Abstract
TH2 clones may produce very variable amounts of IL-4. Among six TH2 clones prepared from homozygous or heterozygous mice in which Gfp replaced the first exon of Il4, a range of patterns of CpG methylation in the Il4/Il13 locus was observed correlating with the degree of expression of IL-4 or green fluorescence protein. Patterns of histone acetylation also showed differences between “high” and “low” TH2 clones. These results indicate that in TH2 cells the Il4 locus may display variable patterns of chromatin accessibility associated with distinct degrees of IL-4 expression. This finding suggests a regulation of IL-4 expression keyed to the function of this cytokine in cell/cell interactions and in the regulation of threshold responses.
Interleukin-4 (IL-4) is a type I cytokine produced by CD4 TH2 cells, NKT cells, basophils, and mast cells. Through its regulation of Ig class switching to IgE and its control of TH2 differentiation, it plays a dominant role in allergic inflammatory conditions (1, 2).
Individual TH2 cells may express IL-4 encoded on one of the two alternative alleles (3–5). We prepared mice in which an “artificial” IL-4 allele was created by replacing a portion of the Il4 gene with a cDNA encoding enhanced green fluorescence protein (GFP) (5). Using CD4 T cells from mice that were heterozygous for Il4 and the “knocked-in” Il4 allele (the Gfp allele), we concluded that the expression of IL-4 or GFP was based on the probability that sets of uniformly differentiated cells would transcribe the gene (5). In recently differentiated TH2 cells, the probability of expression of the two alleles was relatively low and similar; however, clones of TH2 cells prepared from Il4/Gfp CD4 cells had widely different ratios of IL-4 and GFP expression that were stable, suggesting that, with further TH2 differentiation, one or both alleles acquired an increased probability of expression, which then became fixed. We proposed that such differential expression could be caused by differences in accessibility of the Il4 genetic region, such that in TH2 clones in which one allelic product was expressed in a larger proportion of cells than the other that allele would have a more accessible conformation.
Studies of DNase I hypersensitivity (6–9) and methylation (10) of the Il4 gene have revealed substantial differences between TH2- and non-IL-4-producing (TH1) clones. A TH2 clone was shown to have five Il4 DNase I hypersensitive sites (HS I-V) using a promoter probe (7). More recently, a region near HS V, 3′ of exon 4, was observed to become hypersensitive in TH2 cells stimulated with phorbol 12-myristate 13-acetate and ionomycin (8). This site, HS VA, contained Gata3 and NFAT binding elements. Chromatin immunoprecipitation (CHIP) revealed that antibodies to Gata3 and NFATc2 precipitated DNA spanning this region.
Deletion of a highly conserved sequence (CNS-1) in a transgenically expressed human Il4/Il13 intergenic region reduced human IL-4 and IL-13 expression by ≈2/3 (11). CNS-1−/− mice showed attenuated IL-4, IL-13, and IL-5 expression as assessed both in vitro and in vivo (12). This region contains two DNase I HSs expressed in TH2 but not TH1 cells (9).
Expression of a reporter-containing transgene was enhanced if, in addition to the IL-4 promoter, the intergenic region, the second intron hypersensitivity regions, and the region encoding HSs V and VA were included in the construct, suggesting the importance of these sites in the regulation of IL-4 expression (13).
Here we have examined CpG methylation in several TH2 clones, expressing distinctive amounts of IL-4 or GFP, in a series of selected regions across the Il4/Il13 locus, beginning in the Il13 promoter region and ending downstream of the fourth exon of Il4. In addition, we have studied patterns of histone acetylation by using CHIP in some of these regions. These studies reveal that TH2 clones expressing “high” and “low” amounts of IL-4 or GFP show distinct patterns of CpG methylation and histone acetylation.
Materials and Methods
Cell Culture.
Naïve CD4 T cells were purified from lymph nodes of C57BL/6 mice and Gfp homozygous and heterozygous B6/129 mice, as described (5). Single cell cloning was performed (5). For drug treatment, 5-aza-2′-deoxycytidine (Sigma) was added daily during the first 3 days of priming with anti-CD3/anti-CD28 by using irradiated T-depleted spleen cells as antigen-presenting cells.
Flow Cytometry Analysis and Intracellular Staining.
Intracellular GFP and IL-4 expression were analyzed as described (5). To detect nerve growth factor receptor (NGFR), cells were incubated with biotin-labeled anti-human NGFR antibody (PharMingen) in staining buffer for 30 min. After three washes, cells were stained with antigen-presenting cell–streptavidin (PharMingen).
Analysis of Chromosome 11 Copy Number.
