Abstract
cDNA array technology has proven to be a powerful way to monitor global changes in gene expression patterns. Here, we present an approach that extends the current utility of cDNA arrays to allow the evaluation of the relative roles of transcription and mRNA turnover in governing gene expression on a global basis, compared with current individual gene-by-gene analyses. This method, which involves comparison of large-scale hybridization patterns generated with steady-state mRNA versus newly transcribed (nuclear run-on) RNA, was used to demonstrate the importance of mRNA turnover in regulating gene expression following several conditions of stress.
Keywords: transcription, stress response, gene expression
Following exposure to harmful agents such as oxidants, genotoxins, metabolic poisons, or extreme temperatures, mammalian cells implement a series of adaptive responses that are collectively known as the “cellular stress response.” At the center of such adaptation are precise alterations in expression of a multitude of genes. In addition to the long-established transcriptional mechanisms regulating gene expression, the involvement of posttranscriptional gene regulation events, in particular alterations in mRNA stability (reviewed in refs. 1 and 2), is increasingly recognized. In mammalian cells, where mRNA half-life can range from 20 min to 24 h, different processes regulating mRNA turnover have been described over the past two decades, some leading to selective mRNA stabilization, others enhancing mRNA degradation (3, 4). Even small differences in mRNA half-life provide a highly effective means of dramatically altering the abundance of a given mRNA and consequently the amount of protein expressed (5).
Here, we introduce a use of cDNA arrays for the assessment of global gene transcription, using a modified nuclear run-on protocol coupled with analysis of changes in steady-state mRNA levels. Following exposure of human cells to stressful stimuli, we compared the information obtained from cDNA arrays assessing gene transcription with that obtained from cDNA arrays in which steady-state mRNA levels were measured. Through these comparisons, we were able to ascertain the relative importance of transcriptional regulation and mRNA turnover in influencing global gene expression patterns during the cellular stress response. Use of this methodology provided systematic confirmation that changes in transcription influenced the altered expression of many stress-regulated genes, about 47% in the current analysis. However, it also revealed that mRNA stabilization and destabilization significantly influenced the expression of approximately 53% of stress-regulated genes, underscoring the prominent role of mRNA turnover as a major contributor in the implementation of stress-altered gene expression patterns.
Materials and Methods
Cell Culture and Treatment.
H1299 cells were cultured as described (6). Cells were treated with 36 μM prostaglandin A2 (PG), exposed to short-wavelength UV light (UVC) at 20 J/m2, or subjected to heat shock (HS) at 43°C for 2 h. PG and actinomycin D were from Sigma.
Northern and Standard Nuclear Run-On Assays.
Northern blot analysis was performed as described (7). Probes for Northern blots were prepared by random-primed labeling of PCR-amplified, gel-purified cDNA fragments, using Klenow enzyme and [α-32P]dATP. Control hybridizations to detect 18S rRNA [using a radiolabeled oligomer (7)], β-actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were performed to monitor the evenness of loading and transfer of Northern blot samples. For mRNA half-life assessments, actinomycin D (2 μg/ml) was added and total RNA prepared at the times indicated. mRNA half-lives were assessed after measurement of Northern blot signals, normalization to 18S rRNA signals, and plotting on logarithmic scales.
For standard nuclear run-on assay, nascent RNA was obtained through elongation of established transcription complexes in the presence of [α-32P]UTP (7). Radiolabeled RNA [107 counts per minute (cpm) per ml] was hybridized to dot-blotted DNA strips (≈1 μg of purified PCR products per dot) and washed using 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS (37°C, 30 min), 2× SSC containing 200 ng/ml RNase A to degrade unbound RNA and reduce background on the filters (37°C, 30 min), and 1× SSC/0.1% SDS (42°C, 30 min). Analysis of signals on dot-blotted β-actin, GAPDH, and pUC18 control plasmid served to assess the equality and specificity of the nuclear run-on hybridizations.
Array Analysis.
The preparation of total RNA and the synthesis of complementary radiolabeled DNA by reverse transcription in the presence of [α-33P]dCTP have been described (8, 9).
