Abstract
Human complement receptor type 2 (CD21) is the cellular receptor for Epstein–Barr virus (EBV), a human tumor virus. The N-terminal two short consensus repeats (SCR1–SCR2) of the receptor interact with the EBV glycoprotein gp350/220 and also with the natural CD21 ligand C3d. Here we present the crystal structure of the CD21 SCR1–SCR2 fragment in the absence of ligand and demonstrate that it is able to bind EBV. Based on a functional analysis of wild-type and mutant CD21 and molecular modeling, we identify a likely region for EBV attachment and demonstrate that this region is not involved in the interaction with C3d. A comparison with the previously determined structure of CD21 SCR1–SCR2 in complex with C3d shows that, in both cases, CD21 assumes compact V-shaped conformations. However, our analysis reveals a surprising degree of flexibility at the SCR1–SCR2 interface, suggesting interactions between the two domains are not specific. We present evidence that the V-shaped conformation is induced by deglycosylation of the protein, and that physiologic glycosylation of CD21 would result in a more extended conformation, perhaps with additional epitopes for C3d binding.
The spectrum of disease that can be directly linked to Epstein–Barr virus (EBV) infection has dramatically increased since the 1964 discovery of the virus in tumor cells from children with Burkitt's lymphoma (1). The EBV genome can be detected in diverse forms of lymphoma as well as in many epithelial malignancies (2). Leiomyosarcomas in children with AIDS are infected with latent virus, whereas the AIDS-associated lesion, oral hairy leukoplakia, contains actively replicating EBV (2). In many parts of the world, EBV+ Burkitt's lymphoma remains the most common tumor of childhood and nasopharyngeal carcinoma the most prevalent cancer of middle-aged men (3). Acute infectious mononucleosis, a manifestation of primary EBV infection, often is complicated by transient autoimmune phenomena (4), and several recent epidemiologic studies support a role for EBV in the development of autoimmune disease (5, 6).
EBV infection is initiated by specific attachment to human complement receptor type 2 (hCD21) (7–11), a cell surface protein that is highly expressed on B lymphocytes and follicular dendritic cells. The CD21 molecule contains 15 or 16 short consensus repeats (SCR1 to SCR16), domains of ≈60 residues that are found in a large number of cell surface receptors, many with complement regulatory function (12). The major EBV glycoprotein gp350/220 attaches to the two most membrane-distal CD21 repeats, SCR1 and SCR2 (13, 14), and soluble forms of CD21 containing only these two repeats can prevent virus infection (15). The same two-domain fragment is also involved in the interaction with the natural CD21 ligands C3d and CD23 (13, 15, 16). The recent crystal structure of C3d in complex with the N-terminal two repeats of CD21 (17) has provided important information about the structure of CD21 and how it recognizes complement. Surprisingly, the interaction with C3d involves only one CD21 repeat, SCR2, although several earlier studies had implicated both SCR1 and SCR2 in ligand binding (18–22). Perhaps even more surprising, none of the CD21 residues implicated in the interaction were found to contact C3d (17).
Here we present the crystal structure of an unliganded fragment of CD21 (CD21 SCR1–SCR2) that is able to bind EBV. Based on functional data and a three-dimensional model, we identify a region that is most likely to be involved in contacting EBV. We further suggest an explanation for the observed differences in the receptor's binding to EBV and C3d. Our analysis reveals a previously unreported glycan and substantial flexibility at the SCR1–SCR2 interface, discoveries that have important implications for understanding the receptor's likely conformation in solution. The observed structural features suggest that CD21 is a dynamic molecule whose global conformation can be altered by ligand binding to a single domain. We also find that the protein is monomeric in our crystals, suggesting that the dimeric structure of CD21 seen in the CD21:C3d complex (17) is not physiologically relevant.
Methods
Protein Production, Purification, and Crystallization.
The sequence coding for residues 2–130 of CD21 was amplified by PCR and ligated into the vector pPICZαA (Invitrogen) for expression in Pichia pastoris. The residue numbering used here is based on the experimentally determined N-terminal sequence of the mature protein (23) and differs by one residue from that used in the CD21:C3d complex (17). The secreted protein contains a C-terminal factor Xa cleavage site followed by a hexahistidine tag. After methanol induction, the culture supernatant containing the secreted protein was applied to a nickel-chelating column. Bound protein was eluted with 300 mM imidazole, pooled, and deglycosylated with endoglycosidase H (New England BioLabs). Subsequent nickel-affinity and size-exclusion chromatography steps resulted in pure protein that was concentrated to 24 mg/ml for crystallization. Crystals grew after 4 months at 20°C by mixing equal volumes of protein solution and precipitant (30% polyethylene glycol 4000, 100 mM MgCl2, 100 mM CsCl, 100 mM Hepes, pH 7.5) in hanging drops.
Functional Analysis of CD21 SCR1–SCR2: Flow Cytometry.
