Abstract
To identify proteins undergoing glutathionylation (formation of protein-glutathione mixed disulfides) in human T cell blasts, we radiolabeled the glutathione pool with 35S, exposed cells to the oxidant diamide, and analyzed cellular proteins by two-dimensional electrophoresis. One of the proteins undergoing glutathionylation was identified by molecular weight, isoelectric point, and immunoblotting as thioredoxin (Trx). Incubation of recombinant human Trx with glutathione disulfide or S-nitrosoglutathione led to the formation of glutathionylated Trx, identified by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. The glutathionylation site was identified as Cys-72. Glutathionylation of rhTrx abolished its enzymatic activity as insulin disulfide reductase in the presence of NADPH and Trx reductase. Activity was, however, regained with sigmoidal kinetics, indicating a process of autoactivation due to the ability of Trx to de-glutathionylate itself. These data suggest that the intracellular glutathione/glutathione disulfide ratio, an indicator of the redox state of the cell, can regulate Trx functions reversibly through thiol-disulfide exchange reactions.
Thioredoxin (Trx; ref. 1), a ubiquitous redox protein, is an essential cofactor electron donor for ribonucleotide reductase, but also has many other cellular functions, including regulation of transcription factors, apoptosis, and antioxidant activity and can act exogenously as a redox active growth factor (1, 2). The catalytic activity of Trx resides in its active site where the two redox active Cyss (Cys-31 and Cys-34 in human Trx) undergo reversible oxidation/reduction. In addition to the conserved Cys residues in the active site, three additional structural Cys residues (Cys-61, Cys-68, and Cys-72) are present in the structure of human Trx.
The present paper describes the experiments that led us to the conclusion that Trx can undergo glutathionylation in T cells exposed to oxidative stress. Protein glutathionylation, the formation of a disulfide between a Cys in a protein and the Cys in the tripeptide glutathione (GSH), is a modification that can be induced in cells by oxidative stress. Glutathionylation can occur by direct oxidation of a protein and GSH, by a thiol-disulfide exchange between a protein Cys and oxidized glutathione (GSSG), and also with the intermediacy of S-nitrosoglutathione (GSNO; refs. 3 and 4). Glutathionylation of proteins is reversible, as those proteins can be reduced by glutaredoxins (5, 6), and the process serves to regulate protein functions by the redox state of the cell (i.e., by the GSSG/GSH ratio; refs. 7–9).
To demonstrate the glutathionylation of Trx, we first labeled the intracellular GSH pool of the T-cell blasts with [35S]Cys and analyzed the cell lysate by two dimensional electrophoresis under nonreducing conditions. A spot was found in the autoradiographic protein map with IP/Mr corresponding to those of Trx. In the second part of the study, we incubated recombinant human Trx in the presence or absence of GSSG or GSNO to clarify the structural and functional properties of glutathionylated Trx, then we analyzed the protein by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF). Finally, we studied the effect of incubation with GSSG on the activity of Trx coupled to NADPH as a substrate for Trx reductase (TrxR). The results confirm that Trx can undergo glutathionylation, which is able to regulate its enzymatic activity and function.
Materials and Methods
Materials.
GSH, GSSG, GSNO, N-ethylmaleimide (NEM), and diamide were from Sigma. TrxR was prepared from bovine liver as described (10).
T Cell Blasts.
Human peripheral blood mononuclear cells (PBMC) were isolated by using standard Ficoll/Hypaque gradients from buffy coats of healthy donors (kindly provided by the Blood Center of the Hospital of Magenta, Magenta, Italy). PBMC were cultured at 1 × 106 cells per ml for 3 days in RPMI medium 1640 with 10% (vol/vol) FCS and 2 μg/ml phytohemagglutinin (Sigma), then washed and cultured for 3 days with 50 units/ml human recombinant IL-2 (Chiron).
Cell Labeling and Exposure to Diamide.
