Abstract
Tyrosyl–tRNA synthetase (TyrRS) from Escherichia coli was engineered to preferentially recognize 3-iodo-l-tyrosine rather than l-tyrosine for the site-specific incorporation of 3-iodo-l-tyrosine into proteins in eukaryotic translation systems. The wild-type TyrRS does not recognize 3-iodo-l-tyrosine, because of the bulky iodine substitution. On the basis of the reported crystal structure of Bacillus stearothermophilus TyrRS, three residues, Y37, Q179, and Q195, in the l-tyrosine-binding site were chosen for mutagenesis. Thirty-four single amino acid replacements and 16 of their combinations were screened by in vitro biochemical assays. A combination of the Y37V and Q195C mutations changed the amino acid specificity in such a way that the variant TyrRS activates 3-iodo-l-tyrosine 10-fold more efficiently than l-tyrosine. This engineered enzyme, TyrRS(V37C195), was tested for use in the wheat germ cell-free translation system, which has recently been significantly improved, and is now as productive as conventional recombinant systems. During the translation in the wheat germ system, an E. coli suppressor tRNATyr was not aminoacylated by the wheat germ enzymes, but was aminoacylated by the E. coli TyrRS(V37C195) variant with 3-iodo-l-tyrosine. After the use of the 3-iodotyrosyl–tRNA in translation, the resultant uncharged tRNA could be aminoacylated again in the system. A mass spectrometric analysis of the produced protein revealed that more than 95% of the amino acids incorporated for an amber codon were iodotyrosine, whose concentration was only twice that of l-tyrosine in the translation. Therefore, the variant enzyme, 3-iodo-l-tyrosine, and the suppressor tRNA can serve as an additional set orthogonal to the 20 endogenous sets in eukaryotic in vitro translation systems.
To expand the variety of amino acids incorporated into proteins, many efforts have been made to engineer the translation systems. The synthesis of proteins containing unnatural amino acids, alloproteins (1), is expected to advance protein science and technology profoundly (1–6). In normal protein synthesis according to the natural genetic code, the 20 canonical amino acids are assigned to the sense codons through the specific recognition of an amino acid by its cognate aminoacyl–tRNA synthetase (aaRS), followed by the transfer of the amino acid to its corresponding tRNA(s) by the aaRS. Therefore, for the synthesis of alloproteins, the unnatural amino acids should be attached to the tRNA by some means.
One method of attaching an unnatural amino acid to its adapter tRNA is chemical acylation (7), by which the aminoacylated forms of tRNAs corresponding to a special codon, such as amber codons (2, 8, 9), four-base codons (10, 11), and artificial codons with unnatural bases (12, 13), have actually been synthesized before their use in the cell-free translation (2, 4, 8–10, 12, 13) or in Xenopus oocytes (5). This adapter tRNA is engineered so it is not aminoacylated again by any aaRS involved in the translation system, to prevent the misincorporation of a canonical amino acid at the positions for the unnatural amino acid. This manner of using the adapter tRNA only one time, as well as the low productivity of chemical acylation, has limited the yield of the alloproteins with site-specific incorporation of unnatural amino acids.
Some unnatural amino acids, with chemical structures similar to those of the canonical amino acids, are misrecognized by the wild-type aaRSs, and are thus attached to tRNAs more efficiently, as compared with chemical acylation. By using this feature, unnatural amino acids have been incorporated into proteins at the positions for the corresponding canonical amino acids in vivo (1, 3, 14, 15) and in vitro (6, 16). In addition, some nonphysiological conditions have been found to facilitate the aminoacylation of tRNA with unnatural amino acids (13). The site-specific incorporation of an unnatural amino acid has been attempted by using a wild-type aaRS. It was actually incorporated at the desired position in the protein, although the canonical amino acid cognate to the enzyme was also incorporated to some extent at the same position (1, 14, 15, 17).
The engineering of an aaRS is necessary to use the unnatural amino acids with completely different chemical structures from those of the canonical amino acids (18–22). Wang et al. (22) have created two variants of Methanococcus jannaschii tyrosyl–tRNA synthetase (TyrRS) that are highly specific to O-methyl-l-tyrosine (21) and to l-3-(2-naphthyl)alanine (22). These amino acids have actually been incorporated at amber positions in the Escherichia coli translation system by these variant enzymes together with the cognate suppressor tRNATyr. Notably, the M. jannaschii TyrRS does not recognize any E. coli tRNA, and the suppressor tRNA is not recognized by any E. coli aaRS. The employment of such a tRNA⋅aaRS pair that is “orthogonal” to the host system (23–25), along with the engineering of the aaRS amino acid specificity, allows the reacylation of the tRNA with the unnatural amino acid in the translation. This finding promises a large yield of alloproteins containing unnatural amino acids at the desired positions.
This approach, which has been useful in prokaryotic translation, remained to be extended to the eukaryotic system, in which a prokaryotic tRNA and aaRS may be used as an orthogonal pair. E. coli glutaminyl-tRNA synthetase and its specific suppressor tRNA have been reported to be orthogonal to the eukaryotic systems (25, 26). For TyrRS and tRNATyr, the yeast aaRSs hardly aminoacylate the E. coli amber suppressor tRNATyr (27, 28), whereas the yeast tRNAs, including tRNATyr, are not aminoacylated significantly by E. coli TyrRS (29). Thus, these pairs for glutamine and tyrosine are excellent candidates for use in eukaryotic translation.