Four days after the cells were stimulated with anti-CD3, anti-CD28, and antigen-presenting cells, they were put in IL-2-containing medium for 3 days. Cells were harvested for metaphase chromosomes. A spectrum orange-labeled chromosome 11 painting probe was generated by degenerate oligo-primed–PCR from flow-sorted mouse chromosomes (14).
Retroviral Infection.
Infection of T cells with a retrovirus containing Stat6-VT and NGFR was carried out as described (15).
Methylation-Specific PCR.
Genomic DNA was isolated from 1–2 × 106 cells by using a Wizard Genomic DNA Purification Kit (Promega). Bisulfite treatment of DNA was performed by using a CpGenome DNA Modification Kit (Intergen, Purchase, NY). PCR amplification was performed with AmpliTaq Gold polymerase (Applied Biosystems). Primer sequences are shown in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org.
Bisulfite Sequencing.
The primers were 5′-AGG-TGA-TGT-AGG-TAT-TAA-AAT-TAA-ATA-3′ and 5′-TCC-AAA-TAT-CCC-TAA-AAC-TCC-TTA-AAA-AC-3′. PCR products were cloned into T-vector, and individual clones were picked and sequenced.
CHIP.
CHIP analysis was carried out by using a CHIP Assay kit (United States Biochemical). Soluble chromatin was immunoprecipitated with anti-acetylated H3 IgG (United States Biochemical) or control rabbit IgG (Sigma) overnight at 4°C. After deproteination and reversal of cross-links, the presence of selected DNA sequences was assessed by PCR. Comparisons between samples was carried out by diluting the immunoprecipitated product by 4-fold steps and assessing the degree of dilution required to give a comparable PCR product. Primer sequences are shown in Table 4, which is published as supporting information on the PNAS web site.
Real-Time PCR.
Real-time PCR was done as described (5). Primers for c-maf were 5′-GGT-GAT-CCG-ACT-GA-GCA-GAA-3′ and 5′-TCC-GAC-TCC-AGG-ACG-TGT-CT-3′. The probe was 5′-FAM-TAT-GCC-CAG-TCC-TGC-CGC-TTC-AAG-3′.
Results
Stability of TH2 Clones.
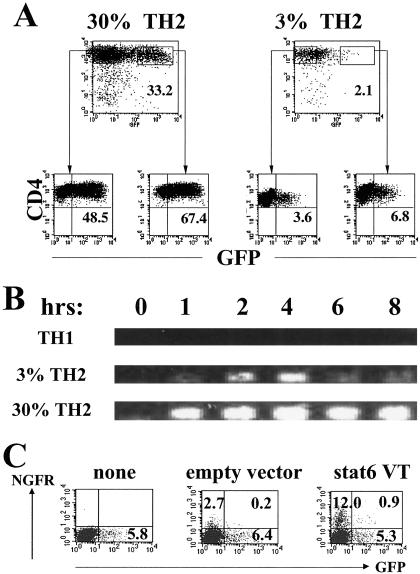
Single naïve CD4+ T cells from Gfp/Gfp mice were cloned in the presence of irradiated T-depleted spleen cells, soluble anti-CD3, anti-CD28, IL-4, anti-IL-12, and anti-IFN-γ. Virtually all of the resultant clones contained cells that became GFP+ when stimulated with immobilized anti-CD3 and anti-CD28; the proportion of GFP+ cells varied from as few as 1–2% to as many as 40% in individual clones. In general, the proportion of cells within a clone that became GFP+ on stimulation was stable.
We chose two TH2 clones for detailed study; one expressed 3–5% GFP+ cells (3% clone) upon stimulation and the other ≈30% GFP+ cells (30% clone) (Fig. 1A). Few, if any, cells from a TH1 clone, prepared in parallel, expressed GFP upon stimulation. No GFP mRNA appeared upon stimulation in the TH1 clone, a modest amount in the 3% TH2 clone and a substantial amount in the 30% TH2 clone (Fig. 1B). Using a chromosome 11-specific probe, all three clones were shown to have two copies of this chromosome per cell.
Fig 1.
Th2 clones from Gfp/Gfp mice. (A) Cells were stimulated with immobilized anti-CD3 and anti-CD28. Twenty-four hours later, they were separated by cell sorting into GFP+ and GFP− populations and cultured for 10 days in IL-2-containing medium. After restimulation with anti-CD3 and anti-CD28, they were analyzed for GFP expression 24 h later. Percentages of GFP-positive cells are indicated. (B) Total RNA was isolated from either unstimulated cells or the cells stimulated by immobilized anti-CD3 and anti-CD28 for indicated periods. Reverse transcription–PCR for GFP mRNA was carried out. The primers were 5′-TGA-ACC-GCA-TCG-AGC-TGA-AG-3′ and 5′-GAT-GTT-GTG-GCG-GAT-CTT-GAA-G-3′. (C) Cells from the 3% Th2 clone were stimulated with anti-CD3 and anti-CD28 in the presence of anti-IL-4, anti-IL-12, and anti-IFN-γ. After 36 h, they were infected with a retrovirus containing Stat6 VT-IRES-NGFR or NGFR only or were uninfected. The cells were maintained under the initial conditions for 3 days. They were washed, placed into IL-2-containing medium for 2 days, and restimulated with anti-CD3 and anti-CD28, and the frequency of GFP+ cells among the NGFR− and NGFR+ populations was determined 24 h later.