Cells were resuspended at 3.3 × 106 cells per ml and maintained throughout the experiment in Hanks' balanced salt solution (HBSS) containing 50 μg/ml cycloheximide. After 20-min preincubation in HBSS/cycloheximide, the cells were incubated with 8 μCi/ml of l-[35S]Cys [specific activity 1,000 Ci/mmol (1 Ci = 37 GBq); Amersham Pharmacia]. After 30 min, the cells were treated for 5 min with 1 mM diamide (Sigma), then collected by centrifugation and either processed for electrophoretic analysis as described below, or the proteins were precipitated with trichloroacetic acid (TCA) at a final concentration of 5%. After five washes with 5% TCA, the TCA-precipitable radioactivity was counted.
Protein Electrophoresis.
The cell pellet (2 × 107 cells per sample) was extracted with chloroform/methanol, resuspended in 8 M urea + 0.5% carrier ampholytes, and run on immobilized pH gradients (IPG; ref. 11) covering, with an exponential course, the pH range 4–10 (12). IPG strips were embedded on 12% polyacrylamide gel slabs for the second-dimensional run in the discontinuous buffer system of Laemmli (13). Electrophoresis was carried out under nonreducing conditions. The slabs either were stained with Coomassie blue and radioactivity was analyzed with a phosphor-imager after drying, or they were used for Western blot analysis.
Western Blot Analysis of Trx.
The slabs were blotted onto a nitrocellulose membrane in the presence of 0.01% SDS. A goat polyclonal antibody against human Trx (14) was used at 1:200 dilution and revealed by enhanced chemiluminescence with a horseradish peroxidase-conjugated mouse anti-goat secondary antibody from Pierce.
Peptide Synthesis.
Cys-containing Trx fragments were synthesized by solid-phase chemistry on an Applied Biosystems 433A synthesizer using N-(9-fluorenylmethoxycarbonyl) as a protective group for the aminic residues and N-[(1H-benzotriazol-1-yl)(dimethylamino)methylene]-N-methylmethanaminium tetrafluoroborate N-oxide and N,N-diisopropylethylamine as activators of carboxylic residues. Peptides were cleaved from the resin with trifluoroacetic acid/thioanisole/water/phenol/ethanedithiol in the ratio 82.5:5:5:5:2.5 (vol/vol), precipitated, and washed with diethylether.
Glutathionylation of Recombinant Trx.
Recombinant human Trx was expressed in E. coli and purified as described (15). The numbering of amino acids is calculated without taking into account the N-terminal methionine, which is removed in rhTrx. Before the experiments, rhTrx was fully reduced as described below. Briefly, 500 μl of the concentrated protein (2.5 mg/ml) were reacted under argon flow with a 20-fold molar excess of DTT in 50 mM Tris/1 mM EDTA, pH 7.4. The reaction was allowed to proceed for 30 min at 37°C. To remove the excess of DTT, samples were desalted on a NAP5-column (Amersham Pharmacia), and the exact concentration of the fully reduced rhTrx was established by measuring the absorbance at 280 nm using an extinction coefficient of 1.48 mg/ml (16). Full reduction of rhTrx was checked by titration of the free -SH groups, as described below.
To induce glutathionylation, rhTrx was incubated with the indicated concentration of GSSG for 2 h at room temperature. When indicated, rhTrx was incubated with 5 mM GSNO under the same experimental conditions.
Mass Spectrometric Analysis of Trx.
Control or glutathionylated Trx was analyzed by a Bruker-Reflex III MALDI-TOF mass spectrometer. A few μl of the sample were mixed with an equal volume of a saturated solution of sinapinic acid or α-ciano-4-hydroxycinnamic acid (Sigma) in acetonitrile/0.1% trifluoroacetic acid 1:3 (vol/vol); 1 μl of the mixture was deposited on the MALDI mass spectrometry target.
Titration of Free -SH Groups with 5′-5′-Dithiobis-2-Nitrobenzoic Acid (DTNB).