In the present study, we created a variant E. coli TyrRS that attaches 3-iodo-l-tyrosine to tRNA more efficiently than l-tyrosine, to construct a eukaryotic translation system in which 3-iodo-l-tyrosine is genetically assigned to the amber codon. Notably, 3-iodo-l-tyrosine is not recognized by the wild-type TyrRSs from prokaryotes or eukaryotes (30–32). We used a wheat germ cell-free translation system to produce the alloproteins; its productivity, as well as that of the E. coli cell-free system (33, 34), has been increased to produce milligram quantities of proteins (35). Finally, the exclusive incorporation of iodotyrosine at an amber site was shown by mass spectrometric analyses of the produced alloproteins.
Materials and Methods
Preparation of the Wild-Type TyrRS and Its Variants.
The DNA fragment encoding E. coli TyrRS was amplified from the genomic DNA by the PCR, using a GeneAmp PCR system 9700 (Applied Biosystems). It was cloned within the NdeI–HindIII sites of the pET26b vector (Novagen) to generate the plasmid pET-TYR-His. TyrRS, with an additional His tag at the C terminus, was overproduced from this plasmid in E. coli BLR(DE3) cells (Novagen). The site-directed mutagenesis of TyrRS was performed by PCR with mutagenic primers, with pET-TYR-His as the template. The sequences of the mutant genes were confirmed by using an ABI PRISM 377 DNA sequencer (Applied Biosystems). The variant enzymes were also overproduced in the BLR(DE3) cells. For rough screening, the wild-type TyrRS and the variant enzymes were purified by chromatography on Ni–nitrilotriacetic acid (NTA) agarose (Qiagen). For further analyses, the purification was carried out by chromatography on Ni-NTA agarose, followed by FPLC using a UNO-Q column (Bio-Rad Laboratories).
In Vitro Analyses of the Aminoacylation and Pyrophosphate-Exchange Reactions.
E. coli tRNATyr was prepared by runoff transcription using T7 RNA polymerase (36). The aminoacylation was performed at 37°C in 50 μl of buffer A (100 mM Tris⋅HCl, pH 7.6/15 mM MgCl2/40 mM KCl/1 mM DTT) containing tRNATyr (60 μM), 5 units/ml of yeast inorganic pyrophosphatase (Sigma), 1 mM ATP, and either l-tyrosine (0.1 or 0.2 mM) or 3-iodo-l-tyrosine (0.2 or 1 mM). TyrRS was used at various concentrations (0.05–5 μM). After a 6-min reaction, the Biomol Green reagent (100 μl; Biomol, Plymouth Meeting, PA) was added to the reaction mixture to stop the reaction and to detect the free phosphates produced by the pyrophosphatase from the pyrophosphates released on the amino acid activation. The absorbance at 630 nm was measured by using a microplate reader, Model 550 (Bio-Rad). A more quantitative assay of the produced free phosphate was performed by using 2-amino-6-mercapto-7-methylpurine ribonucleoside, according to Lloyd et al. (37). The aminoacylation was performed here under the conditions described above, except the concentration of tRNATyr was 0.1 mM. The pyrophosphate-exchange reaction was analyzed according to the method described (38). The reaction was performed in buffer A containing 4 mM ATP, 4 mM 32P-labeled pyrophosphates (New England Nuclear), TyrRS (0.05–3 μM), and either l-tyrosine (3–16 μM) or 3-iodo-l-tyrosine (44–291 μM).
Analysis of the aminoacylated tRNAs was carried out by acidic PAGE (39) using a 7% denaturing gel. On the basis of the above-described aminoacylation assays, the aminoacylation was performed in buffer A containing 4 mM ATP, 50 nM TyrRS, 3 μM tRNATyr, and either l-tyrosine or 3-iodo-l-tyrosine (0.2 mM each) at 37°C for 3 min, which reflected the differences in the initial velocity. The tRNA in the gel was stained with toluidine blue O (Kanto Chemical, Tokyo). The tRNA mixture (37 μM; Sigma) from the wheat germ extract was aminoacylated in buffer A containing 2.5 mM ATP, 0.22 μM E. coli TyrRS, and 6.1 μM 14C-labeled l-tyrosine (16.9 GBq/mmol) at 37°C for 30 min in a 50-μl reaction. The used tRNA fraction (7.4 μg) can accept the labeled l-tyrosine at 3,400 dpm above the background level of this assay (150 dpm).
Cell-Free Translation and Mass Spectrometric Analyses.
The PROTEIOS cell-free protein synthesis kit (Toyobo, Tokyo) was used for the wheat germ cell-free translation. This kit contains the 20 amino acids, each at a concentration of 0.3 mM. 3-Iodo-l-tyrosine was used at the maximum practical concentration (0.6 mM), which was limited by the volume of the stock solution that could be added to the reaction mixture. The dialysis system reaction was performed according to Madin et al. (35). The products were analyzed on 12% NuPAGE Bis–Tris gels with Mes-running buffer (both purchased from Invitrogen). Quantitative analyses were performed by using an image analyzer, FLA-2000 (Fuji), to measure the radioactivity of the products or the fluorescence of the products stained with the Cypro-tangerine protein gel stain (Molecular Probes). Within 3 days, 0.3-μg yields of the ras(Am) products, including 3-iodo-l-tyrosine, were synthesized in a 30-μl reaction mixture. The liquid chromatography electrospray mass spectrometry (LC-MS) analysis and the tandem MS sequencing were performed as described (13). The occupancy of iodotyrosine at the amber position was determined for the ras(Am) products on the basis of the relative abundance in the mass spectrometric analysis and the peak areas in liquid chromatogram by UV detection for the IY and Y fragments.
