1 in line SL.RC (Fig. 1d), consistent with a level often seen in normal human tissues (7, 18). Finally, as a negative control, we examined the nonimprinted gene NAP2, by RT-PCR using a G/A polymorphism we identified at nucleotide 2166, which is within the 3′ UTR, and which was heterozygous in line SL.RC. Sequencing of the cDNA product of this gene showed equal expression of the two alleles, and an identical pattern to genomic DNA (Fig. 1e), as expected for a nonimprinted gene.
Normal Methylation of an ICR in Human EG Cell-Derived Lineages.
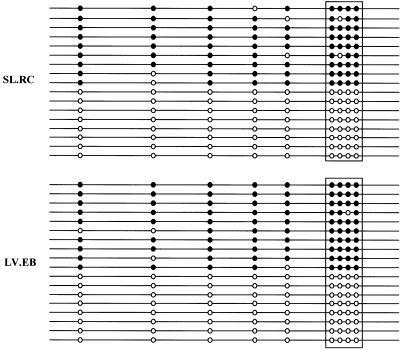
Imprinting is mediated in part by differential methylation of G+C-rich regions termed ICRs. We examined the ICR upstream of the human H19 gene, which when methylated prevents binding of the insulator protein CTCF, allowing access of the H19 enhancer on both alleles to the IGF2 promoter (19), regulating both H19 and IGF2 imprinting (20). We performed bisulfite sequencing analysis of CTCF binding site 1 within the H19 ICR, which shows monoallelic methylation in normal cells and biallelic methylation in cells with loss of imprinting of IGF2 (10). Both EG cell lines examined, SL.RC and LV.EB, showed normal methylation, with equal representation of methylated and unmethylated alleles (Fig. 2). This result was also consistent with the relatively normal imprinting of IGF2 observed in these two EG lines. In summary, human EG cells showed predominantly monoallelic expression of four genes, monoallelic methylation of the H19 ICR, and normal biallelic expression of a nonimprinted gene.
Fig 2.
Normal half-methylation of a CTCF binding site in the H19 ICR, analyzed by bisulfite DNA sequencing. PCR products were cloned and 20 randomly selected clones were sequenced for each EG line. Methylated CpG sites are depicted by filled circles, and the unmethylated CpG sites as open circles. The boxed area represents the CTCF core binding site 1 (ref. 10). (Upper) Line SL.RC. (Lower) Line LV.EB.
An in Vitro Mouse Model of Genomic Imprinting.
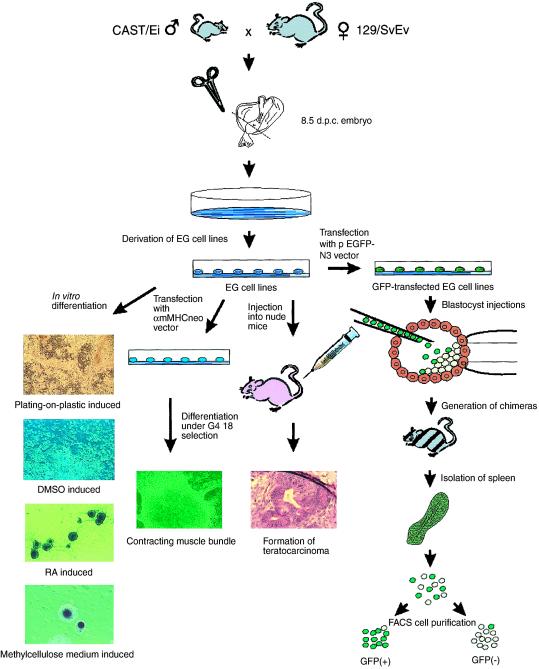
Currently there is no in vitro mouse model using EG or ES cells for the analysis of differentiation-dependent epigenetic modification, which would be a valuable adjunct to studies of the epigenetics of cultured human stem cells. Such a model would require the ability to distinguish parental alleles, so that allele-specific expression could be measured directly, as well as allele-specific methylation or allele-specific chromatin modifications. Two particular advantages of a mouse model are (i) one could isolate populations of pure undifferentiated stem cells; and (ii) one could reintroduce these cells into blastocysts after experimental manipulation, which obviously cannot be done with human cells. We therefore generated EG cell lines from an interspecific mouse cross (129/SvEv × CAST/Ei). The experimental strategy is summarized in Fig. 3, and it allows differentiation in vitro by a variety of mechanisms, including targeted differentiation using a selectable construct, and differentiation in vivo by injection into the blastocyst to generate chimeric mice. Forty EG cell lines were derived from primordial germ cells of 8.5-day embryos, as determined by colony morphology and positive alkaline phosphatase staining (Fig. 3 and data not shown), and four of these lines were characterized in detail (termed SJEG-1, -2, -7, and -15). These EG cell lines formed embryoid bodies after in vitro differentiation (Fig. 3) and teratocarcinomas in nude mice (Fig. 3), and they generated chimeric mice when injected into the blastocyst of C57BL/6 mice, with subsequent germ-line transmission.