The number of free -SH groups of rhTrx was established spectrometrically with DTNB at 412 nm (ɛM = 13.6 M−1·cm−1), as described (17).
Insulin Assay for Trx Enzyme Activity.
The activity of human Trx was measured as its ability to reduce insulin disulfides in the presence of NADPH and calf thymus thioredoxin reductase (CT-TrxR; ref. 10). The reaction mixture containing 100 mM potassium phosphate buffer pH 7.0, 2 mM EDTA, 0.2 mM NADPH, and 0.16 mM insulin. The samples were prepared by mixing 500 μl of the reaction mixture and different concentrations of Trx (2 or 4 μM) in cuvettes. The reactions were started by the addition of 38 nM TrxR, and the absorbance of the samples were read at 340 nm against blank solution, which included all reagents except Trx.
The activity also was tested in the endpoint assay (10) by using a 20-min incubation to determine -SH groups formed in insulin by breaking the reaction with 6 M guanidine-HCl, 1 mM DTNB, and reading at 412 nm.
Tryptic Digestion of rhTrx.
Fully reduced rhTrx in water (8.6 μM) was reacted with 5 mM GSSG for 2 h at room temperature as described above. Iodoacetamide was added to the reaction mixture to a final concentration of 2 mM to block residual Cys. The glutathionylated rhTrx was digested with trypsin (at a trypsin:Trx ratio of 1:10, wt/wt) in a solution containing 80% of acetonitrile and 50 mM ammonium bicarbonate (18). The reaction was allowed to proceed for 10 min at 37°C, and the tryptic peptides were analyzed by MALDI-TOF, as described above. Data were compared with the theoretical molecular weight of the rhTrx tryptic peptides, taking into consideration the possible glutathionylated Cys.
Results
Identification of Trx Among the Glutathionylated Protein in Oxidatively Stressed Lymphocytes.
To identify proteins undergoing S-thiolation in human phytohemagglutinin blasts, cells were labeled with [35S]Cys for 30 min in the presence of cycloheximide, washed, then incubated with 1 mM diamide for 5 min. Previous studies have shown that under these experimental conditions, diamide induces marked 35S incorporation into proteins (19).
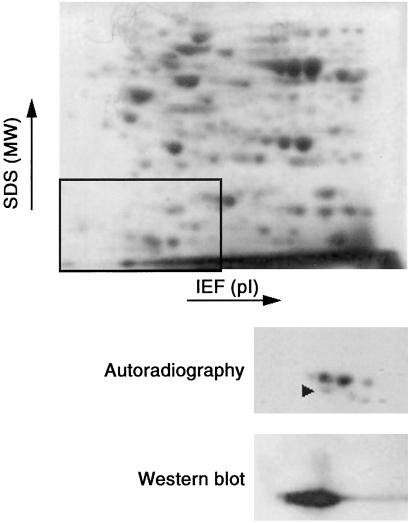
Cell extracts were analyzed by nonreducing two-dimensional gel electrophoresis. The Coomassie blue stain and the autoradiography of gels from diamide-treated cells are in Fig. 1 Top and Middle. Based on molecular weight and pI, we tentatively postulated that one of the radioactively labeled proteins, indicated by an arrow in the autoradiography shown in Fig. 1, might be Trx. To confirm the identity of this spot with Trx, we performed Western blot analysis by using an anti-human Trx antibody. As shown in Fig. 1 Bottom, immunoreactive Trx was identified, corresponding to the radioactive spot.
Figure 1.
Two-dimensional gel electrophoretic analysis of glutathionylated proteins. T cell blasts (106 per sample) were labeled with 35S, then exposed to 1 mM diamide for 5 min; the proteins were separated by isoelectrofocusing (pH range 4–10, left to right) and then by SDS/10% PAGE (migration from top to bottom). All runs were done under nonreducing conditions. Figure shows the portion of the gel where Trx is migrating. (Top) Coomassie blue staining. (Inset) Portion of the gel for which autoradiography and Western blot are shown. (Middle) Autoradiography. (Bottom) Immunoblotting with anti-Trx.