Results
Engineering the Amino Acid Specificity of E. coli TyrRS.
E. coli TyrRS was engineered to recognize 3-iodo-l-tyrosine more efficiently than l-tyrosine, based on the crystal structure of the complex of a tyrosyl–adenylate analogue and Bacillus stearothermophilus TyrRS (40), which is 56% identical to its E. coli counterpart in the amino acid sequence. In this crystal structure, the hydroxyl group of the l-tyrosine moiety of the bound ligand hydrogen bonds with Tyr-34 and Asp-176, and the two meta positions of the tyrosine ring are located in the vicinity of Gly-70, Thr-73, and Asn-123, and of Tyr-34, Gln-173, and Gln-189, respectively. These residues are highly conserved among the prokaryotic TyrRS species. A structure modeling showed that, when one of the meta hydrogens is replaced by iodine, the bulky iodine atom clashes with the main chain at Gly-70, and this steric hindrance cannot be alleviated by mutating the Gly-70, Thr-73, or Asn-123 residue. On the other hand, the steric hindrance caused by a similar substitution at the other meta position may be circumvented by the mutation(s) of Tyr-34, Gln-173, and/or Gln-189, which correspond to Tyr-37, Gln-179, and Gln-195 in E. coli TyrRS, respectively.
We searched for 3-iodo-l-tyrosine-specific variants of E. coli TyrRS in two steps; variants with a single mutation at position 37, 179, or 195 were first prepared to find the mutations with effects that are favorable for 3-iodo-l-tyrosine, and these mutations were then combined in the variant enzymes to enhance their effects. In the first step, the screening of the single-mutation variants was performed on the basis of their activities with l-tyrosine and 3-iodo-l-tyrosine, which were roughly determined by detecting the pyrophosphates released on the amino acid activation. The activities were classified into four levels, according to the minimal enzyme concentrations required for the detection (Table 1) in reactions where the substrate concentration was 0.1 mM for l-tyrosine and 1 mM for 3-iodo-l-tyrosine. This l-tyrosine concentration was an excess, as compared with the reported Km value (30), whereas that for 3-iodo-l-tyrosine was a practical limit imposed by the preparation of the stock solution.
Table 1.
Activities of TyrRS variants with tyrosine and 3-iodo-l-tyrosine
| Enzyme | Amino acid
|
Enzyme | Amino acid
|
||
|---|---|---|---|---|---|
| Tyrosine | 3-Iodo-l-tyrosine | Tyrosine | 3-Iodo-l-tyrosine | ||
| Wild type (Y37, Q179, Q195) | +++ | − | F195 | − | − |
| L195 | ++ | − | |||
| I195 | + | − | |||
| F37 | +++ | − | M195 | + | − |
| H37 | +++ | + | V195 | ++ | − |
| A37 | ++ | +++ | S195 | +++ | + |
| V37 | +++ | +++ | T195 | +++ | − |
| I37 | ++ | +++ | A195 | +++ | + |
| L37 | +++ | +++ | Y195 | ++ | − |
| M37 | +++ | +++ | H195 | +++ | − |
| G37 | ++ | ++ | N195 | ++ | + |
| S37 | +++ | ++ | K195 | − | − |
| D195 | + | − | |||
| A179 | ++ | − | E195 | +++ | − |
| E179 | − | − | C195 | ++ | + |
| N179 | ++ | − | W195 | − | − |
| S179 | ++ | − | R195 | − | − |
| Y179 | − | − | G195 | ++ | − |
The symbols “+++,” “++,” and “+” represent the activities detected with enzyme concentrations of 0.05 μM, 0.5 μM, and 5 μM, respectively, and “−” represents no activity detected with the enzyme, up to a 5 μM concentration.
The Tyr residue at position 37 was mutated to nine different amino acids (Table 1). The Phe and His mutations yielded no more than the lowest level of activity for 3-iodo-l-tyrosine. On the other hand, the mutations to nonpolar amino acids (Ala, Val, Ile, Leu, and Met) caused the highest level of activity for 3-iodo-l-tyrosine. By contrast, the Gly and Ser variants exhibited only an intermediate level of activity for the 3-iodo-l-tyrosine. The level of activity for l-tyrosine was slightly decreased by the Ala, Gly, and Ile mutations, and was not changed by the other six. Thus, for two mutations, Y37A and Y37I, the activity for 3-iodo-l-tyrosine was ranked higher than that for l-tyrosine. Nevertheless, when the two substrates were used at the same concentrations (0.2 mM) in the assay, the activity for l-tyrosine remained higher than that for the analogue, even with the Y37A and Y37I mutations (data not shown).
The Gln-173 residue of Bacillus TyrRS, corresponding to Gln-179 in E. coli TyrRS, is involved in the recognition of the l-tyrosine amino group in the crystal structure (40), and its replacements by other amino acids actually reduce the activity with l-tyrosine (41). We mutated Gln-179 to Ala, and then to several different types of amino acids (Glu, Asn, Ser, and Tyr); all of these mutations severely impaired the activity for l-tyrosine without providing any activity for 3-iodo-l-tyrosine (Table 1). This position is probably also involved in the recognition of the l-tyrosine amino group in E. coli TyrRS. Therefore, we did not further explore the effects of mutations at position 179.