Fig 3.
Experimental design for generating mouse EG cells from an interspecific cross. Day 8.5 (129/SvEv × CAST/Ei)F1 embryos were dissected near the base of the allantois to initiate primordial GC cultures from which EG cell lines were established, which was confirmed by s.c. injection into athymic nude mice to form teratocarcinomas, and by blastocyst injection to generate chimeric mice capable of germ-line transmission. The EG cell lines could be induced to differentiate in vitro by any of several methods, including transfection with a vector selectable after cardiomyocyte differentiation. RA, retinoic acid. In addition, cells differentiated in vivo in chimeric mice could be flow-sorted by prior transfection with a GFP-containing vector.
To distinguish the two alleles of imprinted genes in these EG cell lines, we identified transcribed polymorphisms distinguishing 129/SvEv and CAST/Ei in four imprinted genes, Snrpn, Kvlqt1, Igf2, and H19, as well as the nonimprinted gene L23mrp as a negative control. For each gene, an assay for allele-specific expression was then developed. Snrpn normally shows preferential expression of the paternal allele in mice. In these mouse EG lines, Snrpn showed equal biallelic expression before differentiation, and preferential expression of the paternal allele after differentiation (Fig. 4 a and b). The manner of in vitro differentiation was immaterial for inducing preferential expression of one allele, and similar results were seen after differentiation induced by DMSO (Fig. 4a), after plating in methylcellulose (Fig. 4b), after treatment with retinoic acid (data not shown), or by spontaneous differentiation on plastic in the absence of a feeder cell layer (data not shown). Kvlqt1 also showed biallelic expression before differentiation and preferential expression of the maternal allele after differentiation by these agents (data not shown), the same as is seen in normal mice of this genetic background (21). We also transfected EG cells with a vector containing the neo selectable marker gene under the control of a mouse α-cardiac myosin heavy chain gene promoter (14). Clones of transfected EG cells remained undifferentiated, and differentiation of transfected EG cells under G418 selection produced a network of rhythmically contracting myocyte bundles in culture (Fig. 3). Examination of these cells for allele-specific expression showed preferential allele expression similar to that seen when other differentiation approaches were used, but with a slightly greater ratio of allele-specific expression. For example, Kvlqt1 achieved a 9:1 ratio of maternal to paternal allele expression after cardiac myocyte-specific differentiation in vitro (Fig. 4c). Thus, establishment of imprinting was due to differentiation itself, and not to the specific methods used to induce it.
Fig 4.
Partial imprinting after differentiation induced by the following: DMSO, Snrpn analyzed by SSCP-PCR (a); culture in methylcellulose, Snrpn analyzed by SSCP-PCR (b); or transfection with αmMHCneo and selection for cardiac myocytes, Kvlqt1 analyzed by SSCP-PCR (c). In each case there is equal biallelic expression before differentiation and preferential expression of the paternal (Snrpn) or maternal (Kvlqt1) allele after differentiation.
To verify that the changes in imprinting we observed in vitro also occurred during natural differentiation in vivo, we took advantage of the pluripotency of our EG cell lines to generate mouse chimeras. To purify cells derived from these EG cells after in vivo differentiation in chimeric mice, we first transfected EG cells with a vector containing a modified GFP gene under the control of the cytomegalovirus (CMV) promoter. We then injected the cells into C57BL/6 blastocysts, which were introduced into pseudopregnant mice and allowed to develop to term. Spleens were removed from chimeras, and the EG-derived GFP-positive cells were purified by fluorescence-activated cell sorting (FACS) to 99% homogeneity. Purity of EG-derived cells isolated from the chimeric mice was confirmed by measuring the allele ratio in genomic DNA for polymorphisms that distinguish the two strains (data not shown). Analysis of imprinting of EG-derived cells isolated after in vivo differentiation in chimeric mice indicated that all of the imprinted genes studied showed the same pattern of allele-specific expression found after in vitro differentiation. However, after in vivo differentiation, the degree of allele-specific expression for some genes was nearly complete. Thus, Kvlqt1 showed equal biallelic expression after transfection of the pEGFP-N3 vector and before blastocyst injection, and monoallelic expression of the maternal allele after in vivo differentiation in three different chimeric mice (Fig. 5a). Snrpn exhibited predominant expression of the paternal allele (4:1 ratio) after in vivo differentiation (Fig. 5b). As a control, L23mrp showed equal biallelic expression after in vitro or in vivo differentiation (data not shown). Thus, in vivo differentiation of EG cells, like in vitro differentiation, led to preferential expression of a specific parental allele.