Cells from the 3% and 30% clones were stimulated with immobilized anti-CD3/anti-CD28. Twenty-four hours later, we separated GFP+ and GFP− cells by FACS and cultured the cells in IL-2 for 10 days. Upon restimulation, the frequency of GFP+ cells that appeared in the GFP+ and GFP− sorted populations from the 3% clone were similar to each other as was the case with the sorted populations from the 30% clone (Fig. 1A). Thus, the capacity of cells that had been 100% GFP+ to produce GFP upon restimulation was no greater than that of cells that had been 100% GFP−. This finding implies that although only a minority of the cells from the two TH2 clones express GFP upon challenge, all of the cells have the potential to do so.
The stability of the clones also was tested by carrying out additional rounds of stimulation under TH2-inducing conditions. The two clones maintained their respective low and high proportion of GFP+ cells upon subsequent stimulation, indicating that the degree of expression of GFP was a stable property of the cloned cell populations (data not shown). Retroviral introduction of a constitutively active form of Stat6 (Stat6VT) into the 3% clone also failed to increase the proportion of cells that expressed GFP upon stimulation (Fig. 1C).
Expression of Transcription Factors in the TH2 Clones.
We measured levels of transcription factors to determine whether they might account for differences in GFP expression between the 3% and 30% clones. There was no difference between the 3% and the 30% clones in Gata3 and c-maf mRNA, as measured by real-time PCR. Normalized to actin mRNA, the TH1 clone had a level of expression of 0.04 for Gata3 whereas expression in 3% TH2 clone was 0.81, and in the 30% clone it was 0.84. For c-maf, the values were 0.03 for the TH1 clone, 0.83 for the 3% clone, and 0.88 for the 30% clone. A large number of transcription factors were compared by semiquantitative reverse transcription–PCR (Fig. 6A and Table 5, which are published as supporting information on the PNAS web site). We observed differences between the two TH2 clones and the TH1 clone for CIITA, JNK1, Stat6, and T-bet, with the two former being higher in the TH2 clones and the two latter being higher in the TH1 clone. There were limited differences between the 3% and 30% clones. JNK1 was expressed to a somewhat greater degree in the 30% clone and Stat4 and T-bet to a slightly greater degree in the 3% clone. There was no obvious differences between the three clones in expression of cEBP, CBP, JunB, NFATc2, and Stat6 when analyzed by immunoblotting (Fig. 6B). Thus, there were only modest differences in expression of a limited number of transcription factors between the 3% and 30% TH2 clones.
Methylation Patterns in the Il4/Il13 Genetic Region in the Clones from Gfp/Gfp Mice.
We examined patterns of methylation in the Il4/Il13 genetic region in unstimulated cells from the 3% and 30% TH2 clones and the TH1 clone. DNA was treated with bisulfite, converting unmethylated cytidine to uridine but preserving methylcytidine as cytidine. We then used primers that distinguish uridine (thymidine) and cytidine at sites of CpGs in modified DNA to evaluate the degree of methylation within the Il4/Il13 genetic region. We designate the primers complementary to uridine as “unmethylated” (U) primers and primers complementary to cytidine at these sites as “methylated” (M) primers.
We selected six regions to analyze: a region in the Il13 promoter particularly rich in CpGs (13P), a highly conserved region in the Il13/Il4 intergenic region in which two DNase I HSs have been described (CNS-1), two regions in the second intron of the Il4 gene that contain DNase I HSs (HS II and III), a third region in the second intron, and a region of induced DNase I hypersensitivity (VA) 3′ of the fourth exon of the Il4 gene (Fig. 2A). These were assessed with eight sets of primers, one for 13P, two for CNS-1 (CNS-1A and CNS-1B), one for HS II (ITR-A), two for HS III (ITR-B and ITR-C), one for the third region in the second intron (ITR-D), and one for VA.