For position 195, Gln was replaced by the other 19 amino acids. The variant with Pro at this position could not be prepared in a soluble form. Of the eighteen variants, the Ser, Ala, Asn, and Cys variants exhibited the activity for 3-iodo-l-tyrosine, even though their activity levels were the lowest. These mutations changed the activity for l-tyrosine differently. The activity levels for l-tyrosine of the four 3-iodo-l-tyrosine-accepting variants were either the same as (Ser and Ala) or slightly lower than (Asn and Cys) that of the wild-type enzyme.
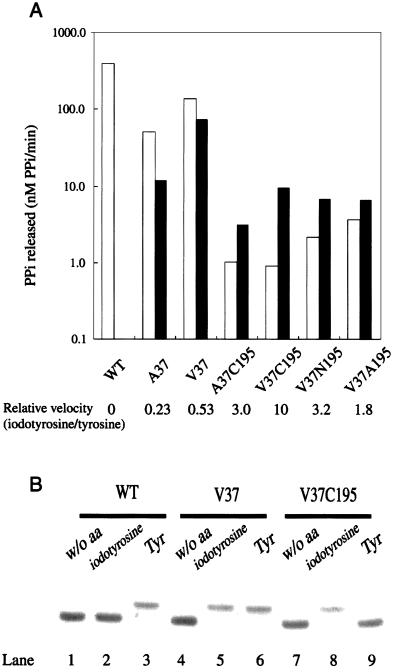
In the second step, we combined the mutations at position 37 (to Ala, Val, Ile, and Leu) and those at position 195 (to Ser, Ala, Asn, and Cys), because their effects were favorable for 3-iodo-l-tyrosine. The 16 possible variant enzyme combinations were examined for the activities for l-tyrosine (0.2 mM) and 3-iodo-l-tyrosine (1 mM). We first used the above concentration (0.1 mM) for l-tyrosine, and found that the activities of several variants were reduced to near the lowest level. To distinguish these activities more clearly, the l-tyrosine concentration was raised to 0.2 mM. Four combinations, A37C195, V37C195, V37N195, and V37A195, showed significant activity for 3-iodo-l-tyrosine and lower activity for l-tyrosine (Table 2). The variants with these double mutations were subjected to a more detailed examination, in which the release of pyrophosphates on amino acid activation was quantified with 0.2 mM substrate concentrations, for both l-tyrosine and 3-iodo-l-tyrosine, as shown in Fig. 1A, where the effects of two single mutations, A37 and V37, were included for comparison. The four variants actually exhibited higher activity for 3-iodo-l-tyrosine than for l-tyrosine, although they did not activate the tyrosine analogue as efficiently as the V37 variant. Among these variants, TyrRS(V37C195) had the highest activity for the analogue relative to that for l-tyrosine. The kinetic parameters for the pyrophosphate-exchange reaction were determined for the wild-type and V37C195 variant TyrRSs (Table 3). Actually, the TyrRS(V37C195) variant can activate 3-iodo-l-tyrosine 10-fold more efficiently than l-tyrosine, whereas the wild-type enzyme does not activate the analogue to a detectable extent.
Table 2.
Activities of TyrRS variants with tyrosine and 3-iodo-l-tyrosine
| Enzyme | Amino acid
|
Enzyme | Amino acid
|
||
|---|---|---|---|---|---|
| Tyrosine | 3-Iodo-l-tyrosine | Tyrosine | 3-Iodo-l-tyrosine | ||
| Wild type (Y37, Q195) | +++ | − | V37 | +++ | +++ |
| A37A195 | − | − | I37A195 | + | + |
| A37C195 | + | ++ | I37C195 | − | − |
| A37N195 | − | − | I37N195 | − | − |
| A37S195 | + | + | I37S195 | − | − |
| V37A195 | + | ++ | L37A195 | + | − |
| V37C195 | + | ++ | L37C195 | + | + |
| V37N195 | + | ++ | L37N195 | + | − |
| V37S195 | ++ | ++ | L37S195 | − | − |
The symbols “+++” and “++” represent the activities detected with enzyme concentrations of 0.05 μM and 0.25 μM, respectively. “+” represents the activity detected with an enzyme concentration of 0.25 μM, although the activity is lower than that represented by “++.” “−” represents no activity detected with an enzyme concentration of up to 0.25 μM.
Figure 1.
Analyses of the activities of the wild-type TyrRS and its variants for l-tyrosine and 3-iodo-l-tyrosine. (A) The initial velocities of the release of pyrophosphates on amino acid activation in the presence of tyrosine (white bars) and 3-iodo-l-tyrosine (black bars). The initial velocity in the presence of 3-iodo-l-tyrosine relative to that in the presence of l-tyrosine is shown for each enzyme at the bottom. (B) Acidic PAGE analysis of the aminoacylation of tRNATyr by the wild-type TyrRS (lanes 1–3), by TyrRS(V37) (lanes 4–6), and by TyrRS(V37C195) (lanes 7–9) in the presence of l-tyrosine (lanes 3, 6, and 9), 3-iodo-l-tyrosine (lanes 2, 5, and 8), or no amino acid (lanes 1, 4, and 7).