Glutathionylation of rhTrx by GSSG.
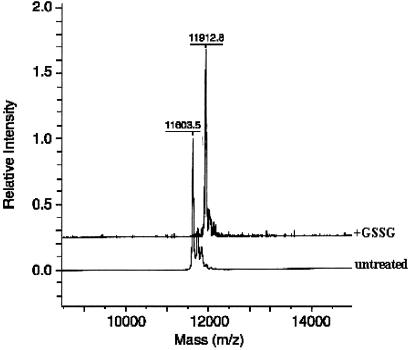
To confirm the susceptibility of Trx to glutathionylation and to identify the modified Cys(s), we used pure rhTrx. The protein was treated with GSSG and then analyzed by MALDI-TOF. rhTrx had the expected molecular weight of 11,606.2 ± 1.9 (n = 3), and incubation with 5 mM GSSG induced the formation of a peak of 11,911.7 ± 1.0, compatible with the addition of one glutathione molecule (305.5 Da). A representative analysis of rhTrx and GSSG-treated rhTrx is shown in Fig. 2. Longer incubation times (overnight) with 5 mM GSSG induced the formation of two peaks compatible with the addition of one or two glutathione molecules (data not shown).
Figure 2.
MALDI identification of glutathionylated Trx. HrTrx (10 pmol) was incubated at room temperature without or with 5 mM GSSG and analyzed by MALDI-TOF MS.
The fact that the extent of glutathionylation depends on the concentration of the oxidant also was seen in separate experiments where rhTrx was treated with 100 mM GSSG for 2 h and the free -SH groups were titrated with DTNB. Incubation of fully reduced rhTrx with GSSG decreased the free -SH groups from five to one.
When rhTrx was incubated with 5 mM GSNO, under the same experimental conditions used with GSSG treatment, only one peak was found, corresponding to the protein with one GSH (data not shown). We did not observe a peak corresponding to the molecular weight of an NO adduct, suggesting that under those experimental conditions, rhTrx is not S-nitrosylated by GSNO, unlike other proteins (20, 21).
Identification of Glutathionylation Sites of Trx.
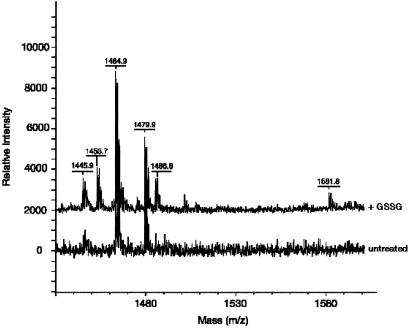
To identify the glutathionylation site(s), we performed tryptic digestion of fully reduced or GSSG-oxidized rhTrx, and analyzed the tryptic fragments by mass spectrometry. However, we could not use standard digestion protocols as they include a reduction of the disulfide bridges in the protein, which would also cleave the mixed disulfide with GSH. Because preliminary experiments have shown that tryptic digestion under nonreducing conditions is not efficient, we used a modified protocol with digestion in organic-aqueous solvents (18) to ensure successful digestion of GSSG-treated rhTrx. The results are shown in Table 1. Two peptides of molecular mass of 1,452.6 Da and 1,580.8 Da are generated by digestion of GSSG-treated rhTrx that are not present in the spectrum of the untreated protein (shown in Fig. 3) and are compatible with glutathionylation of Cys-72.
Table 1.