Radiolabeled, newly transcribed RNA was prepared from 108 nuclei of H1299 cells in a nuclear run-on reaction as described (7), except that [α-33P]UTP was used. Radiolabeled RNA was used at 107 cpm per ml in hybridization buffer [10 mM Tris⋅HCl (pH 7.5)/4× SSC/1 mM EDTA/0.1% SDS/2× Denhardt's solution/40% formamide/100 μg/ml yeast tRNA/20 μg/ml poly(A)/2 μg/ml human Cot-1 DNA]. Hybridizations were carried out for 72 h at 42°C; washes were performed using 2× SSC/0.1% SDS (37°C, 30 min), 2× SSC containing 200 ng/ml RNase A (37°C, 30 min), and 1× SSC/0.1% SDS (42°C, 30 min). cDNA array membranes (www.grc.nia.nih.gov/branches/rrb/dna/array.htm) were visualized for analysis by using a PhosphorImager (Molecular Dynamics), and were quantitated as described (8, 9).
Results and Discussion
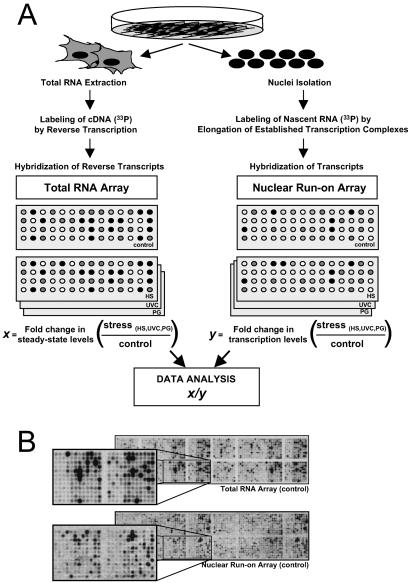
Non-small cell lung carcinoma H1299 cultures were exposed to each of three stresses, HS, UVC, or PG, after which comparisons of expressed transcripts were carried out as depicted in Fig. 1A. First, using total cellular RNA and standard cDNA array methodologies (Fig. 1A Left), we compared steady-state mRNA expression patterns in cells that were either left untreated or exposed to each stress. This comparison of signals (control vs. stress-treated mRNAs) on cDNA arrays was used to ascertain the relative changes in steady-state mRNA levels (x). Following similar stress treatments, we isolated nuclei from H1299 cells and performed a modified nuclear run-on (NRO) protocol (see Materials and Methods) to examine fold differences in nascent, newly transcribed RNA (Fig. 1A Right, y); examples of similar array hybridizations that use nuclear run-on-generated transcripts have been reported (10, 11). Representative Total RNA arrays and nuclear run-on arrays illustrating hybridization signals from untreated populations are shown in Fig. 1B. The cDNA arrays used, encompassing 1,152 genes, have been described (www.grc.nia.nih.gov/branches/rrb/dna/array.htm). It was essential to use cDNA arrays for this analysis, because total RNA signals hybridized to the sense strand, whereas nuclear run-on RNA signals hybridized to the antisense strand. It is important to note that postspliced RNA signals (on total RNA arrays) cannot be directly compared with prespliced RNA (on NRO arrays) to infer decay, because of the differences in length of each run-on transcript and the uneven incorporation of the radiolabel. This problem is circumvented through the comparison of fold differences of each type of RNA signals, as explained below.
Fig 1.
Strategy to study stress-triggered alterations in mRNA turnover by using cDNA arrays. (A) Following treatment of H1299 cells with either UVC (20 J/m2), HS (43°C, 2 h), or PG (36 μM), two sets of radiolabeled probes were prepared: total RNA (Left), isolated 8 h after stimulation, was used to prepare radiolabeled cDNA through reverse transcription in the presence of [α-33P]dCTP; and newly transcribed RNA (Right), prepared 3 h after stimulation, was radiolabeled in a nuclear run-on reaction in the presence of [α-33P]UTP. x, fold difference in signal intensity (stress-treated relative to untreated) for a given gene on total RNA arrays; y, fold difference in signal intensity (stress-treated relative to untreated) for a given gene on nuclear run-on arrays. (B) Representative cDNA arrays to illustrate the hybridization signals corresponding to either steady-state RNA levels (total RNA array) or newly transcribed RNA (nuclear run-on array), in untreated populations. Correlation coefficients serving to evaluate the internal reproducibility of the signals on total RNA arrays and nuclear run-on arrays were 0.99 in each case (not shown). The array (ref. 8; www.grc.nia.nih.gov/branches/rrb/dna/array.htm) includes cDNA segments typically comprising the 3′ UTR and coding region. cDNA fragments range from 500 to 2,000 bp.