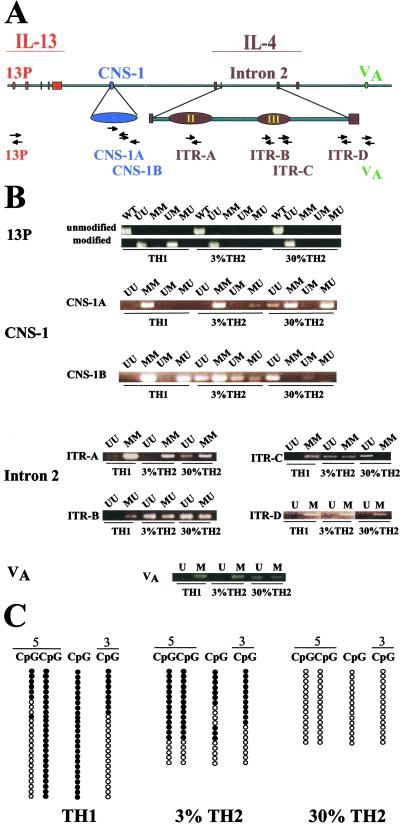
Fig 2.
Methylation pattern of clones from Gfp/Gfp mice. (A) Map of the murine Il4/Il13 locus showing the six regions and eight sets of primer pairs used to study the methylation pattern of the clones. The red area indicates the Il13 promoter and the four Il13 exons. The expanded blue region is the highly conserved Il4/Il13 intergenic CNS-1 region. The brown area indicates the four Il4 exons; the expanded region is Il4 intron 2. II and III indicate the second and the third DNase I HSs in murine Il4 intron 2 (HS II and HS III). The green VA indicates the inducible DNase I HS 5 kb downstream of the fourth exon of Il4. The arrow sets indicate the positions of the sequences to which the primer pairs are complementary. The designation of primer pairs is listed below the arrow sets. (B) Methylation patterns of the clones as measured by methylation-specific PCR. (C) Methylation patterns in CNS-1B by sequencing. Among the four CpGs shown, the first two CpGs were in the sequence complementary to the 5′ CNS-1B primer and the last in the sequence complementary to the 3′ CNS-1B primer. The third CpG lies between these sequences. •, methylated CpGs; ○, unmethylated CpGs.
The results obtained with U and M primers are presented in Fig. 2B. Differences between the TH1 clone and the two TH2 clones were observed by using primer sets specific for 13P, CNS-1A, CNS-1B, ITR-B, ITR-C, and VA.
A striking difference between the 3% and 30% TH2 clones was observed in CNS1-B. The 30% TH2 clone gave a product only with the UU primer set, implying that the three CpGs complementary to the primers were all unmethylated; by contrast, all possible patterns were observed in the 3% clone.
To study the CNS-1B methylation pattern more quantitatively, bisulfite-treated DNA was amplified with primers complementary to sequences upstream of 5′ CNS-1B and downstream of 3′ CNS-1B. A series of molecular clones were sequenced. All 15 sequences from the 30% TH2 clone had Ts rather than Cs at the three CpG sites complementary to the 5′ CNS-1B primer and the 3′ CNS-1B primer (Fig. 2C). In addition, all had T at a CpG site between the sequences complementary to the two primers. Thus, the sequences are consistent with the PCR finding that UU was the major product at CNS-1B. For the 3% TH2 clone, 11/19 sequences would be scored as MM in the PCR method, five as UU, and three as MU; no clones corresponding to the UM product were observed. Of the 11 clones that corresponded to MM sequences, seven had Cs at the intervening CpG site and four had Ts, indicating a further degree of complexity. Thus, direct sequencing confirmed the striking difference between the 3% and 30% TH2 clones. All 26 TH1 sequences had Cs at the second CpG in 5′ CNS-1B and in the intervening CpG, a pattern quite different from seen in either the 3% or 30% TH2 clones.
Other notable differences between the 3% and 30% TH2 clones were observed at CNS1-A (the 3% clone yielded products with the MM and MU primer sets whereas the 30% clone yielded products with the MM, MU, and UU primer sets), ITR-A (the 3% clone yielded a product with the MM primer set only whereas the 30% clone yielded products with the UU and MM primer sets), ITR-C (the 3% clone yielded products with the MM and UU primer sets whereas the 30% clone yielded products only with the UU primer set), and VA [the 3% clone yielded a product with the 5′ M primer (there is no CpG complementary to the 3′ primer) whereas the 30% clone yielded products with the primer sets involving both 5′U and 5′M].
These results are summarized in Table 1. Table 1 makes clear that the 30% and 3% TH2 clones are distinguishable at the level of CpG methylation and that both differ from the TH1 clone.
Table 1.