Table 3.
The kinetic parameters for the PPi exchange of tyrosine and 3-iodo-l-tyrosine by the wild-type TyrRS and the VC mutant enzyme
| Enzyme | Tyrosine
|
3-Iodo-l-tyrosine
|
||||
|---|---|---|---|---|---|---|
| kcat, s−1 | Km, μM | kcat/Km, M−1s−1 | kcat, s−1 | Km, μM | kcat/Km, M−1s−1 | |
| Wild-type TyrRS | 12 (0.4) | 5.3 (0.7) | 2.3 × 106 | ND | ND | ND |
| TyrRS(V37C195) | 0.045 (0.005) | 140 (20) | 3.2 × 102 | 0.43 (0.08) | 130 (20) | 3.3 × 103 |
The kinetic parameters were obtained from Lineweaver–Burk plots. The obtained data are the averages of three independent assays. Standard deviations between assays are shown in the parentheses. ND, not determined because of low activity.
To determine whether TyrRS(V37C195) can attach 3-iodo-l-tyrosine to tRNATyr, we analyzed the aminoacylation in the presence of either l-tyrosine or 3-iodo-l-tyrosine at the same concentration (0.2 mM), by acidic polyacrylamide gel electrophoresis (Fig. 1B). When the wild-type enzyme and l-tyrosine were involved in the reaction, the aminoacylated form of the tRNA was detected on the gel (lane 3). By contrast, when the wild-type enzyme and 3-iodo-l-tyrosine were involved, only the uncharged form was detected (lane 2). TyrRS(V37), which activates both l-tyrosine and 3-iodo-l-tyrosine, can attach both of these amino acids to the tRNA (lanes 5 and 6). For TyrRS(V37C195), the aminoacylated form was detected in the presence of 3-iodo-l-tyrosine, whereas only the uncharged form was detected in the presence of l-tyrosine (lanes 8 and 9). Thus, the double mutation, Tyr-37 to Val and Gln-195 to Cys, has conferred the specific 3-iodo-l-tyrosylation on E. coli TyrRS.
Site-Specific Incorporation of 3-Iodo-l-tyrosine into a Protein in a Eukaryotic Cell-Free Protein Synthesis System.
We produced an alloprotein containing 3-iodo-l-tyrosine at the desired position by cell-free translation using a wheat germ extract, which was performed in two manners: a batch reaction, which is convenient for use, but stops within a few hours, and a more productive reaction in a dialysis system, which continues for days (35). The concentration of 3-iodo-l-tyrosine was maximized up to a practical limit for the use in the cell-free translation (0.6 mM), which is twice the concentration of l-tyrosine provided in the translation kit. It has been reported that the TyrRS⋅tRNATyr pair from E. coli hardly cross-acylates with the eukaryotic pairs (27, 29). In addition, our preliminary in vitro aminoacylation assay results showed that E. coli TyrRS cannot aminoacylate a wheat germ tRNA mixture beyond the background level of the assay, suggesting that the E. coli enzyme does not misrecognize either tRNATyr or any other tRNA species present in the extract.
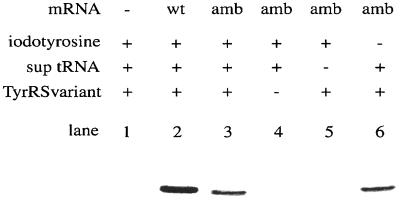
The tyrosine codon at position 32 in the synthetic human c-Ha-Ras gene (42) was replaced by an amber codon (6), which was to be assigned to 3-iodo-l-tyrosine. The ras(WT) or ras(Am) mRNA was translated in the batch reaction, and then the products, labeled with 14C-Leu, were analyzed by SDS/PAGE. The Ras protein of the expected length was produced from both mRNAs, in the presence of 3-iodo-tyrosine, TyrRS(V37C195), and a suppressor tRNATyr from E. coli (Fig. 2, lanes 2 and 3), whereas it was not produced without the mRNA (lane 1). The band intensity of the ras(Am) products on the gel was 30% of that of the ras(WT) products. This amber suppression did not occur without either TyrRS(V37C195) or the suppressor tRNA (lanes 4 and 5). On the other hand, the suppression was observed with a comparable efficiency even in the absence of 3-iodo-l-tyrosine, when the suppressor tRNA and the variant TyrRS were involved in the translation (lane 6). This finding implied that, in the absence of the competing 3-iodo-l-tyrosine, l-tyrosine instead was recognized by TyrRS(V37C195) to a significant extent, and was then incorporated at position 32, causing the amber suppression. We supposed that when l-tyrosine and 3-iodo-l-tyrosine coexist in the translation, 3-iodo-l-tyrosine is preferentially incorporated into the Ras protein at the amber position.
Figure 2.
Synthesis of Ras proteins in the wheat germ cell-free translation. An autoradiogram of an SDS/polyacrylamide gel, with the products labeled with [14C]leucine, is shown. The ras(WT) mRNA, the ras(Am) mRNA, the amber suppressor tRNATyr, and TyrRS(V37C195) involved in the translation are denoted by “wt,” “amb,” “sup tRNA,” and “TyrRS variant,” respectively, on the top of the autoradiogram.