Examples of genes with different x and y values are shown in Table 1. By comparing xi (Fold change in steady-state levels) and yi (Fold change in transcription levels) for each given gene (i), we obtained scores (Score, right column) for each gene and treatment [HS(xi/yi), UVC(xi/yi), PG(xi/yi)]. Such scores reflected the relative contribution of mRNA turnover on the stress-triggered modifications in gene expression. A complete list of x values, y values, and scores can be found in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. According to xi and yi values, we classified the genes on the cDNA array into five groups. Group I comprises genes whose expression do not change with stress (841 genes, approximately 73% of genes on this array). This gene group exhibits scores approximating 1, as x ≈ 1, y ≈ 1, and hence x/y ≈ 1. Included in Group I is also a small set of genes exhibiting no change in steady-state levels, whose transcription either increased (y > 2) or decreased (y < 0.7). Group II comprises genes whose total mRNA levels increase after stress (typically x > 2 with each stress) and their transcription rates also increase (y > 1.5); consequently, their scores remain close to 1. This group constitutes 8.5% of genes (98 genes) on the array. Group III encompasses genes whose total mRNA levels decrease following stress (typically x < 0.6) and their transcription rates are similarly reduced (y < 0.7); accordingly, their scores are also close to 1. This group comprises 4% of genes (46 genes) on the cDNA array. By contrast, Group IV genes showed sizeable increases in total RNA abundance (x > 2), while their transcription rates remained unchanged (y ≈ 1, and occasionally lower than 1). Because mRNA abundance increases while transcription remains largely unchanged, these genes are considered to be up-regulated through mRNA stabilization. This group comprises 3% of the genes on the array (34 genes), and their scores are typically >2. Conversely, Group V genes show marked decreases in steady-state levels (x < 0.6), while their transcription rates are typically unchanged (y ≈ 1, and occasionally higher than 1). These genes, comprising approximately 11.5% of the total (133 genes), are down-regulated through mRNA destabilization; they exhibit scores <0.6.
Table 1.
Classification of array genes according to stress-triggered changes in expression
| Gene
|
Total RNA array, fold change in steady-state levels (stress/ control = x) | Nuclear run-on array, fold change in transcription levels (stress/ control = y) | Score (x/y) | Percent of total genes