Summary of methylation and histone acetylation patterns of the clones from Gfp/Gfp mice
| Clone
|
13P
|
CNS | ITR | VA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | 1B | AcH | A | AcH | B | C | AcH | D | AcH | |||
| TH1 | UU | MM | MM | 0 | MM | 0 | MU | MM | 0 | M | M | 0 |
| UM | MU | |||||||||||
| 3% TH2 | UU | MM | MM | +1 | MM | 0 | UU | UU | +1 | M | M | 0 |
| MU | UU | MU | MM | |||||||||
| UM | ||||||||||||
| MU | ||||||||||||
| 30% TH2 | UU | MU | UU | +3 | MM | +3 | UU | UU | +3 | M | U | +2 |
| MM | UM | UU | MU | M | ||||||||
| UU | ||||||||||||
The histone acetylation patterns (AcH) were scored as 0 (no product) +1 (product with undiluted DNA), +2 (product with a ¼ dilution of DNA), or +3 (product with a  dilution of DNA) (Fig. 4).
dilution of DNA) (Fig. 4).
Methylation Patterns in the Il4/Il13 Genetic Region in Clones from Il4/Gfp Mice.
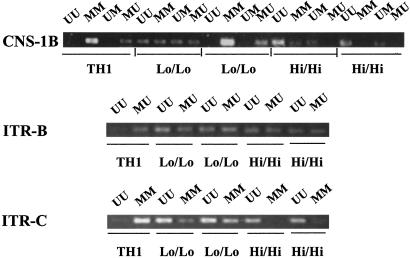
To determine whether the association of patterns of methylation and level of expression of GFP in clones from Gfp/Gfp mice were also found in other settings, we examined a group of five CD4 clones from Il4/Gfp donors, one TH1 clone and four TH2 clones. The four TH2 clones were selected to express either low levels of IL-4 and GFP (<7% for IL-4, <6% for GFP) or high levels of IL-4 and GFP (>55% for IL-4, >35% for GFP) upon challenge. We studied methylation patterns with primer sets complementary to CNS-1B, ITR-B, and ITR-C (Fig. 3). There was a good correlation in methylation patterns between the Gfp/Gfp TH1 and the Il4/Gfp TH1 clone, between the 3% clone and the low/low Il4/Gfp TH2 clones, and between the 30% clone and the high/high Il4/Gfp TH2 clones.
Fig 3.
Methylation pattern of TH1 and low and high TH2 clones from Il4/Gfp mice. Three sets of primers, CNS-1B, ITR-B, and ITR-C, were used to study the methylation pattern of the clones from Il4/Gfp mice by methylation-specific PCR.
5′-Aza-2-Deoxycytidine Treatment Increases Degree of Expression of IL-4 by 3% Clone.
To test whether the degree of methylation controls levels of expression of GFP in Th2 clones, we treated the 3% and 30% TH2 clones and the Gfp/Gfp TH1 clone with 2, 0.3, or 0 μM 5′-aza-2-deoxycytidine during a 4-day stimulation with soluble anti-CD3 and anti-CD28. After a 2-day “rest” in IL-2, the cells were stimulated with phorbol 12-myristate 13-acetate and ionomycin. The untreated cells retained their characteristic level of Gfp expression. Cells from the 3% clone increased expression to 12% treated with 0.3 μM drug and to 26% when treated with 2 μM drug. The 30% Th2 clone showed modest differences and the TH1 clone did not acquire the capacity to express Gfp (Table 2). These results suggest that altering the pattern of CpG methylation in the 3% clone allows substantial increase in expression of GFP. The lack of further increase in the 30% clone suggests that these cells are close to maximal in expression.
Table 2.
Effect of 5′-aza-2′-deoxycytidine treatment on GFP expression
| Treatment | TH1 | 3% TH2 | 30% TH2 |
|---|---|---|---|
| 0 | 0 | 4% | 37% |
| 0.3 μM | 0 | 12% | 32% |
| 2 μM | 0 | 26% | 43% |
Cells were cultured with 5-aza-2′-deoxycytidine during the priming period. They were then washed and maintained in IL-2-containing, drug-free medium for 2 days. Cells were restimulated with phorbol 12-myristate 13-acetate and ionomycin for 24 h, and percent of GFP-expressing cells was measured with a FACScan.
Patterns of Histone Acetylation in Clones from Gfp/Gfp Mice.
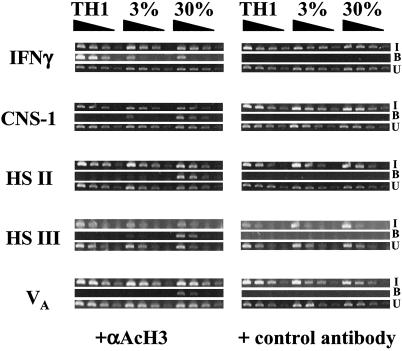
We evaluated histone acetylation in unstimulated clones, using CHIP with antibodies to acetylated histone H3 and then carried out PCRs for sequences in the CNS-1, HS II, HS III, and VA regions as well as for a region in the first DNase I HS in the IFNg gene. The immunoprecipitated IFNg PCR product could be detected at a 16-fold greater dilution in the extract from the TH1 clone than in the 3% or 30% clone extracts (Fig. 4).