Attachment of CD21 SCR1–SCR2 to EBV was analyzed by virus binding inhibition by using a CD21+ B cell line (7). Cell surface CD21 expression was confirmed by mAb staining (7, 24). EBV was prepared and concentrated as described (7). Attachment of EBV to CD21 was detected with anti-gp350/220 mAb 2L10 (IgG2a, gift of Gary Pearson, Georgetown University, Washington, DC), which does not block virus attachment, using UPC10 (IgG2a, ICN-Cappel) as a control, followed by FITC-conjugated goat (Fab′)2 anti-mouse IgG (BioSource International, Camarillo, CA) (25). Virus was used at a concentration 10-fold in excess of cell surface saturation based on prior staining. Serial dilutions (20–100 μg/ml) of purified proteins (SCR1–SCR2 glycoform, deglycosylated SCR1–SCR2, and CD46 SCR1–SCR4, a related SCR protein that does not bind EBV gp350/220) were incubated in the presence or absence of EBV for 1 h on ice. The cells were then combined with the respective EBV–SCR1–SCR2 mixtures, incubated for an additional hour on ice, washed three times, stained, and analyzed on a FACScan (Becton Dickinson) to detect bound EBV.
Glycosylation Mutant.
CD21mut (hCD21 with 66-NKTI replacing 66-NKYS) was synthesized (26), sequenced, and then cloned into the expression vector πH3M (27). L cells transfected with either CD21 or CD21mut together with the vector pSV2-neo (28) or with vector alone (neo) were selected with Geneticin (800 μg/ml) (GIBCO), stained with rabbit anti-CD21 (29), and sorted twice (EPICS 750 flow cytometer, Coulter) to obtain pure populations. The transfectants were stained with three anti-CD21 mAbs OKB7 (Ortho Diagnostics), HB5 (American Type Culture Collection), and B2 (Coulter) compared with relevant isotype-matched controls (23, 24), and glycosylation was documented by immunoblot (not shown). To assess virus binding, FITC-labeled EBV (FITC-EBV) was prepared (7), incubated with receptor-bearing L cells or controls for 45 min on ice, washed twice in cold media, and analyzed by cytometry (7). The ability of L cells bearing CD21 or CD21mut to bind C3d was concurrently evaluated. Sheep erythrocytes (E) coated with 7S antibody and human C3d (EAC1-C3d) were prepared (29, 30). Cells were incubated with C3d-coated E for 20 min at 37°C. Raji, a CD21+ (CR1/CR3−) Burkitt lymphoma line (7), was used to standardize the assay. Raji cells bound >4 EAC1-C3d/cell, whereas a CD21− T cell line, CEM, bound none (not shown). L cells that bound >4 E per cell were enumerated, and photomicrographs of typical fields were obtained by using a Zeiss MC100 camera and Plus-X Pan 125 film (Kodak).
Structure Determination.
The CD21 SCR1–SCR2 crystals are triclinic (a = 31.60 Å, b = 33.27 Å, c = 42.11 Å, α = 91.51°, β = 111.44°, γ = 117.81°) and contain one polypeptide chain per asymmetric unit. Crystals were cryoprotected with 15% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at beamline X25 of the National Synchrotron Light Source and processed with hkl (31). The structure was determined by using the molecular replacement method (32) and the published coordinates for the SCR1 and SCR2 domains of liganded CD21 (17) (Table 1). Molecular replacement solutions were obtained separately for each domain, with initial R factors of 48.5% and 50.2% for SCR1 and SCR2, respectively (6 to 3.5 Å). The solutions were adjusted by subsequent rounds of model building in o (33) and crystallographic refinement with x-plor (34), finally yielding a model with excellent refinement statistics (Table 1). The refined model contains three cis-prolines (Pro-20, Pro-87, and Pro-120) and 146 water molecules. The remaining residues of the factor Xa site and the hexahistidine tag are disordered. Coordinates and structure factors have been deposited in the Protein Data Bank (ID code ). Figures were prepared with ribbons (35) and grasp (36).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Resolution range, Å | 20–1.8 |
| Completeness, % | 88.4 (55.8) |
| Total reflections | 42,404 |
| Unique reflections | 11,265 |
| Rmerge, % | 10.5 (10.0) |
| I/σ | 13.8 (6.9) |
| Refinement statistics | |
| Rcryst, %; working set | 17.9 (29.4) |
| Rcryst, %; free set | 23.4 (30.6) |
| rms deviation bond lengths, Å | 0.007 |
| rms deviation bond angles, deg. | 1.34 |
| Number of water molecules | 146 |
Data were collected at 100 K with synchrotron radiation at 1.1 Å. Values in parentheses refer to the outermost 0.1-Å resolution shell.
Rmerge = ∑hkl|I − 〈I〉 | I ∑hklI, where I is the intensity of a reflection hkl, and 〈I〉 is the average over symmetry-related observations of hkl.
Rcryst = ∑hkl | Fobs − Fcalc |/∑hklFobs, where Fobs and Fcalc are observed and calculated structure factors, respectively. No I/σI cutoff was used. Free set contains 10% of reflections. Values in parentheses have been calculated with water molecules omitted from the model.
Superpositions and Modeling.