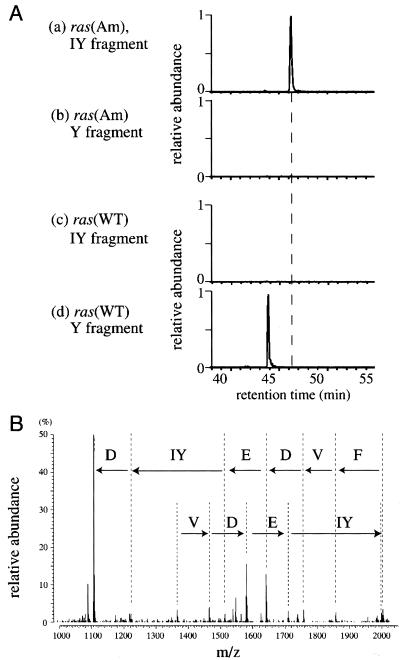
To test this possibility, the ras(Am) and ras(WT) products were analyzed by LC-MS. The Ras proteins were produced in the presence of 3-iodo-l-tyrosine, the suppressor tRNA, and TyrRS(V37C195); the cell-free translation was performed in the dialysis system, to prepare sufficient amounts of the products for the analysis. The purified Ras proteins were digested with Achromobacter Protease I (Lys-C), and the fragments were subjected to LC-MS. Two possible fragments that correspond to the region from Ser-17 to Lys-42 contain iodotyrosine and tyrosine at position 32 (designated as the IY and Y fragments, respectively). Therefore, the amino acids incorporated at position 32 were analyzed on the basis of the mass chromatograms for the IY and Y fragments along the retention time in LC (Fig. 3A).
Figure 3.
The LC-MS analysis of Ras proteins synthesized in the wheat germ cell-free translation. The Y fragment consists of amino acid residues 17–42, SALTIQLIQNHFVDEYDPTIEDSYRK, of the Ras protein. Tyr-32, underlined in the sequence, was replaced by iodotyrosine in the IY fragment. (A) Mass chromatograms of the fragments from the ras(Am) products (charts a and b) and the ras(WT) products (charts c and d) for the theoretical value (the average mass of the [M + 2H]2+ ion) of the IY fragment (charts a and c) or the Y fragment (charts b and d). The relative abundance of the ion with the theoretical average mass value of the indicated fragment was detected by a mass spectrometer, and was plotted against the LC retention time. One unit for the relative abundance is common between charts a and b, and between charts c and d. (B) The tandem mass spectrum of the IY fragment. The partial sequence in the direction from the N terminus to the C terminus reads as Phe, Val, Asp, Glu, iodotyrosine, and Asp in this order.
In the analysis of the ras(Am) products, the IY fragment was strongly observed (chart a). Therefore, the incorporation of 3-iodo-l-tyrosine was efficient in the presence of l-tyrosine in the reaction. The partial sequence of this IY fragment, determined by tandem MS, confirmed that iodotyrosine was actually incorporated at position 32 (Fig. 3B). On the other hand, no Y fragment was detected (chart b), indicating that 3-iodo-l-tyrosine suppressed the misincorporation of l-tyrosine at position 32. Further analysis of LC-MS data showed that any other canonical amino acids were not incorporated at this position (data not shown). The occupancy of iodotyrosine at position 32 was thus estimated to be higher than 95% by the present LC-MS analysis. In contrast, for the ras(WT) products synthesized in the presence of 3-iodo-l-tyrosine, the Y fragment was observed as expected (chart d), but the IY fragment was not detected (chart c). This observation demonstrates that the endogenous TyrRS in the wheat germ extract does not recognize 3-iodo-l-tyrosine, and TyrRS(V37C195) does not recognize the endogenous tRNATyr, because either misrecognition would incorporate 3-iodo-l-tyrosine in place of Tyr.
Discussion
In the present study, we created an E. coli TyrRS variant that is specific to 3-iodo-l-tyrosine, to produce an alloprotein site-specifically containing 3-iodo-l-tyrosine in a eukaryotic cell-free translation system. Here we report of a set comprising an unnatural amino acid, a variant aaRS, and a suppressor tRNA that is orthogonal to the 20 canonical sets of a eukaryotic cell-free system; similar sets were previously developed in the E. coli cell system for O-methyl-l-tyrosine and l-3-(2-naphthyl)alanine by Wang et al. (21, 22).
Wang et al. obtained M. jannaschii TyrRS variants specific to the tyrosine derivatives, by screening a large library of enzyme variants, each with several random mutations at certain positions in the substrate-binding site; these mutated positions were determined on the basis of the crystal structure of B. stearothermophilus TyrRS. We also used this structure for determining the mutated positions in E. coli TyrRS. However, the effects of the mutations at these positions were examined separately from those of the mutations at the other positions. By combining the effects of two mutations that were found to be more favorable for 3-iodo-l-tyrosine than l-tyrosine, we finally obtained a useful TyrRS variant.