|
Full gene name
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HS | UVC | PG | HS | UVC | PG | HS | UVC | PG | |||
| Group I. mRNA expression unchanged | Scores ≈1 | 73% | |||||||||
| PALM | 0.88 | 1.14 | 1.16 | 1.00 | 1.11 | 0.89 | 0.88 ± 0.13 | 1.03 ± 0.10 | 1.30 ± 0.13 | Paralemmin | |
| IL14 | 1.08 | 0.97 | 1.00 | 0.81 | 0.83 | 0.96 | 1.33 ± 0.14 | 1.16 ± 0.07 | 1.04 ± 0.30 | IL-14 | |
| PTPN13 | 1.13 | 1.47 | 1.25 | 1.28 | 1.46 | 1.34 | 0.88 ± 0.18 | 1.01 ± 0.13 | 0.93 ± 0.10 | ESTs, similar to APO-1/CD95 (Fas)-associated phosphatase | |
| CCNE1 | 1.18 | 1.30 | 1.30 | 1.34 | 1.18 | 0.95 | 0.88 ± 0.15 | 1.1 ± 0.36 | 1.37 ± 0.25 | Cyclin E | |
| Group II. mRNA levels increased through enhanced transcription | Scores ≈1 | 8.5% | |||||||||
| HSP70 | 2.86 | 1.36 | 3.00 | 2.94 | 1.61 | 2.78 | 0.97 ± 0.05 | 0.84 ± 0.08 | 1.07 ± 0.13 | Heat shock 70-kDa protein | |
| CASP2 | 1.60 | 2.85 | 2.06 | 1.45 | 1.40 | 1.63 | 1.11 ± 0.12 | 2.03 ± 0.10 | 1.26 ± 0.18 | Caspase 2 | |
| IGFBP4 | 2.43 | 1.76 | 3.16 | 2.30 | 2.15 | 2.61 | 1.06 ± 0.12 | 0.82 ± 0.12 | 1.21 ± 0.16 | IGFP-binding protein | |
| GGCX | 1.90 | 2.61 | 1.81 | 2.00 | 1.74 | 1.60 | 0.95 ± 0.19 | 1.50 ± 0.31 | 1.13 ± 0.23 | γ-glutamyl carboxylase | |
| Group III. mRNA levels decreased through reduced transcription | Scores ≈1 | 4% | |||||||||
| RPL7A | 0.58 | 0.67 | 0.38 | 0.58 | 0.63 | 0.52 | 1.00 ± 0.17 | 1.06 ± 0.24 | 0.73 ± 0.13 | Ribosomal protein L7a | |
| FRK | 0.72 | 0.78 | 0.47 | 0.73 | 0.67 | 0.65 | 0.98 ± 0.09 | 1.16 ± 0.09 | 0.72 ± 0.11 | Fyn-related kinase | |
| NDUOR1 | 0.68 | 0.73 | 0.69 | 0.73 | 0.62 | 0.73 | 0.93 ± 0.05 | 1.17 ± 0.11 | 0.94 ± 0.05 | ESTs, similar to NADH-ubiquinone oxidoreductase PDSW subunit | |
| ST3GALVI | 0.65 | 0.75 | 0.73 | 0.83 | 0.56 | 0.71 | 0.78 ± 0.03 | 1.33 ± 0.11 | 1.02 ± 0.07 | α-2,3-sialyltransferase | |
| Group IV. mRNA levels increased through mRNA stabilization | Scores > 2 | 3% | |||||||||
| p21 | 2.35 | 3.05 | 2.88 | 1.41 | 0.92 | 1.02 | 1.66 ± 0.24 | 3.31 ± 0.12 | 2.81 ± 0.31 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | |
| DEFB2 | 2.39 | 3.89 | 2.45 | 0.82 | 1.06 | 1.14 | 2.91 ± 0.55 | 3.66 ± 0.48 | 2.14 ± 0.34 | Defensin beta 2 | |
| PRKCSH | 1.74 | 2.83 | 2.05 | 1.39 | 1.01 | 1.11 | 1.25 ± 0.36 | 2.79 ± 0.25 | 1.84 ± 0.14 | Protein kinase C substrate 80K-H | |
| TRADD | 2.89 | 3.84 | 3.73 | 0.72 | 1.05 | 1.05 | 4.00 ± 0.21 | 3.65 ± 0.71 | 3.55 ± 0.52 | TNFRSF1A-associated via death domain; TRADD | |