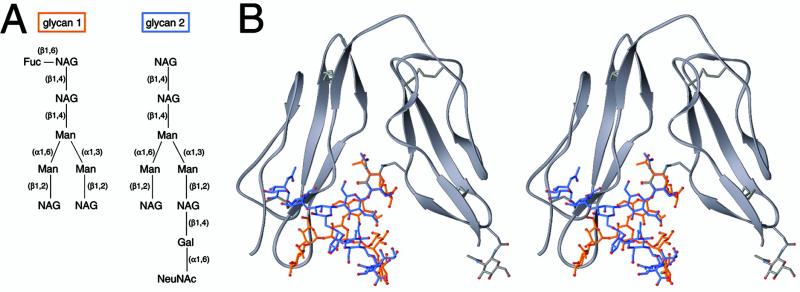
The superpositions (see Figs. 2b and 4) are based on the Cα atoms of SCR2 (60 atoms, rms deviation = 0.46 Å). The models (see Figs. 3 and 5) were obtained by using the known structures of two glycans. Glycan-1 is from the FcRn/heterodimeric Fc complex (Protein Data Bank ID code ; ref. 37). Glycan 2 is from human ZAG (Protein Data Bank ID code ; ref. 38), which was purified from human serum and therefore has a typical mammalian glycosylation pattern. The glycans were treated as rigid bodies and manually superimposed with CD21. To prepare the model shown in Fig. 3, the terminal N-acetylglucosamine (NAG) residue of each glycan was superimposed onto NAG107. The model shown in Fig. 5 was created by superimposing the Asn linked to glycan-2 onto Asn-66 and Asn-101 of hCD21.
Fig 2.
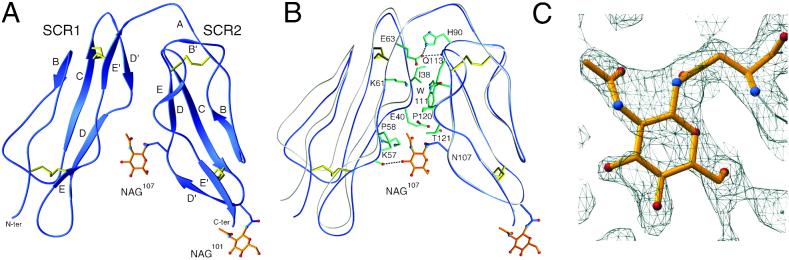
Structure and conformation of CD21 SCR1–SCR2. (A) Ribbon drawing of the crystallized protein, with β-strands labeled. Disulfide bonds and NAG residues are shown in yellow and orange, respectively. (B) Interface between domains SCR1 and SCR2. Amino acids (single-letter code) that participate in the contact are shown in green. Hydrogen bonds and salt bridges are represented with dashed lines. A comparison with the structure of liganded CD21 SCR1–SCR2 (gray) (17) reveals substantial interdomain flexibility despite extensive interface contacts. (C) Final 2 Fobs − Fcalc electron density map at 1.8-Å resolution, contoured at 0.7 σ, and centered at NAG107.
Fig 4.
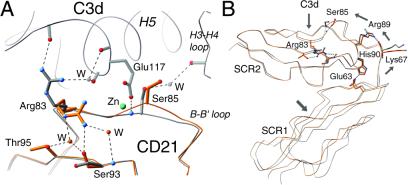
Changes in CD21 structure upon interaction with its ligand C3d. (A) Superposition of nonliganded CD21 SCR1–SCR2 (orange) with the CD21 SCR1–SCR2–C3d complex (gray) (17). Binding of C3d introduces a main-chain shift in the B–B′ loop of SCR2, displacing Ser-85 and unlocking the Arg-83 side chain. The interaction primarily involves the base of C3d helix H5. The C3d/CD21 interface contains a nonphysiologic zinc ion (cyan), which may distort the interaction between CD21 and C3d somewhat because the zinc-coordinating C3d residue Glu-117 might otherwise be available to form a salt bridge with Arg-83 or interact with other CD21 residues. W denotes water molecules. (B) Differences in interdomain orientation between the unliganded (orange) and liganded (gray) forms of CD21. The view is the same as in A; the tracing for C3d has been omitted for clarity. The small changes at the C3d binding site lead to a different interdomain orientation. Hydrogen bonds are indicated with dashed lines. Arrows indicate the directions of main-chain movements.
Fig 3.
Glycosylation at the SCR1/SCR2 interface and interdomain orientation. (A) Schematic drawings of two glycans whose linkages and composition are similar to those that exist in native hCD21. The atomic structures of both glycans are known (see Methods). (B) Stereoview of glycans 1 (orange) and 2 (blue) modeled onto NAG107 of CD21 (see Methods). In both cases the distal ends of the carbohydrate chains clash severely with residues at the base of SCR1. The “true” glycans are likely to be even more bulky, and therefore can be expected to clash even more severely with the protein.
Fig 5.