Besides these TyrRS variants, the TyrRSs from yeast and E. coli have been engineered for their amino acid specificity. A variant TyrRS from E. coli that recognizes azatyrosine 1.5-fold better than tyrosine has been isolated through a genetic selection from the variants in which the amino acid replacements are not confined to particular positions (43). A yeast TyrRS variant that recognizes 3-iodo-l-tyrosine has been generated by replacing Tyr-43 with Gly (32), but it still recognizes l-tyrosine more efficiently than 3-iodo-l-tyrosine. Tyr 43 in yeast TyrRS corresponds to Tyr-37 in the E. coli counterpart, whose mutations to several types of amino acids were found, in the present study, to evoke various levels of misrecognition of 3-iodo-l-tyrosine. In addition, the amino acid residue, corresponding to Tyr-37 in the E. coli enzyme, of the M. jannaschii TyrRS has been reported to be mutated in its variants specific to O-methyl-l-tyrosine (21) and l-3-(2-naphthyl)alanine (22). This position thus appears to be involved in the discrimination between l-tyrosine and its derivatives. In the E. coli TyrRS, an additional mutation at the second position, position 195, contributed to converting the specificity of the variant enzyme in favor of 3-iodo-l-tyrosine. Similarly, the specificity of the yeast G43 mutant TyrRS could be further changed by an additional mutation at the corresponding position in favor of 3-iodo-l-tyrosine.
The engineering of TyrRS allows the reacylation of the uncharged suppressor tRNA in the cell-free translation. In fact, it was estimated that a single tRNA molecule participates more than twice in the insertion of 3-iodo-l-tyrosine into alloproteins on the ribosome, as judged by the amount of the suppressor tRNA relative to that of the produced alloproteins. On the other hand, in the previous report, no more than 10% of the aminoacylated forms of suppressor tRNAs used in the cell-free translation was estimated to contribute to the final protein yield (6). In the E. coli cells developed by Wang et al. to incorporate either O-methyl-l-tyrosine or l-3-(2-naphthyl)alanine, the reacylation of the uncharged suppressor tRNAs is expected to occur naturally, thus allowing efficient alloprotein production. The advantage of a cell-free system, as has been claimed (16), is that the possibilities of toxicity and difficulties in cellular uptake of unnatural amino acids can probably be circumvented. For such unnatural amino acids that are difficult to use in vivo, it is necessary to obtain their specific aaRS variants by screening on the basis of an in vitro assay, as in the present study.
The usefulness of the incorporation of novel substructures into proteins has been demonstrated with various unnatural amino acids (1–6). Iodine incorporation may facilitate the process of an x-ray crystallographic analysis of proteins, as unnatural nucleotides with iodine atoms have been used for the structure determination of a protein–nucleic acid complex (44). It was recently shown that a phosphorylated 3-iodo-l-tyrosine residue [pTyr(3-I)] in a peptide derived from a growth factor receptor is selectively accepted by an engineered Src homology 2 (SH2) domain, whereas the wild-type SH2 domain binds to a phosphorylated l-tyrosine residue, but not to pTyr(3-I) (K. Saito and S.Y., unpublished data). Thus, 3-iodo-l-tyrosines inserted within cellular signaling proteins may be used to modulate the communication between the proteins.
Eukaryotic translation systems are advantageous for incorporating unnatural amino acids into eukaryotic proteins, because some of them are not properly folded into their native conformation in prokaryotic systems, and this difficulty may be alleviated in eukaryotic translation systems (35). Toward alloprotein synthesis in mammalian cells, nonsense suppression systems have been constructed by the coexpression of a prokaryotic pair of glutaminyl-tRNA synthetase (GlnRS)⋅tRNAGln in the cell (26) or by importing the aminoacylated amber suppressor E. coli tRNATyr into the cell (45). By using these techniques, the variety of translation systems, together with aaRS engineering, must be further expanded to include mammalian cells.
Acknowledgments
We thank Dr. T. Masaki (Ibaraki University) for providing the Achromobacter Protease I used for the LC-MS analysis.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- LC-MS
liquid chromatography-electrospray mass spectrometry
- TyrRS
tyrosyl–tRNA synthetase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Koide H, Yokoyama S, Kawai G, Ha J M, Oka T, Kawai S, Miyake T, Fuwa T, Miyazawa T. Proc Natl Acad Sci USA. 1988;85:6237–6241. doi: 10.1073/pnas.85.17.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noren C J, Anthony-Cahill S J, Griffith M C, Schultz P G. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 3.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht S M. Acc Chem Res. 1992;25:545–552. [Google Scholar]
- 5.Nowak M W, Kearney P C, Sampson J R, Saks M E, Labarca C G, Silverman S K, Zhong W, Thorson J, Abelson J N, Davidson N, et al. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- 6.Yabuki T, Kigawa T, Dohmae N, Takio K, Terada T, Ito Y, Laue E D, Cooper J A, Kainosho M, Yokoyama S. J Biomol NMR. 1998;11:295–306. doi: 10.1023/a:1008276001545. [DOI] [PubMed] [Google Scholar]