Fig 4.
Pattern of histone acetylation of clones from Gfp/Gfp mice. Cross-linked DNA-protein complexes were immunoprecipitated with control antibody (rabbit IgG) or anti-acetylated-histone H3. Primers complementary to the first DNase I HS of the murine Ifng gene and to CNS-1, HS II, HS III, and VA in the Il4/Il13 gene were used. Semiquantitative PCR was carried out on 4-fold serial dilutions of DNA. I, input fractions; B, bound fractions; U, unbound fractions.
At CNS-1, the TH1 clone displayed no detectable immunoprecipitated product whereas the 30% clone had a 16-fold greater degree of precipitation than did the 3% clone. At HS III, the TH1 clone had no immunoprecipitated product, the 3% clone had a weak signal with the undiluted sample, and the PCR product of the 30% clone could be detected at a 16-fold dilution. At HS II and VA, neither the TH1 clone or the 3% clone had a detectable product; the 30% clone displayed a product with a 16-fold diluted sample (HS III) or a 4-fold-diluted sample (VA). These patterns are summarized in Table 1 and correlate well with differences in methylation patterns in these three regions between the three clones.
Methylation Patterns in Developing TH1 and TH2 Cells.
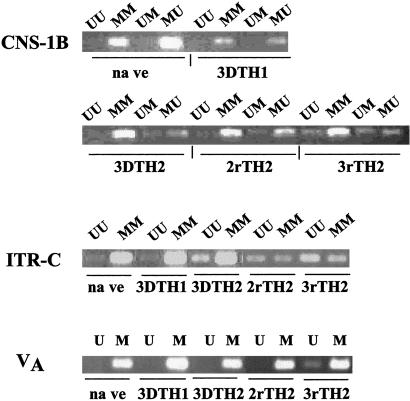
To relate the patterns of methylation seen among TH1 and TH2 clones to that in normal CD4 T cells undergoing TH1 and TH2 differentiation, we primed naïve C57BL/6 CD4 T cells with immobilized or soluble anti-CD3 and anti-CD28. For this purpose, we chose three sets of primer pairs, those complementary to CNS-1B, ITR-C, and VA (Fig. 5).
Fig 5.
Methylation patterns of freshly sorted naïve CD4 cells and developing TH1 and TH2 cells. Naïve CD4 T cells were prepared by sorting for CD44dull/CD62Lbright CD4+ C57BL/6 lymph node T cells. 3DTH1 and 3DTH2 cells were prepared from naïve cells by activation with immobilized anti-CD3 and anti-CD28 under TH1 or TH2 conditions for 3 days; 2rTH2 and 3rTH2 cells activation with soluble anti-CD3, anti-CD28, and antigen-presenting cells, two and three rounds, respectively.
Naïve T cells had the same pattern of methylation at these three sites as did the TH1 clone (Figs. 2B and 3). Within 3 days of TH2 priming of naïve cells, a substantial UU product is observed at ITR-C; the pattern is similar to that of the 3% clone and the low/low Il4/Gfp clones. At CNS-1B, it is only after three rounds of priming that a changed pattern of methylation is observed, resembling that of the 3% clone and the low/low Il4/Gfp clones. At VA, a changed pattern of methylation is also not observed until after the third round of priming; the pattern resembles that seen in the 30% clone.
Overall, the results indicate that naïve T cells undergoing TH2 differentiation progressively develop patterns of methylation resembling the 3% and the low TH2 clones and, in one case, going beyond that observed in the 30% clone. What is striking is that some changes occur very early in TH2 differentiation whereas others are considerably delayed.
Discussion
CD4 TH2 cell clones display distinctive patterns of CpG methylation and histone acetylation in the Il4 genetic region. The degree of demethylation is associated with the frequency of cells within a clone that will produce GFP or IL-4 upon stimulation with immobilized anti-CD3 and anti-CD28 or with phorbol 12-myristate 13-acetate and ionomycin. This association was observed in clones from Gfp/Gfp donors (one low TH2 clone and one high TH2 clone) and in clones from Il4/Gfp mice (two low TH2 clones and two high TH2 clones). By contrast, we found no correlation between transcription factor expression, including Gata3 and c-maf, and degree of GFP expression between the TH2 clones.
We analyzed the clones from the Gfp/Gfp mice in the greatest detail. The probability of cells within a clone that expressed or failed to express GFP to subsequently express GFP upon challenge was similar, suggesting that the cells in the clone had a uniform probability of expressing GFP. The methylation status of the Il4 genetic region was studied in these clones by using eight primer pairs, representing six regions, all but one chosen based on prior evidence of either DNase I hypersensitivity or the presence of a CpG island. These clones displayed great complexity in their patterns of methylation.