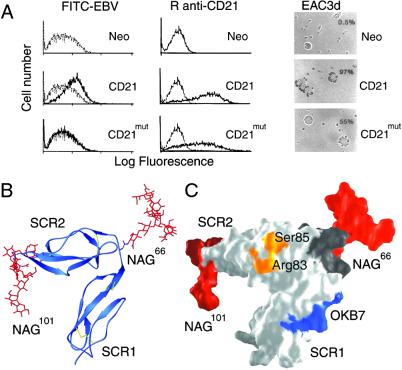
Functional and structural characterization of a hCD21 glycosylation mutant, CD21mut. Mutation to Asn-66 causes glycosylation at this position as occurs naturally in mCD21, which does not bind EBV. (A) Comparison of ligand binding. Shown is binding of FITC-labeled EBV (FITC-EBV, Left), monospecific rabbit anti-CD21 antibody (R anti-CD21, Center), and C3d (EAC3d, Right) to L cells transfected with control vector (Neo), CD21, or CD21mut. (Left) Flow cytometry showing binding of FITC-EBV (dark line) to L cells transfected with CD21 but not to cells transfected with CD21mut or the control vector Neo. FITC-labeled avidin (gray line) was used as a control. (Center) Flow cytometry showing binding of R anti-CD21 antibody to CD21 and CD21mut (dark lines) but not to L cells transfected with vector alone. Preimmunization rabbit serum (gray line) was used as a control. (Right) Photomicrographs (magnification ×137) showing EAC3d binding to CD21 and CD21mut, but not to L cells transfected with vector alone. The percentage of cells binding >4 EAC3d in a field of 200 cells is indicated in the upper right corner and coordinates with receptor distribution and density on the respective transfectants. (B) Ribbon drawing of hCD21 SCR1–SCR2 with modeled murine glycosylation pattern. mCD21 has glycans at positions Asn-66 and Asn-101, and a large structurally known glycan structure (red; glycan-2 from Fig. 3A) was modeled onto each of these amino acids (see Methods). (C) Surface representation of the structure shown in B. The primary C3d-binding residues Arg-83 and Ser-85 (see Fig. 4A) are shown in orange, the OKB7 epitope (residues 8–15) is in blue, the linker region (residues 63–70) is in dark gray, and the two glycans are shown in red. The glycans are distant from the C3d binding site and most likely prevent EBV and OKB7 binding by steric hindrance.
Results
Functional Assessment.
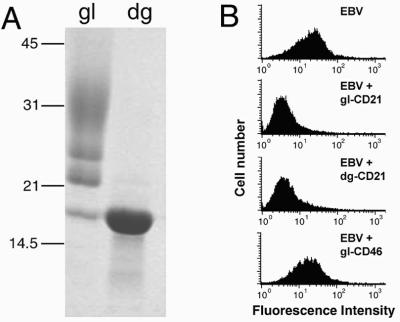
The purified CD21 SCR1–SCR2 protein was extensively glycosylated, with three major glycoforms ranging in molecular mass from ≈22 kDa to ≈35 kDa as well as a nonglycosylated fragment of ≈16 kDa (Fig. 1A, lane gl). Treatment with endoglycosidase H yielded a homogeneous species (Fig. 1A, lane dg) that was used for crystallization. Both glycosylated and deglycosylated SCR1–SCR2 fragments prevented attachment of EBV to a CD21-expressing B cell line in a dose-dependent and parallel manner, whereas glycosylated human CD46, a structurally related protein (39), did not interfere with this interaction (Fig. 1B). These results demonstrate that the expressed CD21 fragment is functional and that its ability to bind to EBV is independent of glycosylation. We note that CD46 requires glycosylation for its interaction with measles virus (40). In that case, glycosylation is thought to help maintain the conformation of the receptor (39).
Fig 1.
Biochemical and functional characterization of CD21 SCR1–SCR2. (A) SDS/PAGE showing purified glycosylated (gl) and deglycosylated (dg) protein. The expressed fragment has glycosylation sites at Asn-101 and Asn-107. (B) EBV binding (block) to CD21-expressing B cells (Top) can be inhibited by glycosylated (gl-CD21, second from Top) or deglycosylated (dg-CD21, third from Top) CD21 SCR1–SCR2, but not by glycosylated CD46 SCR1–SCR4 (gI-CD46, Bottom), a closely related protein that does not bind EBV (see Methods).
Overall Structure and Conformation.
The crystallized protein consists of two concatenated SCR domains, SCR1 and SCR2 (Fig. 2A). Each of these assumes the typical SCR fold, with five short β strands, a small hydrophobic core, and disulfide bridges at either end of the domain. Strand A is absent in SCR1. The two domains assemble into a compact, V-shaped structure that exhibits extensive side-by-side interactions (Fig. 2B). SCR1 residue Ile-38 and SCR2 residues Trp-111 and Pro-120 form a small hydrophobic cluster at the center of the interface. Pro-120 is in the cis configuration in our structure, although it was reported as a trans-proline in liganded CD21 (17). The two domains are also linked via a Glu-63–His-90 salt bridge near the interdomain linker.
The folded-back arrangement of the two domains, which is most likely a result of the unusually long linker connecting SCR1 and SCR2, is dramatically different from more extended conformations adopted by many other SCR-containing structures (41). On the other hand, the conformation is similar to that seen in the structure of CD21 SCR1–SCR2 in complex with C3d (17) (Fig. 2B). Because the crystal forms and the packing forces for the liganded and unliganded structures of CD21 SCR1–SCR2 are entirely different this similarity would suggest that the folded-over conformation is an inherent property of CD21 SCR1–SCR2. However, both structures were obtained by using glycosylated protein that was treated with endoglycosidase H before crystallization and therefore retains only a single NAG residue at each site. Two key observations indicate that the conformation observed in both structures is at least partially a result of this nonphysiologic minimal glycosylation pattern.