- 7.Hecht S M, Alford B L, Kuroda Y, Kitano S. J Biol Chem. 1978;253:4517–4520. [PubMed] [Google Scholar]
- 8.Bain J D, Diala E S, Glabe C G, Dix T A, Chamberlin A R. J Am Chem Soc. 1989;111:8013–8014. [Google Scholar]
- 9.Short G F, 3rd, Golovine S Y, Hecht S M. Biochemistry. 1999;38:8808–8819. doi: 10.1021/bi990281r. [DOI] [PubMed] [Google Scholar]
- 10.Murakami H, Hohsaka T, Ashizuka Y, Sisido M. J Am Chem Soc. 1998;120:7520–7529. [Google Scholar]
- 11.Moore B, Persson B C, Nelson C C, Gesteland R F, Atkins J F. J Mol Biol. 2000;298:195–209. doi: 10.1006/jmbi.2000.3658. [DOI] [PubMed] [Google Scholar]
- 12.Bain J D, Switzer C, Chamberlin A R, Benner S A. Nature (London) 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 13.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, et al. Nat Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 14.Koide H, Yokoyama S, Katayama Y, Muto Y, Kigawa T, Kohno T, Takusari H, Oishi M, Takahashi S, Tsukumo K, et al. Biochemistry. 1994;33:7470–7476. doi: 10.1021/bi00189a055. [DOI] [PubMed] [Google Scholar]
- 15.Kiick K L, Weberskirch R, Tirrell D A. FEBS Lett. 2001;502:25–30. doi: 10.1016/s0014-5793(01)02657-6. [DOI] [PubMed] [Google Scholar]
- 16.Kigawa T, Yamaguchi-Nunokawa E, Kodama K, Matsuda T, Yabuki T, Matusda N, Ishitani R, Nureki O, Yokoyama S. J Struct Funct Genomics. 2001;2:27–33. doi: 10.1023/a:1013203532303. [DOI] [PubMed] [Google Scholar]
- 17.Furter R. Prot Sci. 1998;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kast P, Hennecke H. J Mol Biol. 1991;222:99–124. doi: 10.1016/0022-2836(91)90740-w. [DOI] [PubMed] [Google Scholar]
- 19.Sharma N, Furter R, Kast P, Tirrell D A. FEBS Lett. 2000;467:37–40. doi: 10.1016/s0014-5793(00)01120-0. [DOI] [PubMed] [Google Scholar]
- 20.Döring V, Mootz H D, Nangle L A, Hendrickson T L, de Crecy-Lagard V, Schimmel P, Marliere P. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Brock A, Herberich B, Schultz P G. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Brock A, Schultz P G. J Am Chem Soc. 2002;124:1836–1837. doi: 10.1021/ja012307j. [DOI] [PubMed] [Google Scholar]
- 23.Ohno S, Yokogawa T, Fujii I, Asahara H, Inokuchi H, Nishikawa K. J Biochem (Tokyo) 1998;124:1065–1068. doi: 10.1093/oxfordjournals.jbchem.a022221. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Magliery T J, Liu D R, Schultz P G. J Am Chem Soc. 2000;122:5010–5011. [Google Scholar]
- 25.Kowal A K, Kohrer C, RajBhandary U L. Proc Natl Acad Sci USA. 2001;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drabkin H J, Park H J, RajBhandary U L. Mol Cell Biol. 1996;16:907–913. doi: 10.1128/mcb.16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards H, Schimmel P. Mol Cell Biol. 1990;10:1633–1641. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards H, Trezeguet V, Schimmel P. Proc Natl Acad Sci USA. 1991;88:1153–1156. doi: 10.1073/pnas.88.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakasugi K, Quinn C L, Tao N, Schimmel P. EMBO J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calendar R, Berg P. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]
- 31.Scherberg N H, Seo H, Hynes R. J Biol Chem. 1978;253:1773–1779. [PubMed] [Google Scholar]
- 32.Ohno S, Yokogawa T, Nishikawa K. J Biochem (Tokyo) 2001;130:417–423. doi: 10.1093/oxfordjournals.jbchem.a003001. [DOI] [PubMed] [Google Scholar]
- 33.Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 34.Kim D-M, Swartz J R. Biotechnol Bioeng. 2001;74:309–316. [PubMed] [Google Scholar]
- 35.Madin K, Sawasaki T, Ogasawara T, Endo Y. Proc Natl Acad Sci USA. 2000;97:559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giege R, Yokoyama S. J Mol Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd A J, Thomann H U, Ibba M, Söll D. Nucleic Acids Res. 1995;23:2886–2892. doi: 10.1093/nar/23.15.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekine S, Shimada A, Nureki O, Cavarelli J, Moras D, Vassylyev D G, Yokoyama S. J Biol Chem. 2001;276:3723–3726. doi: 10.1074/jbc.C000756200. [DOI] [PubMed] [Google Scholar]
- 39.Varshney U, Lee C P, RajBhandary U L. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 40.Brick P, Bhat T N, Blow D M. J Mol Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- 41.Lowe D M, Winter G, Fersht A R. Biochemistry. 1987;26:6038–6043. doi: 10.1021/bi00393a014. [DOI] [PubMed] [Google Scholar]
- 42.Ha J-M, Ito Y, Kawai G, Miyazawa T, Miura K, Ohtsuka E, Noguchi S, Nishimura S, Yokoyama S. Biochemistry. 1989;28:8411–8416. doi: 10.1021/bi00447a021. [DOI] [PubMed] [Google Scholar]
- 43.Hamano-Takaku F, Iwama T, Saito-Yano S, Takaku K, Monden Y, Kitabatake M, Söll D, Nishimura S. J Biol Chem. 2000;275:40324–40328. doi: 10.1074/jbc.M003696200. [DOI] [PubMed] [Google Scholar]
- 44.Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Nature (London) 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- 45.Kohrer C, Xie L, Kellerer S, Varshney U, RajBhandary U L. Proc Natl Acad Sci USA. 2001;98:14310–14315. doi: 10.1073/pnas.251438898. [DOI] [PMC free article] [PubMed] [Google Scholar]