List of tryptic fragments of glutathionylated rhTrx
| Experimental m/z | Calculated m/z | Tryptic fragment |
|---|---|---|
| 1148.7 | 1148.5 | 72–80 |
| 1165.7 | 1165.6 | 85–95 |
| 1221.8 | 1221.6 | 82–93 |
| 1258.8 | 1258.7 | 94–104 |
| 1292.6 | 1292.6 | 72–81* |
| 1336.8 | 1336.6 | 8–20 |
| 1453.7 | 1453.5 | 72–80† |
| 1463.9 | 1463.8 | 36–47 |
| 1479.9 | 1479.8 | 36–47* |
| 1581.8 | 1581.6 | 72–81‡ |
| 1606.9 | 1606.9 | 81–95 |
| 2148.1 | 2148.1 | 85–104 |
| 2149.3 | 2149.1 | 1–20 |
| 2942.6 | 2942.4 | 8–35 |
rhTrx was treated with GSSG 5 mM for 2 h at room temperature and subjected to tryptic digestion; the fragments were analyzed by MALDI-TOF. Glutathionylated fragments are shown in italics.
Methionine oxidized to sulfoxide (MSO).
Fragment 1148.7 + one GSH moiety (305 kDa).
Fragment 1276.6 and + one GSH moiety.
Figure 3.
Tryptic fragments obtained from the digestion of untreated or GSSG-treated rhTrx. Only the portion of the mass spectrum comprising the glutathionylated fragments is shown.
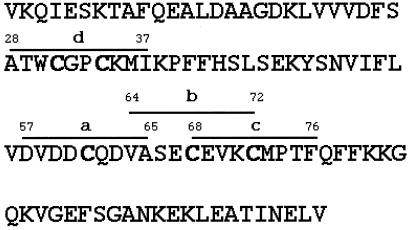
To understand whether the susceptibility of Cys-72 was due to the presence of specific flanking amino acids or to the three-dimensional structure of the protein, we synthesized four peptides homologous to the corresponding rhTrx fragments, and that contained at least one Cys residue (Fig. 4). These four peptides were reacted with 5 mM GSSG and analyzed by mass spectrometry. The results, shown in Table 2, indicate that all of the Cys-residues can be glutathionylated when they are not involved in the three-dimensional structure of the protein. It is of interest to note that only one glutathione molecule is attached to the peptide 28–37 which comprises the two Cys of the active site of Trx.
Figure 4.
Design of synthetic peptides spanning Cys-containing regions of rhTrx. The four synthetic peptides (a, b, c, and d) are shown in relation to the sequence of rhTrx. To have a single Cys for each peptide (with the exception of the active site Cys-31 and -34) in peptide b, a Ser was substituted for Cys-72, and in peptide c, a Ser was substituted for Cys-68.
Table 2.
Synthetic peptides
| Peptide sequence* | Calculated m/z | Experimental m/z | Experimental m/z + GSSG |
|---|---|---|---|
| a | 979.4 | 979.4 | 1306.5† |
| b (C72S) | 951.4 | 951.5 | 1256.6 |
| c (C68S) | 1041.5 | 1041.5 | 1369.0† |
| d | 1109.5 | 1109.5 | 1414.5 |
From Fig. 4.
Corresponding to the form + 1 Na+.
Effect of Glutathionylation on Trx Activity.
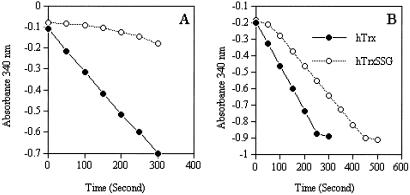
To investigate the effect of glutathionylation on the enzymatic activity of Trx, we incubated Trx with 5 mM GSSG, removed GSSG by desalting on a Sephadex G25 NAP5 column, and then evaluated disulfide reduction with insulin as a substrate. Glutathionylation reduced the activity of Trx to 63% of the control using an endpoint assay of formed -SH groups in insulin by a 20-min incubation. The attachment of a GSH residue on the -SH group of Cys-72, which is located in the active site surface of Trx, would be expected to abolish the activity of Trx-S2 as a substrate for TrxR. To study this further, we used an assay where the kinetics of insulin disulfide reduction was followed in time as the oxidation of NADPH at 340 nm (Fig. 5). As can be seen from Fig. 5 A and B, the initial rate of the reaction with Trx-SG was almost zero, and the activity then was gradually regained. Thus, glutathionylation operated as a reversible regulatory device of the activity.