A strategically placed glycan at the SCR1–SCR2 interface.
Although both structures contain a NAG moiety attached to Asn-101 at the base of SCR2 (NAG101), our analysis reveals the presence of a previously unreported second NAG moiety lodged at the SCR1–SCR2 interface (NAG107) (Fig. 2A). This carbohydrate was presumably present but not seen in the structure of the CD21 SCR1–SCR2/C3d complex (17); however, it is well defined in our final electron density map at 1.8-Å resolution (Fig. 2C). Although covalently attached to Asn-107 in SCR2, NAG107 primarily interacts with residues in SCR1 (Fig. 2B). Its N-acetyl group forms extensive van der Waals contacts (distances <4.5 Å) with Pro-58 and the hydrophobic portions of the side chains of Glu-40 and Lys-57. In addition, the O3 hydroxyl group of NAG107 is hydrogen-bonded to the main-chain carbonyl of Asp-56 in SCR1, and the glycan also has additional water-mediated hydrogen bonds to both domains. Thus, we consider NAG107 an integral part of the CD21 SCR1–SCR2 structure. Furthermore, its strategic location at the SCR1–SCR2 interface suggests that a more complex physiologic glycan at this position might be incompatible with the V-shaped conformation observed in both CD21 structures. In fact, molecular modeling experiments show that even moderately sized branched carbohydrates would sterically interfere with residues in SCR1 in the folded-over conformation and likely result in a more open conformation of CD21 SCR1–SCR2 (Fig. 3). This finding is consistent with solution studies, which also suggest a more extended conformation of this CD21 fragment (42).
Flexibility at the SCR1–SCR2 interface.
A superposition of the unliganded and liganded structures shows that CD21 is substantially more compact in the absence of C3d (Fig. 2B). The “span” between SCR1 and SCR2 at the base of the molecule increases by almost 2 Å in the liganded protein, indicating that the two domains are able to move with respect to each other, despite substantial inter-domain contacts of a mixed hydrophilic and hydrophobic nature (Fig. 2B). It is therefore evident that inter-domain flexibility is an integral property of the CD21 SCR1–SCR2 fragment, just as it is for several other SCR-containing proteins (39, 43–45).
Binding to C3d.
SCR domains are involved in diverse cellular processes, and they also serve as receptors for a large number of pathogens (41). Flexibility between domains and the requirement of more than one domain for many interactions are hallmarks of many SCR-containing proteins, but the importance of these properties for ligand binding is unclear. The availability of high-resolution crystal structures for CD21 SCR1–SCR2 in its free and C3d-bound (17) forms presents a unique opportunity to analyze, in atomic detail, the structural changes that occur upon complement ligand binding to an SCR domain. A detailed comparison of the two structures reveals two salient differences: local induced-fit movements occur at the C3d binding site of SCR2 (Fig. 4A), and global changes are seen in interdomain orientation between SCR1 and SCR2 (Fig. 4B). These two changes appear to be linked allosterically through a network of intramolecular contacts.
Local changes.
CD21 residues Arg-83 and Ser-85, located in a loop between β-strands B and B′ in SCR2, play a central role in the interaction with C3d (17) (Fig. 4A). Arg-83 protrudes from the loop, occupying an oxyanion hole formed by carbonyl oxygens of residues 116, 117, and 119 at the base of helix H5 of C3d, whereas the Ser-85 amide group is hydrogen-bonded to Glu-117 in H5 (Fig. 4A). In addition, the Ser-85 hydroxyl group forms a water-mediated hydrogen bond with the H3–H4 loop of C3d. Remarkably, the Arg-83 side chain is tucked away into a pocket in the unliganded structure, with its guanidinium group fastened to the protein surface by a hydrogen bond network involving residues Ser-93 and Thr-95 in the adjacent β-strand C (Fig. 4A). Ser-85, on the other hand, protrudes farther from the protein surface in the unliganded structure, and if this position were maintained its side chain would sterically interfere with the Glu-117 side chain upon C3d binding. Thus, the interaction with C3d displaces Ser-85 and surrounding residues in the B–B′ loop, and these structural changes could help to “unlock” the Arg-83 side chain, perhaps by altering the water structure that surrounds its guanidinium group. Binding of C3d, therefore, does not occur through a simple docking mechanism involving two static surfaces but involves induced fit movements at the CD21 ligand binding site.
Global changes.
Although the more “open” conformation of liganded CD21 (Fig. 4B) could be a result of crystal packing forces, it is far more likely that it is related to C3d binding because the key regions involved in the observed changes do not participate in crystal contacts in either structure. More importantly, the local ligand-induced changes in the B–B′ loop of CD21 offer a straightforward explanation for how they might effect global changes between the two domains. The movement of Ser-85 in response to C3d binding (Fig. 4A) alters, among others, the positions of the nearby residues Arg-89 and His-90, which follow Ser-85 in sequence and are also part of the SCR2 B–B′ loop. The Arg-89 side chain is hydrogen-bonded to the main-chain carbonyl of linker residue Lys-67, and His-90 forms a salt bridge with Glu-63 in strand E′ of SCR1 (Fig. 4B). Upon formation of the complex with C3d, both Arg-89 and His-90 move 0.5–0.8 Å away from the C3d binding site, pulling the linker and the attached SCR1 domain with it (Fig. 4B). Thus, although direct binding to C3d involves only residues in SCR2, these residues are connected through a hydrogen bond network to SCR1.