Fig 5.
Nearly complete imprinting of EG cells after differentiation in vivo in mouse chimeras. EG cells were transfected with the pEGFP-N3 vector and injected into blastocysts of C57BL/6 mice, and spleen cells from the chimeras were purified by fluorescence-activated cell sorting. (a) Kvlqt1 analyzed by SSCP-PCR. (b) Snrpn analyzed by SNuPE. (c) Igf2 analyzed by SSCP-PCR; the upper band is a nonspecific product of SSCP as indicated in the parental genomic DNA lanes.
However, in mouse EG cells, Igf2 and H19 did not show a normal imprinting pattern after differentiation. Both genes exhibited equal biallelic expression in undifferentiated EG cells, and preferential expression (3:1) of one allele of Igf2 after differentiation, but the normal pattern of maternal and paternal alleles was reversed. Real-time PCR indicated that both alleles showed increased expression but that this activation was limited in the paternal allele. Furthermore, Igf2 showed monoallelic expression of the maternal allele in one chimeric mouse and nearly monoallelic expression (>10:1) in two others (Fig. 5c). This reversal of Igf2 imprinting in mouse EG cells is similar to the reversal of Igf2 imprinting (in both undifferentiated and differentiated cells) observed earlier in mouse ES cells by Dean et al. (4), although those authors did not see the consistent transition from biallelic to monoallelic expression on differentiation reported here. We also found that H19 also showed monoallelic expression of the paternal allele in chimeric mice, the same allele preferentially expressed after in vitro differentiation (data not shown). Consistent with that result, the H19 ICR was unmethylated before differentiation and methylated on the maternal allele after differentiation (data not shown).
Discussion
In summary, we have observed monoallelic expression and methylation of imprinted genes in human EG cell derivatives. Three of four imprinted genes showed monoallelic expression, and the fourth, IGF2, showed partially relaxed imprinting but nevertheless at a ratio of 4:1 to 5:1, consistent with a level often seen in normal human cells (7, 18). Nevertheless it may be important to ascertain IGF2 imprinting status before transplantation of a given EG-derived cell line. In addition, DNA methylation at the H19 ICR was normal. We cannot exclude the possibility that the imprinting pattern is reversed in human EG cells, even though the imprinted genes show preferential allelic expression and methylation. However, this question could not be addressed, because of an explicit ban on any examination of the parental tissues of the human EG cells, imposed by the Institutional Review Board. It will be useful to examine parental origin if this ban is lifted. Our results with human EG cells also indicate an important difference in the timing of epigenetic erasure between humans and mice. We found that in human EG cells derived from the gonadal ridge, the epigenetic mark and preferential expression were not erased, a surprising result given mouse studies that suggest these signals are lost by the time of primordial GC colonization of the gonadal ridge. Another contrast between the present work and earlier studies is the absence of epigenetic instability or epigenetic heterogeneity in human EG-derived cells, compared with the mouse. All of the lines showed the same pattern of monoallelic expression and methylation even after separate or repeated culture, and there were no significant differences among the lines. This relative stability may reflect differences between human and mouse cells, or between human EG and human ES cells. It will therefore be important to revisit this issue in ES cells derived from human blastocysts. However, the present study suggests that there may be no significant epigenetic barrier to human EG cell-derived tissue transplantation.