| TIMP | 1.83 | 5.53 | 2.68 | 0.71 | 0.83 | 1.05 | 2.57 ± 0.20 | 6.66 ± 0.92 | 2.55 ± 0.52 | Tissue inhibitor of metalloproteinase 1 | |
| ZNF24 | 2.64 | 3.74 | 2.81 | 1.29 | 0.80 | 0.99 | 2.04 ± 0.91 | 4.67 ± 0.15 | 2.83 ± 0.73 | ESTs, weakly similar to zinc finger protein ZNF191 | |
| FLOT1 | 1.75 | 2.38 | 2.13 | 1.01 | 0.70 | 0.70 | 1.73 ± 0.51 | 3.40 ± 0.27 | 3.04 ± 0.72 | Flotillin 1 | |
| NFIC | 1.64 | 2.71 | 2.44 | 1.30 | 1.00 | 1.05 | 1.26 ± 0.08 | 2.71 ± 0.14 | 2.32 ± 0.13 | Nuclear factor I/C (CCAAT-binding transcription factor) | |
| Group V. mRNA levels decreased through mRNA destabilization | Scores < 0.6 | 11.5% | |||||||||
| TFDP1 | 0.52 | 0.54 | 0.58 | 1.48 | 1.22 | 1.07 | 0.35 ± 0.02 | 0.44 ± 0.02 | 0.54 ± 0.03 | Transcription factor DP-1 | |
| ADRB2 | 0.59 | 0.52 | 0.55 | 0.97 | 0.85 | 0.97 | 0.60 ± 0.01 | 0.61 ± 0.02 | 0.56 ± 0.11 | Adrenergic, β-2-, receptor, surface | |
| Cyclin D1 | 0.75 | 0.61 | 0.63 | 1.19 | 1.15 | 1.62 | 0.63 ± 0.03 | 0.53 ± 0.11 | 0.38 ± 0.06 | Cyclin D1 | |
| EDNRA | 0.50 | 0.46 | 0.52 | 1.27 | 1.19 | 0.98 | 0.39 ± 0.03 | 0.38 ± 0.15 | 0.53 ± 0.02 | Endothelin receptor type A | |
| UBL1 | 0.50 | 0.50 | 0.56 | 1.43 | 1.10 | 1.07 | 0.34 ± 0.01 | 0.45 ± 0.12 | 0.52 ± 0.05 | Ubiquitin-like 1 (sentrin) | |
| PGY1 | 0.51 | 0.91 | 0.43 | 1.30 | 1.23 | 1.07 | 0.39 ± 0.04 | 0.73 ± 0.14 | 0.40 ± 0.10 | P-glycoprotein 1/MDR1 | |
| USP8 | 0.55 | 0.73 | 0.54 | 1.30 | 1.22 | 1.17 | 0.42 ± 0.04 | 0.59 ± 0.20 | 0.46 ± 0.07 | Ubiquitin specific protease 8 | |
| RPIA | 0.45 | 0.59 | 0.52 | 1.35 | 1.21 | 1.07 | 0.33 ± 0.06 | 0.48 ± 0.09 | 0.48 ± 0.03 | Ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) | |
Group I, genes whose steady-state mRNA levels are unchanged following stress (0.6 < x < 2); genes exhibit no changes in transcription (y ≈ 1), and scores are consequently ≈1. Group II, genes displaying elevated steady-state mRNA levels following stress (x > 2), and enhanced transcription rates (y > 1.5); gene scores are typically ≈1. Group III, genes with reduced steady-state mRNA levels following stress (x < 0.6), and diminished transcription rates (y < 0.7); gene scores are ≈1. Group IV (mRNA stabilization group), genes displaying elevated steady-state mRNA levels following stress (x > 2), but exhibiting unchanged transcription rates (0.7 < y < 1.5); genes present scores >2. Group V (mRNA destabilization group), genes with diminished steady-state mRNA levels following stress (x < 0.6), but presenting unchanged transcription rates (0.7 < y < 1.5); genes present scores <0.6. Scores are shown as means ± SEM from three independent experiments.