For several reasons, the possibility that the crystallized CD21–C3d complex provides an incomplete view of the physiologic interactions has to be considered. First, our modeling studies suggest that a fully glycosylated SCR1–SCR2 fragment assumes a more extended conformation (Fig. 3), which may result in additional contacts between C3d and CD21. Second, the CD21–C3d complex was crystallized in the presence of 200 mM Zn2+ (17). As a result the C3d/CD21 interface contains two nonphysiologic Zn2+ ions, which are contacted by residues that could otherwise engage in alternate interactions. One Zn2+ ion appears to be close (≤4 Å) only to two lysine side chains; one of these, Lys-112 of C3d, could form a salt bridge with CD21 residue Glu-73 in the absence of zinc. Similarly, the only side chain contacting the other Zn2+ ion, C3d residue Glu-117 (Fig. 4A), is close to Arg-83 of CD21 (Fig. 4B). Third, the CD21/C3d interface is surprisingly small for an interaction with nanomolar affinity (42). Although the buried surface area of the complex was reported as 1,400 Å2 (17), our own calculations using the published coordinates (Protein Data Bank ID code ), the program surface (46), and a standard probe radius of 1.4 Å show that only 790 Å2 are buried in the CD21:C3d complex. This area is considerably smaller than the interface between CD21 SCR1 and SCR2 (1,150 Å2) and only slightly larger than a presumably nonphysiologic crystal contact between C3d and an adjacent CD21 molecule (550 Å2). Thus, additional contacts between CD21 and C3d may exist in a more physiologic setting.
Binding to EBV.
Beyond the finding that SCR1 and SCR2 are required for attachment (13, 15), little is known about the interaction of CD21 with EBV. Murine CD21 (mCD21) does not bind EBV (29), despite an overall sequence identity of 65% with hCD21 (47). Unlike hCD21, however, mCD21 contains an N-linked glycosylation site within its SCR1–SCR2 linker sequence (66-NKSI versus 66-NKYS). Although the prominent location of a glycan at the mCD21 interdomain linker has long been suspected to account for why mCD21 does not bind EBV (18, 19, 47), it is unclear whether this glycan alters the conformation of CD21 SCR1–SCR2 (and therefore the structure of the EBV binding site) or simply prevents EBV binding through steric hindrance.
To directly test the role of the glycan in EBV binding, we introduced the murine glycosylation site into hCD21 by replacing the human 66-NKYS sequence with 66-NKTI. This approach eliminates the unknown mCD21 structure as a variable and simply tests the effect of this additional glycan on the known hCD21 structure. Except for the Thr replacing a Ser, the substituted sequence is identical to the mCD21 linker sequence 66-NKSI. The Ser → Thr-68 substitution is unlikely to induce structural changes as the corresponding residue in hCD21 (Tyr-68) is fully solvent exposed and faces away from the protein. Notably, the mutated protein (CD21mut) completely lost the ability to bind EBV (Fig. 5A Left) and also failed to recognize OKB7, a mAb that blocks EBV attachment (48) (not shown). In contrast, CD21mut retains the ability to bind to human C3d (Fig. 5A Right), as rosette formation correlated with the level of CD21 expression on the respective cell lines. Thus, glycosylation at Asn-66 prevents EBV binding but not binding to C3d.
These mutagenesis data alone do not explain whether the glycan at Asn-66 prevents EBV binding by inducing structural changes in CD21 or simply by altering the accessibility of the EBV binding site. To reproduce the glycosylation of mCD21, we modeled carbohydrates with typical human glycosylation patterns onto Asn-66 and Asn-101 of hCD21 SCR1–SCR2 (Fig. 5B). Our model shows that both glycans are located opposite to the region that interacts with C3d (Fig. 5C), and therefore would not be expected to interfere with C3d binding. Most importantly, the glycan at Asn-66 is unlikely to affect the overall conformation of the SCR1–SCR2 fragment as it faces away from the receptor (Fig. 5 B and C) and has only minimal contacts with protein residues. Therefore, the most likely interpretation of our data is that the introduced glycan prevents access of mAb OKB7 and EBV to CD21 through steric hindrance rather than through inducing a conformational change. We note that our findings differ from earlier studies suggesting that the inability of mCD21 to bind EBV is related to conformational differences between mCD21 and hCD21 (19). Ser-15 of hCD21 was specifically implicated in EBV binding and also in interdomain contacts between SCR1 and SCR2 (19). This residue is distant from the interdomain interface and thus unlikely to play a role in determining the molecule's overall conformation. However, Ser-15 is part of the epitope for OKB7, a mAb that blocks EBV attachment (18–20, 48), and therefore Ser-15 may be located in the greater vicinity of the EBV binding site (Fig. 5C). Our model shows that an extensive glycan structure at Asn-66 would make access of the large EBV gp350/220 protein to this area difficult.