Because of the inherent technical, legal, and ethical limitations of human stem cell research, we also sought to develop a model system using mouse EG cells that would allow one to investigate in detail the nature and consequence of epigenetic marks in these pluripotent cells in an undifferentiated state, as well as through their differentiation both in vitro and in vivo. For these experiments, we chose to isolate EG cells from 8.5-day embryos, on the basis of mouse studies indicating that imprinting erasure is completed after primordial germ cell migration to the gonadal ridge. We were excited to observe biallelic expression of imprinted genes before differentiation, but preferential allele expression after differentiation. Clearly, the mouse EG cells examined here, while pluripotent and capable of forming teratocarcinomas, chimeric mice, and eventual germ-line transmission through germ cells, are not themselves fully reprogrammed germ cells. They must lie somewhere along a pathway of epigenetic erasure that is not yet complete, and in which germ-line reprogramming has not yet fully occurred. Thus, these cells may harbor a latent mark that allows the establishment of monoallelic expression after differentiation. The nature of the mark is unknown, but could include unknown sites of DNA methylation, or histone acetylation or methylation. We hypothesize that once these cells are differentiated, additional chromatin modifications or proteins that recognize these latent marks are present, allowing gene silencing to take place. These cells should serve as a valuable model system to understand epigenetic modifications in stem cells that arise by in vitro culture or after introduction in vivo into the blastocyst or after tissue transplantation.
The ability of mouse EG cells to acquire allele-specific expression on differentiation may also be of value in understanding developmental and tumor-specific modification of genomic imprinting. For example, some genes show tissue-specific imprinting (22, 23), indicating that a latent mark is present early in development, but that mark is manifest only in certain tissues. Similarly, these EG cells harbor a latent mark that is manifest as monoallelic expression after differentiation. While it would be difficult to identify such a latent mark in a whole animal, and the development-specific changes that permit allele-specific silencing, a cell culture system would be ideal for such a purpose. For example, we and our collaborators recently identified BORIS, a paralogue of CTCF that interacts with ICRs (24). BORIS normally shows testis-specific expression and is hypothesized to be involved in epigenetic reprogramming. Whereas CTCF is expressed in both undifferentiated and differentiated mouse EG cells, BORIS is expressed only in the undifferentiated cells (R. S. Lee and A.P.F., unpublished observations). Thus, we can determine what effect ectopic BORIS expression in differentiated EG cells may have on imprinted genes—e.g., by erasing epigenetic marks. Furthermore, many cancers show loss of imprinting (LOI), which in some cases can be reestablished on exposure to certain drugs (25). Similarly, imprinted chromosomes can lose allele-specific expression when transferred to embryonal carcinoma cells, but they regain monoallelic expression on differentiation (26). The mouse EG cells derived here will be valuable experimental tools to investigate this process of differentiation-dependent allele-specific silencing.
Acknowledgments
This work was supported by National Institutes of Health Grant CA54358 (to A.P.F.). Funding for the derivation of the human EG-derived cells in this study was provided by Geron, Inc. Under a licensing agreement between Geron and Johns Hopkins University, the University, M.J.S., and J.D.G. are entitled to a share of sales royalty from Geron. The University, M.J.S., and J.D.G. own stock in Geron, the sale of which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by the University in accordance with its conflict of interest policies. H.U. is a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations
EG, embryonic germ
ES, embryonic stem
ICR, imprinting control region
RT, reverse transcriptase