This complexity was also observed when patterns of histone H3 acetylation were examined. Essentially no DNA from the four Il4 regions tested was immunoprecipitated with anti-acetylated histone H3 from an extract of cross-linked TH1 cells. By contrast the high (30%) TH2 clone yielded substantially more immunoprecipitated product at the four regions than did the low (3%) TH2 clone.
The finding that low TH2 clones from Il4/Gfp donors had similar patterns of methylation in three regions (CNS-1B, ITR-B, and ITR-C) to that of the 3% TH2 clone from a Gfp/Gfp mouse and that the high clones from heterozygotes had a pattern similar to that of the 30% clone from the homozygous donor, implies that the degree of GFP and/or IL-4 expression and methylation patterns of the Il4 gene are linked. The complexity was also observed in analysis of differentiating TH2 cells.
These results indicate that the Il4 gene can exist in a series of distinct chromatin conformations associated with distinctive probabilities of expression of IL-4 or GFP. That is, the Il4 gene does not appear to exist simply in an on or off form in which all TH2 cells are very similar to one another. Rather, the substantial variability in likelihood of expression of IL-4 is associated with (and presumably caused by) distinctive states of chromatin structure.
What is particularly interesting is that, in long-term clones of TH2 cells, the patterns of expression are fixed and can only be modestly altered by further TH2 type stimulation, including culture with anti-CD3 and anti-CD28 in the presence of IL-4, anti-IL-12, and anti-IFN-γ. In addition, we found that even the introduction of a retroviral construct expressing constitutively active Stat6 (Stat6VT) failed to alter the pattern of IL-4 expression in a long-term clone.
We believe these results shed substantial light on the problem of monoallelism in IL-4 expression by TH2 cells. As we have previously reported (5), recently primed TH2 cells have a uniform, but low, probability of expressing either Il4 allele whereas long-term T cell clones may express, stably, one or the other allelic product in a larger proportion of their cells. That is, with additional priming, one or the other allele acquires a greater probability of expression. Here we have shown that clones in which IL-4 or GFP are expressed in a higher proportion of the cells display a greater degree of demethylation and that key areas have a greater degree of H3 histone acetylation. Although we have not yet been able to directly link a given allele to a given pattern of methylation or acetylation, the differences between low and high TH2 clones would presumably be reflected in similar differences at the level of low and high alleles in Il4/Gfp TH2 cells.
In support of a process in which cells can progressively acquire differential accessibility of the Il4 locus as they undergo further TH2 differentiation is the observation that after 3 days of TH2 priming, we see a change in methylation pattern only in ITR-C; changes in CNS-1B and VA are observed only after three rounds of TH2 priming. We have recently observed increased restriction enzyme sensitivity at a RsaI site very close to ITR-C at 3 days of priming. Lee et al. (16) have recently reported progressive demethylation in the 5′ end of the IL-4 gene as the duration of Th2 priming increased.
What biological value might such probabilistic expression have? In general, IL-4 and many type I cytokines do not function as endocrine hormones. For the latter, because most of the action is at a distance from the hormone-producing cells, the response depends on the ambient concentration of the hormone. Thus, regulation is aimed at controlling this concentration. By contrast, IL-4 functions in the context of the interaction of T cells with partner cells, such as dendritic cells, macrophages, and B cells. In these interactions, mediated by T cell receptor recognition of peptide/class II MHC molecule complexes on the target cell, and characterized by the formation of an immunological synapse (17) into which IL-4 is secreted (18), what is important is the capacity of the T cell to deliver IL-4 to its partner cell. One means through which the intensity of stimulation of partner cells could be controlled would be by regulating the proportion of uniformly differentiated TH2 cells that actually made IL-4. This would be particularly useful in situations in which the behavior of the partner cell being regulated was a quantal one, as best illustrated by Ig class switching in B cells. Under such circumstances, controlling the frequency of “productive” interactions between TH2 T cells and B cells might allow a “smoother” regulation of the number of class switching events than controlling the level of expression of IL-4 in every TH2 cell interacting with a B cell. Under those situations, because the effect measured is essentially “all or none,” it is likely that small changes in the amount of IL-4 made per cell would lead to major shifts in the proportion of B cells that switch. By contrast, if the fraction of TH2/B cell interactions in which IL-4 is actually made can be continuously varied, than the proportion of B cells that switch can be more smoothly controlled.