Discussion
The high-resolution crystallographic analysis of unliganded CD21 provides important information about its structure, conformation, and function in ligand recognition. Although the protein is highly compact when crystallized in its deglycosylated form, the conspicuous location of a previously unreported glycan and the observed flexibility between SCR1 and SCR2 suggest that the fully glycosylated protein would assume a more extended conformation, perhaps with additional contacts to C3d. Previous studies have implicated both SCR1 and SCR2 in binding to C3d (18–21), and in fact most SCR-containing proteins appear to require at least two consecutive domains for interaction with ligands. Although only SCR2 contacts C3d in the complex, it was hypothesized that inter-domain packing between SCR1 and SCR2 was necessary for stabilization of the C3d binding site (17). Precisely how this occurred was unclear because the regions that contact SCR1 and C3d are located on opposite faces of SCR2. Our analysis suggests a mechanism by which residues within SCR1 and the linker region indirectly stabilize the C3d binding site through an extensive network of interactions. Interestingly, these same interactions may be used to propagate induced-fit movements in the ligand binding site into larger conformational changes in a multidomain fragment.
Our combined functional analysis and molecular modeling data provide insights into the interaction of CD21 with EBV. We suggest that EBV interacts with CD21 through an epitope that is at least partially occluded by the glycan present in mCD21, and that this epitope is distant from the C3d binding site. Thus we predict the EBV binding site to be close to the linker region and the C-terminal region of SCR1.
The current work will advance efforts to design novel antiviral agents that specifically block binding of EBV gp350 to this receptor. Prevention of EBV transmission would have a major impact on international health as >90% of adults are seropositive. However, the virus life cycle is complex and critical immune targets appear to include viral oncoproteins, which complicates strategies to develop an attenuated vaccine. A sophisticated understanding of each component of the EBV-CD21 interaction, at the molecular level, is needed to provide new and creative strategies for eliciting protective immunity.
Acknowledgments
We thank members of our laboratories for helpful discussions and in particular Michael Gill for help with preparation of some of the figures. This research was supported by the Swiss National Science Foundation (to A.E.P.), National Institutes of Health Award AI45716 and the Milton Foundation at Harvard Medical School (to T.S.), and National Institutes of Health Awards DE12186 and K24 and the Center for AIDS Research (to J.D.F.). J.D.F. is an established Investigator of the American Heart Association.
Abbreviations
CD21, complement receptor type 2
hCD21, human CD21
mCD21, murine CD21
EBV, Epstein–Barr virus
NAG, N-acetylglucosamine
SCR, short consensus repeat
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Epstein M. A., Barr, Y. M. & Achong, B. G. (1964) Lancet 1, 702-703. [DOI] [PubMed] [Google Scholar]
- 2.Rickinson A. B. & Kieff, E. (2001) in Field's Virology, ed. Fields, B. N. (Lippincott, Philadelphia), pp. 2575–2627.
- 3.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, (1997) IARC Monographs on the Evaluation of Carcinogenic Risks to Humans (International Agency for Research on Cancer, Lyon, France), Vol. 70.
- 4.Chang R. S., (1980) Infectious Mononucleosis (G. K. Hall Medical Publishers, Boston).
- 5.James J. A., Kaufman, K. M., Farris, A. D., Taylor-Albert, E., Lehman, T. J. & Harley, J. B. (1997) J. Clin. Invest. 100, 3019-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ascherio A., Munger, K. L., Lennette, E. T., Spiegelman, D., Hernan, M. A., Olek, M. J., Hankinson, S. E. & Hunter, D. J. (2001) J. Am. Med. Soc. 286, 3083-3088. [DOI] [PubMed] [Google Scholar]
- 7.Fingeroth J. D., Weis, J. J., Tedder, T. F., Strominger, J. L., Biro, P. A. & Fearon, D. T. (1984) Proc. Natl. Acad. Sci. USA 81, 4510-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frade R., Barel, M., Ehlin-Henriksson, B. & Klein, G. (1985) Proc. Natl. Acad. Sci. USA 82, 1490-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemerow G. R., Wolfert, R., McNaughton, M. E. & Cooper, N. R. (1985) J. Virol. 55, 347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemerow G. R., Mold, C., Schwend, V. K., Tollefson, V. & Cooper, N. R. (1987) J. Virol. 61, 1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner J., Weis, J., Fearon, D., Whang, Y. & Kieff, E. (1987) Cell 50, 202-213. [DOI] [PubMed] [Google Scholar]