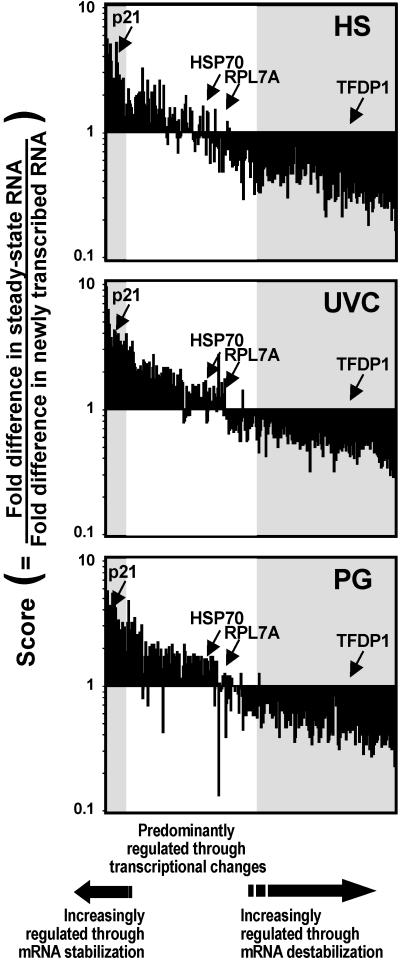
Depicted in the histograms of Fig. 2 are the 311 genes belonging to Groups II–V (whose expression was altered by stress, 132 up-regulated, 179 down-regulated) aligned on the basis of their score values. Genes are arranged from highest (Fig. 2, left) to lowest (right) average score, calculated from the three stresses. Each bar represents a different gene, and the position of a given gene is the same in the three graphs. The shaded region on the left of each graph highlights genes with the highest scores (arbitrarily set at >2, on average), whose stress-mediated up-regulation involves mRNA stabilization. The shaded region on the right highlights genes with the lowest scores (arbitrarily set at <0.6, on average), whose stress-mediated down-regulation is significantly influenced by mRNA destabilization. The remaining ≈46% of stress-regulated genes, with scores between 0.6 and 2, are genes whose expression is largely due to alterations in gene transcription. Importantly, arrangement of the genes on the basis of their scores reveals a smooth continuum, as for the majority of genes, both changes in transcription and turnover contribute to their altered expression. Scores therefore provide an indication of the relative contribution of transcriptional and posttranscriptional mechanisms of gene regulation: for a given stress-regulated gene, the closer the score is to 1, the more transcription contributes to changes in its expression; the lower the score, the greater mRNA destabilization contributes to decreasing its expression; the higher the score, the greater mRNA stabilization contributes to elevating its expression.
Fig 2.
Global assessment of stress-triggered alterations in mRNA turnover relative to stress-triggered alterations in gene transcription. Alignment of the stress-regulated 311 genes according to their score values, beginning with the genes with highest average scores on the left, then genes with progressively lower average scores toward the right. Shaded regions on the left highlight genes with scores >2 (encoding stress-stabilized mRNAs); shaded regions on the right highlight genes with scores <0.6 (encoding stress-destabilized mRNAs). Middle, white regions encompass genes with scores close to 1, whose stress-regulated expression is primarily modulated by changes in transcription. Representative genes from Groups II–V are shown.
Moreover, in the three graphs, each gene occupies the same position, yet the global profiles remain relatively constant among the three stresses. Thus, a gene with a high score after a given stress generally exhibits high scores with the other two stresses. Similarly, low-score genes under one stress condition typically present low scores under the other two stress conditions, and intermediate-score genes (≈1) are also observed consistently among the three stresses. Although some exceptions were noted (e.g., CASP2, NF1C, etc.), gene scores generally showed similar trends—that is, all three were higher than 2, all three were lower than 0.6, or all three were ≈1. These findings illustrate the notion that, in general, genes that are transcriptionally regulated by a given stress are also likely to be transcriptionally regulated by another stress. Genes whose mRNAs are subject to regulation by altered turnover following exposure to a given stress are also likely to undergo altered mRNA turnover after exposure to a different stress. In addition, these findings underscore the view that distinct stress agents can share similar pathways for regulating the expression of a given gene. A corollary of this hypothesis is that a given stress-regulated gene may exert a similar function in the cell, regardless of the stress, and therefore its regulation may be shared among the different stressful stimuli.
Fig. 3 depicts standard Northern blots and nuclear run-on assays that were carried out to validate the accuracy of the gene expression changes identified through cDNA array analysis. Northern blotting served to corroborate changes in total RNA arrays and standard nuclear run-on assays served to confirm the changes in transcription as assessed in nuclear run-on arrays. As with differences in signal intensity on Northern blots, which are generally more pronounced than those observed on standard cDNA arrays (12), nuclear run-on assays also showed more accentuated differences than those of nuclear run-on arrays. In keeping with the nuclear run-on array data, the transcription rates of genes in Groups I, IV, and V remained unchanged following stress (Fig. 3).
Fig 3.
Validation of stress-triggered changes in steady-state and nascent mRNA levels. H1299 cells, treated as described in Fig. 1, were collected at various time intervals for either Northern blot (Left) or nuclear run-on (Right) analyses. Control hybridizations to detect 18S rRNA, β-actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were performed to monitor the evenness of loading and transfer of Northern blot samples, whereas control hybridizations to dot-blotted β-actin, GAPDH, and insert-less plasmid pUC18 served to assess the equality and specificity of the nuclear run-on hybridizations.