Figure 5.
Effect of glutathionylation of thioredoxin on its activity. Trx in TE buffer was incubated with 5 mM GSSG for 2 h, GSSG was removed on a Sephadex G25 column, and insulin disulfide reductase activity was determined with 38 nM CT-Trx plus 2 μM Trx (A) or with 4 μM Trx (B). Activity was followed by oxidation of NADPH in the spectrophotometer.
Discussion
This study shows that Trx can be glutathionylated under conditions of oxidative stress. Glutathionylation was confirmed by incubating rhTrx with GSSG at a concentration (5 mM) that can be observed in vivo under conditions of oxidative stress or in cellular compartments such as the endoplasmic reticulum where the GSH/GSSG ratio can be 1 (22).
The mass spectrometric analysis showed that, at a GSSG concentration of 5 mM, only one GSH can be attached to rhTrx at Cys-72. This higher susceptibility of Cys-72 to oxidation is very likely caused by the fact that it is easily accessible in the active site surface of the three-dimensional structure of Trx (23). In fact, when we synthesized peptides reproducing partial Trx sequences and treated them with GSSG, we found that all Cys with the exception of one of those in the CGPC active site were equally susceptible to glutathionylation. Thus the susceptibility of Cys-72 appears to be observed only within the entire protein with its three-dimensional structure. Further supporting the concept that Cys-72 is the Cys residue of rhTrx most exposed to oxidizing agents, we noted that iodoacetamide treatment of rhTrx under nondenaturing conditions alkylated only Cys-72 (data not shown). Alkylation under nondenaturing conditions has been used to determine oxidant-sensitive Cys (also in terms of accessibility in the native structure; refs. 24 and 25). Thus, in the case of Trx, susceptibility to glutathionylation is mainly due to the three-dimensional structure rather than to the chemical-physical properties of the amino acids in the vicinity, as has been suggested for other proteins (26). A glutathione adduct on Cys-72 also might be favored by the primary structure, however, as this amino acid is next to a basic one, Lys-71, which might stabilize the adduct by interacting electrostatically with the γ-glutamyl group of GSH (27).
Interestingly, Cys-72 is also involved in the dimerization of Trx (23, 28), which may occur under oxidative conditions or spontaneous oxidation, confirming that Cys-72 can be easily oxidized. The fact that the same Cys-72 of Trx can be involved in both the dimerization and in the glutathionylation of Trx suggests that glutathionylation prevents dimer formation. In fact, we found that in the absence of GSSG, the mass spectra of rhTrx revealed some Trx dimers, whereas in GSSG-treated rhTrx, no dimer was detectable (data not shown).
Analysis by MALDI-TOF of rhTrx exposed to GSSG for longer periods (overnight) showed that two GSH residues can be attached to a single rhTrx molecule. This finding would imply that one of the two Cys that can undergo glutathionylation is Cys-72 and the other is either Cys-61 or Cys-68. In fact, a mixed disulfide between glutathione and one of the Cys of the CGPC active site (Cys-31, Cys-34) would be highly unstable (25). In agreement with this hypothesis, we observed that even when a synthetic peptide containing the CGPC active site was treated with GSSG, no adduct was ever formed with two GSH molecules.