GFP, green fluorescent protein
SSCP, single-stranded conformational polymorphism
SNuPE, single-nucleotide primer extension
See commentary on page 10243.
References
- 1.Lee J., Inoue, K., Ono, R., Ogonuki, N., Kohda, T., Kaneko-Ishino, T., Ogura, A. & Ishino, F. (2002) Development (Cambridge, U.K.) 129, 1807-1817. [DOI] [PubMed] [Google Scholar]
- 2.Labosky P. A., Barlow, D. P. & Hogan, B. L. (1994) Development (Cambridge, U.K.) 120, 3197-3204. [DOI] [PubMed] [Google Scholar]
- 3.Tada T., Tada, M., Hilton, K., Barton, S. C., Sado, T., Takagi, N. & Surani, M. A. (1998) Dev. Genes Evol. 207, 551-561. [DOI] [PubMed] [Google Scholar]
- 4.Dean W., Bowden, L., Aitchison, A., Klose, J., Moore, T., Meneses, J. J., Reik, W. & Feil, R. (1998) Development (Cambridge, U.K.) 125, 2273-2282. [DOI] [PubMed] [Google Scholar]
- 5.Humpherys D., Eggan, K., Akutsu, H., Hochedlinger, K., Rideout, W. M., 3rd, Biniszkiewicz, D., Yanagimachi, R. & Jaenisch, R. (2001) Science 293, 95-97. [DOI] [PubMed] [Google Scholar]
- 6.Shamblott M. J., Axelman, J., Wang, S., Bugg, E, M., Littlefield, J. W., Donovan, P. J., Blumenthal, P. D., Huggins, G. R. & Gearhart, J. D. (1998) Proc. Natl. Acad. Sci. USA 95, 13726-13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui H., Horon, I. L., Ohlsson, R., Hamilton, S. R. & Feinberg, A. P. (1998) Nat. Med. 4, 1276-1280. [DOI] [PubMed] [Google Scholar]
- 8.Ravenel J. D., Broman, K. W., Perlman, E. J., Niemitz, E. L., Jayawardena, T. M., Bell, D. W., Haber, D. A., Uejima, H. & Feinberg, A. P. (2001) J. Natl. Cancer Inst. 91, 698-703. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto K., Azuma, C., Koyama, M., Ohashi, K., Kamiura, S., Nobunaga, T., Kimura, T., Tokugawa, Y., Kanai, T. & Saji, F. (1995) Nat. Genet. 9, 109-110. [DOI] [PubMed] [Google Scholar]
- 10.Cui H., Niemitz, E. L., Ravenel, J. D., Onyango, P., Brandenburg, S. A., Lobanenkov, V. V. & Feinberg, A. P. (2001) Cancer Res. 61, 4947-4950. [PubMed] [Google Scholar]
- 11.Buehr M. & McLaren, A. (1993) Methods Enzymol. 225, 58-76. [DOI] [PubMed] [Google Scholar]
- 12.Szabo P. & Mann, J. R. (1994) Development (Cambridge, U.K.) 120, 1651-1660. [DOI] [PubMed] [Google Scholar]
- 13.Allen N. D., Barton, S. C., Hilton, K., Norris, M. L. & Surani, M. A. (1994) Development (Cambridge, U.K.) 120, 1473-1482. [DOI] [PubMed] [Google Scholar]
- 14.Klug M. G., Soonpaa, M. H., Koh, G. Y. & Field, L. J. (1996) J. Clin. Invest. 98, 216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer-Sam J. (1994) PCR Methods Appl. 3, S48-S50. [DOI] [PubMed] [Google Scholar]
- 16.Shamblott M. J., Axelman, J., Littlefield, J. W., Blumenthal, P. D., Huggins, G. R., Cui, Y., Cheng, L. & Gearhart, J. D. (2001) Proc. Natl. Acad. Sci. USA 98, 113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rainier S., Johnson, L. A., Dobry, C. J., Ping, A. J., Grundy, P. E. & Feinberg, A. P. (1993) Nature (London) 362, 747-749. [DOI] [PubMed] [Google Scholar]
- 18.Sakatani T., Wei, M., Katoh, M., Okita, C., Wada, D., Mitsuya, K., Meguro, M., Ikeguchi, M., Ito, H., Tycko, B. & Oshimura, M. (2001) Biochem. Biophys. Res. Commun. 283, 1124-1130. [DOI] [PubMed] [Google Scholar]
- 19.Holmgren C., Kanduri, C., Dell, G., Ward, A., Mukhopadhya, R., Kanduri, M., Lobanenkov, V. & Ohlsson, R. (2001) Curr. Biol. 11, 1128-1130. [DOI] [PubMed] [Google Scholar]
- 20.Ohlsson R., Renkawitz, R. & Lobanenkov, V. (2001) Trends Genet. 17, 520-527. [DOI] [PubMed] [Google Scholar]
- 21.Jiang S., Hemann, M. A., Lee, M. P. & Feinberg, A. P. (1998) Genomics 53, 395-399. [DOI] [PubMed] [Google Scholar]
- 22.Lee M. P., Hu, R. J., Johnson, L. A. & Feinberg, A. P. (1997) Nat. Genet. 15, 181-185. [DOI] [PubMed] [Google Scholar]
- 23.Kishino T., Lalande, M. & Wagstaff, J. (1997) Nat. Genet. 15, 70-73. [DOI] [PubMed] [Google Scholar]
- 24.Loukinov D. I., Pugacheva, E., Vatolin, S., Pack, S. D., Moon, H., Chernukhin, I., Mannan, P., Larsson, E., Kanduri, C., Vostrov, A. A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barletta J. M., Rainier, S. & Feinberg, A. P. (1997) Cancer Res. 57, 48-50. [PubMed] [Google Scholar]
- 26.Mitsuya K., Meguro, M., Sui, H., Schulz, T. C., Kugoh, H., Hamada, H. & Oshimura, M. (1998) Genes Cells 3, 245-255. [DOI] [PubMed] [Google Scholar]