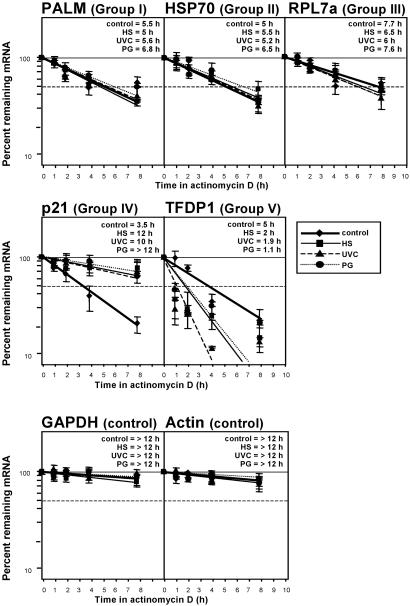
As further confirmation of cDNA array results, direct mRNA half-life assessments in each gene expression category (Groups I–V, Table 1; Fig. 3) were carried out using kinetic studies of mRNA decay. Depicted in Fig. 4 are representative mRNA half-life determinations obtained by an actinomycin D-based approach. As anticipated, genes belonging to Groups I–III, whose stress-regulated expression was predicted not to be influenced by altered mRNA turnover, indeed showed comparable mRNA half-lives under all treatment conditions (Fig. 4). By contrast, the mRNA encoding p21 (representative from Group IV) displayed a longer half-life following exposure to stress than under untreated conditions; the p21 mRNA was thus stabilized by stress, as anticipated (Fig. 4). Similarly, the TFDP1 mRNA (encoding a representative gene from Group V) was destabilized with stress, as it exhibited the expected reduction in half-life following stress treatments (Fig. 4). These findings further validate the usefulness of this methodology in predicting the relative contribution of mRNA turnover to influencing gene expression changes, such as those occurring during the stress response.
Fig 4.
Validation of stress-regulated changes in mRNA half-life. Following treatment of H1299 cells as described in Fig. 1, actinomycin D (2 μg/ml) was added and total RNA collected at various time intervals. Half-lives of mRNAs encoding genes of interest from each gene expression group (Table 1) were assessed after measuring Northern blot signals, normalizing them to 18S rRNA signals, and plotting them on a logarithmic scale. mRNA half-life is defined as the time period required to achieve a reduction of 50% (dashed horizontal line) of transcript. Data represent the means ± SEM of four independent experiments.
Changes in mRNA stability are regulated by the combined action of cis elements and transacting factors. The cis elements are specific mRNA sequences typically present in the 3′ untranslated region (UTR) of labile transcripts. The best characterized sequences, containing an abundance of adenine and uracil (AU-rich elements or AREs), are found in many mRNAs including those that encode cytokines, growth factors, proto-oncogenes, and cell cycle regulatory proteins (13–15). A number of transacting factors (notably RNA-binding proteins) have been shown to selectively bind to AREs and other instability-conferring sequences within labile mRNAs. Among them, HuR and hnRNP D/AUF1 have been most extensively investigated in their role as regulators of mRNA stability (6, 16–18). As anticipated, sequence analysis of stress-stabilized and stress-destabilized mRNAs (Table 1, Fig. 2) revealed numerous AREs, although no specific common features could be identified (data not shown). The absence of discernable shared elements is not unexpected because the RNA sequence requirements for the formation of mRNA–protein complexes are generally more flexible than those governing, for example, DNA–transcription factor interactions. Thus, a given RNA-binding protein can bind to a collection of RNA substrates of related sequences with varying relative affinities; conversely, a given mRNA sequence can likewise be the target of many RNA-binding proteins in vitro. Our findings highlight the complexity of mRNA half-life regulation in response to cell stimulation, which implicates many RNA-binding proteins and mRNA instability determinants (11, 19).