The fact that Trx can be glutathionylated raises the question of how this fact affects its activities. We investigated whether glutathionylation affected its enzymatic activity. Glutathionylated rhTrx lost all activity initially, but gradually, the activity was restored with time. This was probably due to the fact that Trx can reduce mixed disulfides (29), though not as efficiently as glutaredoxin, and its residual activity could reactivate itself according to the proposed scheme:
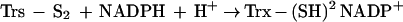 |
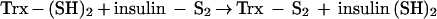 |
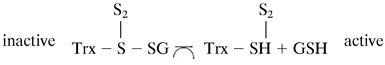 |
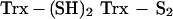 |
The finding that Trx can be regulated by glutathionylation indicates the existence of crosstalk between the glutathione/glutaredoxin system and the Trx system through which Trx activity can be influenced by the GSH/GSSG ratio, an indicator of the redox state of the cell. The observation that GSNO also can induce glutathionylation of Trx with the same efficiency as GSSG suggests that, through the formation of GSNO, NO can influence Trx activity.
Although we demonstrate the occurrence of Trx glutathionylation in living cells exposed to an exogenous oxidant in the present work, other reports show that S-thiolathion of enzymes, namely glyceraldehyde-3-phosphate dehydrogenase, with loss of enzyme activity, can take place during physiologic oxidative stress such as that induced by monocyte activation (30). Therefore, it will be important to evaluate whether Trx is glutathionylated under pathological conditions resulting in oxidative stress.
It should be noted that although glutathionylation reduces the enzymatic activity of Trx, it could have more subtle effects on other activities of this protein, altering its affinity for interacting proteins or even generating new activities. For instance, in a protein related to Trx, sharing its insulin disulfide reducing activity, glycosylation inhibiting factor/macrophage migration inhibitory factor, S-thiolation caused conformational changes and generation of a growth factor like bioactivity (31). Trx has many biological activities, whose mechanisms are largely obscure: it acts extracellularly as a cytokine, chemokine, or growth factor (32–34) and intracellularly as a regulatory protein that influences several regulatory proteins such as NF-κB (35) and ASK1 (36), and it is efficiently secreted through a still unknown mechanism. The relevance of the posttranslational modification of Trx reported here warrants further investigation.
Acknowledgments
P.G. was supported by a Beaumont–Bonelli fellowship from the Nobel Foundation. S.C. is a fellow of the Monzino Foundation, Milano. V.B. is an Assistant Telethon Scientist and was supported by Grants 466/bi and TCP.01010.TEL01 from Telethon-Italy.
Abbreviations
- Trx
thioredoxin
- GSH
glutathione
- GSSG
glutathione disulfide
- GSNO
S-nitrosoglutathione
- MALDI-TOF
matrix-assisted laser desorption ionization–time-of-flight
- TrxR
Trx reductase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 9617.
References
- 1.Arner E S, Holmgren A. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 2.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Huang F L, Huang K P. J Biol Chem. 2001;276:3098–3105. doi: 10.1074/jbc.M008260200. [DOI] [PubMed] [Google Scholar]
- 4.Klatt P, Pineda M D, Perez-Sala D, Lamas S. Biochem J. 2000;349:567–578. doi: 10.1042/0264-6021:3490567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 6.Holmgren A, Aslund F. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 7.Brigelius R. In: Oxidative Stress. Sies H, editor. London: Academic; 1985. pp. 243–272. [Google Scholar]