We suggest that continuous variability in the probabilistic expression of IL-4 could achieve such control. Such continuous variation could be accounted for if the Il4 gene were subject to a wide range of degrees of accessibility and thus to very variable probabilities of expression, given the ambient concentration of transcription factors. If the acquisition of such patterns of accessibility were stochastic but dependent on the “intensity” of the stimuli that cause TH2 differentiation, then one could easily have a situation in which the two alleles in an individual cell had distinctive patterns of accessibility and thus distinctive probabilities of being expressed. Further, the likelihood that there would be more cells that produce IL-4 would increase as the intensity of the differentiation signal increased, which would presumably occur in circumstances, such as helminthic infection, in which robust TH2 differentiation occurred. Our results demonstrating that the patterns of CpG methylation and histone acetylation are variable and associated with distinctive likelihoods of IL-4 (or GFP) expression are consistent with this model.
Supplementary Material
Acknowledgments
We thank Shirley Starnes for editorial assistance, Calvin Eigsti for cell sorting, Dr. Howard A. Young for mouse IFN-g genomic sequences, Drs. Michelle T. McMurry and Michael S. Krangel for CHIP assay protocol, Dr. Gilles Foucras for chromosome analysis, Dr. Kenneth M. Murphy for GFP-RV, Dr. Uli Schindler for pcDNA3-Stat6 VT, Dr. William C. Sha for NGFR-RV, Dr. Anjana Rao for anti-NFATc2 antibody, and Dr. Gary Felsenfeld and Dr. Stephen Smale for helpful discussions.
Abbreviations
GFP, green fluorescence protein
HS, hypersensitive site
CHIP, chromatin immunoprecipitation
NGFR, nerve growth factor receptor
References
- 1.Seder R. A. & Paul, W. E. (1994) Annu. Rev. Immunol. 12, 635-673. [DOI] [PubMed] [Google Scholar]
- 2.Nelms K., Keegan, A. D., Zamorano, J., Ryan, J. J. & Paul, W. E. (1999) Annu. Rev. Immunol. 17, 701-738. [DOI] [PubMed] [Google Scholar]
- 3.Bix M. & Locksley, R. M. (1998) Science 281, 1352-1354. [DOI] [PubMed] [Google Scholar]
- 4.Riviere I., Sunshine, M. J. & Littman, D. R. (1998) Immunity 9, 217-228. [DOI] [PubMed] [Google Scholar]
- 5.Hu-Li J., Pannetier, C., Guo, L., Lohning, M., Gu, H., Watson, C., Assenmacher, M., Radbruch, A. & Paul, W. E. (2001) Immunity 14, 1-11. [DOI] [PubMed] [Google Scholar]
- 6.Henkel G., Weiss, D. L., McCoy, R., Deloughery, T., Tara, D. & Brown, M. A. (1992) J. Immunol. 149, 3239-3246. [PubMed] [Google Scholar]
- 7.Agarwal S. & Rao, A. (1998) Immunity 9, 765-775. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal S., Avni, O. & Rao, A. (2000) Immunity 12, 643-652. [DOI] [PubMed] [Google Scholar]
- 9.Takemoto N., Koyano-Nakagawa, N., Yokota, T., Arai, N., Miyatake, S. & Arai, K. (1998) Int. Immunol. 10, 1981-1985. [DOI] [PubMed] [Google Scholar]
- 10.Bird J. J., Brown, D. R., Mullen, A. C., Moskowitz, N. H., Mahowald, M. A., Sider, J. R., Gajewski, T. F., Wang, C. R. & Reiner, S. L. (1998) Immunity 9, 229-237. [DOI] [PubMed] [Google Scholar]
- 11.Loots G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288, 136-140. [DOI] [PubMed] [Google Scholar]
- 12.Mohrs M., Blankespoor, C. M., Wang, Z. E., Loots, G. G., Afzal, V., Hadeiba, H., Shinkai, K., Rubin, E. M. & Locksley, R. M. (2001) Nat. Immunol. 2, 842-847. [DOI] [PubMed] [Google Scholar]
- 13.Lee G. R., Fields, P. E. & Flavell, R. A. (2001) Immunity 14, 447-459. [DOI] [PubMed] [Google Scholar]
- 14.Liyanage M., Coleman, A., duManoir, S., Veldman, T., McCormack, S., Dickson, R. B., Barlow, C., WynshawBoris, A., Janz, S., Wienberg, J., et al. (1996) Nat. Genet. 14, 312-315. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J., Guo, L., Watson, C. J., Hu-Li, J. & Paul, W. E. (2001) J. Immunol. 166, 7276-7281. [DOI] [PubMed] [Google Scholar]
- 16.Lee D. U., Agarwal, S. & Rao, A. (2002) Immunity 16, 649-660. [DOI] [PubMed] [Google Scholar]
- 17.Shaw A. S. & Allen, P. M. (2001) Nat. Immunol. 2, 575-576. [DOI] [PubMed] [Google Scholar]
- 18.Poo W. J., Conrad, L. & Janeway, C. A., Jr. (1988) Nature (London) 332, 378-380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.