In summary, using cDNA arrays, we have developed a powerful approach to systematically investigate the contribution of gene transcription and mRNA turnover on the pattern of expressed genes. By comparing stress-triggered changes in steady-state mRNA levels with changes in gene transcription rates, we were able to ascertain the extent to which stress-regulated modifications in gene expression were subject to transcriptional and posttranscriptional control. Indeed, transcriptional regulation affects a significant group of genes in response to UVC, PG, and HS. However, mRNA stabilization was found to be a major contributor for the increased expression of many genes and mRNA destabilization an essential means of decreasing the expression of an even larger group of genes. The conclusions drawn through use of this approach greatly strengthen the view that changes in mRNA turnover constitute a major regulatory mechanism in the control of gene expression. As illustrated for the stress response, this methodology can be applied to the large-scale investigation of mRNA turnover events that underlie an increasingly recognized number of physiologic and pathologic situations.
Supplementary Material
Acknowledgments
We are grateful to C. Cheadle and D. Teichberg for their assistance with the array analysis, and we thank N. J. Holbrook, P. J. Morin, and E. Westin for helpful discussions.
Abbreviations
HS, heat shock
PG, prostaglandin A2
UVC, short-wavelength UV light
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sachs A. B. (1993) Cell 74, 413-421. [DOI] [PubMed] [Google Scholar]
- 2.Brennan C. M. & Steitz, J. A. (2001) Cell. Mol. Life Sci. 58, 266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peltz S. W., Brewer, G., Bernstein, P., Hart, P. A. & Ross, J. (1991) Crit. Rev. Eukaryotic Gene Expression 1, 99-126. [PubMed] [Google Scholar]
- 4.Wilusz C. J., Wormington, M. & Peltz, S. W. (2001) Nat. Rev. Mol. Cell Biol. 2, 237-246. [DOI] [PubMed] [Google Scholar]
- 5.Ross J. (1995) Microbiol. Rev. 59, 423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S., Wang, W., Wilson, G. M., Yang, X., Brewer, G., Holbrook, N. J. & Gorospe, M. (2000) Mol. Cell. Biol. 20, 7903-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorospe M., Wang, X. & Holbrook, N. J. (1998) Mol. Cell. Biol. 18, 1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayne M., Cheadle, C., Soldan, S. S., Cermelli, C., Yamano, Y., Akhyani, N., Nagel, J. E., Taub, D. D., Becker, K. G. & Jacobson, S. (2001) J. Virol. 75, 11641-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka T. S., Jaradat, S. A., Lim, M. K., Kargul, G. J., Wang, X., Grahovac, M. J., Pantano, S., Sano, Y., Piao, Y., Nagaraja, R., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 9127-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meininghaus M., Chapman, R. D., Horndasch, M. & Eick, D. (2000) J. Biol. Chem. 275, 24375-24382. [DOI] [PubMed] [Google Scholar]
- 11.Keene J. (2000) Proc. Natl. Acad. Sci. USA 98, 7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi M., Miura, K., Iwao, H. & Yamanaka, S. (2001) Genomics 71, 34-39. [DOI] [PubMed] [Google Scholar]
- 13.Miller A. D., Curran, T. & Verma, I. M. (1984) Cell 36, 51-60. [DOI] [PubMed] [Google Scholar]
- 14.Shaw G. & Kamen, R. (1986) Cell 46, 659-667. [DOI] [PubMed] [Google Scholar]
- 15.Chen C. Y. & Shyu, A.-B. (1995) Trends Biochem. Sci. 20, 465-470. [DOI] [PubMed] [Google Scholar]
- 16.Ma W. J., Cheng, S., Campbell, C., Wright, A. & Furneaux, H. (1996) J. Biol. Chem. 271, 8144-8151. [DOI] [PubMed] [Google Scholar]
- 17.Wang W., Furneaux, H., Cheng, H., Caldwell, M. C., Hutter, D., Liu, Y., Holbrook, N. J. & Gorospe, M. (2000) Mol. Cell. Biol. 20, 760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W., Lin, S., Caldwell, M. C., Furneaux, H. & Gorospe, M. (2000) EMBO J. 19, 2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guhaniyogi J. & Brewer, G. (2001) Gene 265, 11-23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.