- 8.Inoue M. In: Glutathione: Chemical, Biochemical, and Medical Aspects. Dolphin D, Avramovic O, Poulson R, editors. 3B. New York: Wiley; 1989. pp. 613–644. [Google Scholar]
- 9.Cotgreave I A, Gerdes R G. Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 10.Luthman M, Holmgren A. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
- 11.Gianazza E. In: 2-D Proteome Analysis Protocols. Link A J, editor. Totowa, NJ: Humana; 1998. [Google Scholar]
- 12.Gianazza E. J Chromatogr. 1995;705:67–87. doi: 10.1016/0021-9673(94)01251-9. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura H, Vaage J, Valen G, Padilla C A, Bjornstedt M, Holmgren A. Free Radical Biol Med. 1998;24:1176–1186. doi: 10.1016/s0891-5849(97)00429-2. [DOI] [PubMed] [Google Scholar]
- 15.Ren X, Bjornstedt M, Shen B, Ericson M L, Holmgren A. Biochemistry. 1993;32:9701–9708. doi: 10.1021/bi00088a023. [DOI] [PubMed] [Google Scholar]
- 16.Engstrom N E, Holmgren A, Larsson A, Soderhall S. J Biol Chem. 1974;249:205–210. [PubMed] [Google Scholar]
- 17.Zhong L, Arner E S, Holmgren A. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell W K, Park Z Y, Russell D H. Anal Chem. 2001;73:2682–2685. doi: 10.1021/ac001332p. [DOI] [PubMed] [Google Scholar]
- 19.Ghezzi P, Romines B, Fratelli M, Eberini I, Gianazza E, Casagrande S, Laragione T, Mengozzi M, Herzenberg L A, Herzenberg L A. Mol Immunol. 2001;38:773–780. doi: 10.1016/s0161-5890(01)00114-6. [DOI] [PubMed] [Google Scholar]
- 20.So H S, Park R K, Kim M S, Lee S R, Jung B H, Chung S Y, Jun C D, Chung H T. Biochem Biophys Res Commun. 1998;247:809–813. doi: 10.1006/bbrc.1998.8788. [DOI] [PubMed] [Google Scholar]
- 21.Spencer N Y, Zeng H, Patel R P, Hogg N. J Biol Chem. 2000;275:36562–36567. doi: 10.1074/jbc.M005347200. [DOI] [PubMed] [Google Scholar]
- 22.Hwang C, Sinskey A J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 23.Weichsel A, Gasdaska J R, Powis G, Montfort W R. Structure (London) 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- 24.Kim J R, Yoon H W, Kwon K S, Lee S R, Rhee S G. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 25.Kallis G B, Holmgren A. J Biol Chem. 1980;255:10261–10265. [PubMed] [Google Scholar]
- 26.Hess D T, Matsumoto A, Nudelman R, Stamler J S. Nat Cell Biol. 2001;3:E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- 27.Mallis R J, Poland B W, Chatterjee T K, Fisher R A, Darmawan S, Honzatko R B, Thomas J A. FEBS Lett. 2000;482:237–241. doi: 10.1016/s0014-5793(00)02022-6. [DOI] [PubMed] [Google Scholar]
- 28.Gasdaska J R, Kirkpatrick D L, Montfort W, Kuperus M, Hill S R, Berggren M, Powis G. Biochem Pharmacol. 1996;52:1741–1747. doi: 10.1016/s0006-2952(96)00595-3. [DOI] [PubMed] [Google Scholar]
- 29.Mannervik B, Axelsson K, Sundewall A C, Holmgren A. Biochem J. 1983;213:519–523. doi: 10.1042/bj2130519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravichandran V, Seres T, Moriguchi T, Thomas J A, Johnston R B., Jr J Biol Chem. 1994;269:25010–25015. [PubMed] [Google Scholar]
- 31.Watarai H, Nozawa R, Tokunaga A, Yuyama N, Tomas M, Hinohara A, Ishizaka K, Ishii Y. Proc Natl Acad Sci USA. 2000;97:13251–13256. doi: 10.1073/pnas.230445397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini R, Howard O M, Dong H F, Oppenheim J J, Bizzarri C, Sergi R, Caselli G, Pagliei S, Romines B, Wilshire J A, et al. J Exp Med. 1999;189:1783–1789. doi: 10.1084/jem.189.11.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura H, Nakamura K, Yodoi J. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 34.Pekkari K, Gurunath R, Arner E S, Holmgren A. J Biol Chem. 2000;275:37474–37480. doi: 10.1074/jbc.M001012200. [DOI] [PubMed] [Google Scholar]
- 35.Schenk H, Klein M, Erdbrugger W, Droge W, Schulze-Osthoff K. Proc Natl Acad Sci USA. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]