- 12.Reid K. B. M. & Day, A. J. (1989) Immunol. Today 10, 177-180. [DOI] [PubMed] [Google Scholar]
- 13.Lowell C. A., Klickstein, L. B., Carter, R. H., Mitchell, J. A., Fearon, D. T. & Ahearn, J. M. (1989) J. Exp. Med. 170, 1931-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carel J. C., Myones, B. L., Frazier, B. & Holers, V. M. (1990) J. Biol. Chem. 265, 12293-12299. [PubMed] [Google Scholar]
- 15.Moore M. D., Cannon, M. J., Sewall, A., Finlayson, M., Okimoto, M. & Nemerow, G. R. (1991) J. Virol. 67, 6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubry J. P., Pochon, S., Gauchat, J. F., Nueda-Marin, A., Holers, V. M., Graber, P., Siegfried, C. & Bonnefoy, J. Y. (1994) J. Immunol. 152, 5806-5813. [PubMed] [Google Scholar]
- 17.Szakonyi G., Guthridge, J. M., Li, D., Young, K., Holers, V. M. & Chen, X. S. (2001) Science 292, 1725-1728. [DOI] [PubMed] [Google Scholar]
- 18.Molina H., Brenner, C., Jacobi, S., Gorka, J., Carel, J. C., Kinoshita, T. & Holers, V. M. (1991) J. Biol. Chem. 266, 12173-12179. [PubMed] [Google Scholar]
- 19.Martin D. R., Yuryev, A., Kalli, K. R., Fearon, D. T. & Ahearn, J. M. (1991) J. Exp. Med. 174, 1299-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina H., Perkins, S. J., Guthridge, J. M., Gorka, J., Kinoshita, T. & Holers, V. M. (1995) J. Immunol. 154, 5426-5436. [PubMed] [Google Scholar]
- 21.Prodinger W. M., Schwendinger, M. G., Schoch, J., Kochle, M., Larcher, C. & Dierich, M. P. (1998) J. Immunol. 161, 4604-4610. [PubMed] [Google Scholar]
- 22.Clemenza L. & Isenman, D. E. (2000) J. Immunol. 165, 3839-3848. [DOI] [PubMed] [Google Scholar]
- 23.Fingeroth J. D., Clabby, M. L. & Strominger, J. L. (1988) J. Virol. 62, 1442-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fingeroth J. D., Diamond, M. E., Sage, D. R., Hayman, J. & Yates, J. L. (1999) J. Virol. 73, 2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dezube, B., Zambela, M., Sage, D. R., Wang, J.-F. & Fingeroth, J. D. (2002) Blood, in press. [DOI] [PubMed]
- 26.Gillam S. & Smith, M. (1979) Gene 8, 81-87. [DOI] [PubMed] [Google Scholar]
- 27.Seed B. & Aruffo, A. (1987) Proc. Natl. Acad. Sci. USA 84, 3365-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern P. J. & Berg, P. (1982) J. Mol. Appl. Genet. 1, 327-341. [PubMed] [Google Scholar]
- 29.Fingeroth J. D., Benedict, M. A., Levy, D. N. & Strominger, J. L. (1989) Proc. Natl. Acad. Sci. USA 86, 242-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross G. D. & Polley, M. J. (1976) Scand. J. Immunol. 5, 99-111. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 33.Jones T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 34.Brünger A. T., Kuriyan, J. & Karplus, M. (1987) Science 235, 458-460. [DOI] [PubMed] [Google Scholar]
- 35.Carson M. (1987) J. Mol. Graph. 5, 103-106. [Google Scholar]
- 36.Nicholls A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 37.Martin W. L., West, A. P., Jr., Gan, L. & Bjorkman, P. J. (2001) Mol. Cell 7, 867-877. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez L. M., Chirino, A. J. & Bjorkman, P. J. (1999) Science 283, 1914-1919. [DOI] [PubMed] [Google Scholar]
- 39.Casasnovas J. M., Larvie, M. & Stehle, T. (1999) EMBO J. 18, 2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maisner A., Alvarez, J., Liszewski, M. K., Atkinson, D. J., Atkinson, J. P. & Herrler, G. (1996) J. Virol. 70, 4973-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stehle T. & Larvie, M. (2002) in Innate Immunity, ed. Hoffmann, J. A. (Humana, New York), in press.
- 42.Guthridge J. M., Rakstang, J. K., Young, K. A., Hinshelwood, J., Aslam, M., Robertson, A., Gipson, M. G., Sarrias, M. R., Moore, W. T., Meagher, M., et al. (2001) Biochemistry 40, 5931-5941. [DOI] [PubMed] [Google Scholar]
- 43.Barlow P. N., Steinkasserer, A., Norman, D. G., Kieffer, B., Wiles, A. P., Sim, R. B. & Campbell, I. D. (1993) J. Mol. Biol. 232, 268-284. [DOI] [PubMed] [Google Scholar]
- 44.Henderson C. E., Bromek, K., Mullin, N., Smith, B. O., Uhrin, D. & Barlow, P. N. (2001) J. Mol. Biol. 307, 323-339. [DOI] [PubMed] [Google Scholar]
- 45.Smith B. O., Mallin, R. L., Krych-Goldberg, M., Wang, X., Hauhart, R., Bromek, K., Uhrin, D., Atkinson, J. P. & Barlow, P. N. (2002) Cell 108, 769-780. [DOI] [PubMed] [Google Scholar]
- 46.Collaborative Computing Project No. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 47.Fingeroth J. D. (1990) J. Immunol. 144, 3458-3467. [PubMed] [Google Scholar]
- 48.Siaw M. F., Nemerow, G. R. & Cooper, N. R. (1986) J. Immunol. 136, 4146-4151. [PubMed] [Google Scholar]