Abstract
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) proteins expressed on the surface of P. falciparum-infected erythrocytes undergo antigenic variation by switching the gene expressed within a repertoire of approximately 50 var genes per haploid genome. The switching of PfEMP1 plays an important role in the survival and pathogenesis of the parasite. To understand how a parasite switches its var gene expression in human infections, we investigated the composition and change of var gene transcripts during the acute phase of well-defined laboratory-induced P. falciparum infections in naïve human hosts. Multiple var transcripts, with the same dominant transcript, were identified in samples collected after three to four asexual-parasite cycles in two volunteers infected with cloned 3D7 P. falciparum via mosquito bites. A major change in composition and frequency of var gene transcripts was observed between the culture used to infect the mosquitoes and the parasites recovered from the infected volunteers. A further change was seen when infected blood from a mosquito-infected volunteer was either passaged to other volunteers or cultured in vitro. The diversity of var transcripts did not increase with time. The results suggest that the switch of var gene expression is reinitiated after mosquito transmission and that var genes may rapidly switch from the first gene expressed after liver stage, but subsequent switching occurs at a much lower rate.
Sequestration and recrudescence in Plasmodium falciparum infections are attributed to P. falciparum erythrocyte membrane protein 1 (PfEMP1), a set of variant proteins expressed on the infected-erythrocyte surface and encoded by about 50 var genes (1–3). Different PfEMP1 types mediate cytoadhesion of infected erythrocytes to cell surface ligands in microvasculature lining (3, 4), the placenta (5), and other cells, including uninfected erythrocytes (6, 7). Parasites have the ability to switch expressed var genes to alter PfEMP1 serotypes and adhering specificities of infected erythrocytes (8, 9). The cytoadherance of parasitized erythrocytes to receptors such as intercellular adhesion molecule-1 in brain capillaries and chondroitin sulfate A in the placenta is associated with manifestations of severe disease such as cerebral malaria (10) and pregnancy-associated malaria (5). Therefore, understanding of the mechanism that controls the expression of particular PfEMP1 types is of significant medical importance.
During a P. falciparum infection, patients develop an antibody response specific for the particular PfEMP1 type expressed by the parasites. These antibodies are protective against infections of parasites expressing the homologous PfEMP1 type (11, 12). Recrudescent parasites need to switch to a new PfEMP1 type, thus escaping newly generated type-specific anti-PfEMP1 antibody. Mathematical modeling suggests that this interaction of PfEMP1 expression and specific anti-PfEMP1 antibodies is the mechanism that establishes chronic malaria infections with the typical recrudescence pattern seen not only for P. falciparum but also for most species of malaria in their natural host (13–15). PfEMP1 variation parallels that of variant surface glycoproteins (VSG) in trypanosomes where the sequential expression of VSG types after the generation of specific antibodies results in cyclical population changes (16).
In trypanosomes, the expressed vsg genes are located in specific expression sites. These sites are always located at the end of a chromosome and the expressed vsg genes are the last genes before the telomere. The direction of vsg gene transcription is from the centromere toward the telomere and the transcription is controlled by elements upstream from the vsg gene. Several potential expression sites are present, with all but one silenced. Switching vsg expression involves changing the active expression site, as well as recombination to move individual vsg genes into expression sites (17). Different vsg expression sites are used by blood-stage parasites and the metacylic stage, the form of trypansome present in the salivary glands of tsetse flies.
Many, but not all, P. falciparum var genes encoding PfEMP1 are also located in the subtelomeric region of the chromosomes (18). Apart from this similarity with vsg genes in trypanosomes, little is known about the regulation of PfEMP1 expression, although a recent study using a reporter gene indicates that both the upstream and the intron regions within var genes are involved in silencing of var genes (19).
In this paper, we examine the sequence of PfEMP1 types transcribed in a series of human volunteers infected with the 3D7 cloned line of P. falciparum. We studied the var genes transcribed in (i) cultured parasites used to infect mosquitoes; (ii) two volunteers infected with these mosquitoes; (iii) cultured blood derived from these volunteers; and (iv) a second set of volunteers infected by blood passage from one of the mosquito infected volunteers. The results indicate that there is a single dominant var type transcribed early in a mosquito-initiated infection, which rapidly switches to other types. Subsequent switching of different PfEMP1 types proceeds more slowly.
Patients, Materials, and Methods
Samples of a P. falciparum 3D7 Parasite Lineage.
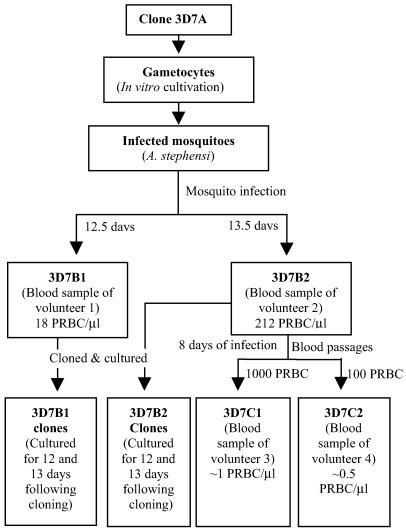
Several human trials have been conducted in which volunteers with no prior history of malaria infection were challenged with 3D7A, a cloned line of P. falciparum (20). Infected blood samples from these defined infections were stored during the trials. We obtained consent from the volunteers and ethics approval from the Bancroft Centre Research Ethics Committee for using these samples to investigate var gene transcripts. The source of samples is illustrated in Fig. 1 and described in detail below. (i) 3D7A, the parent clone, was used to infect Anopheles stephensi by Richard Carter's group at the Institute of Cell, Animal and Population Biology, University of Edinburgh. Before infecting mosquitoes, the parasites had been cultured in vitro for a total of 39 cycles since cloning. The 3D7A sample used in this paper to assess premosquito var gene transcription was an aliquot of the culture taken during the last 18 cycles of culture. (ii) 3D7B1 and 3D7B2 were collected from volunteers 1 and 2, respectively, at 12.5 and 13.5 days after receiving 3D7A through 8 to 9 infectious mosquito bites. Their parasitemias were 18 and 212/μl, respectively, when the samples were taken. The infected blood was cryopreserved after removing white blood cells (20). (iii) 3D7C1 was collected from volunteer 3, 8 days after receiving 1,000 viable 3D7B2-infected erythrocytes via i.v. injection. Parasitemia at time of collection was estimated as 1/μl (20). (iv) 3D7C2: Volunteer 4 was enrolled in a trial during which the volunteer was infected several times with a small inoculum (100 viable 3D7B2-infected erythrocytes) and treated, at each time, 8 days after infection (D. J. Pombo, G. Lawrence, C. Hirunpetcharat, C. Rzepczyk, M. Bryden, N. Cloonan, K. Anderson, Y. Mahakunkijcharoen, S. Elliott, D. P. Eisen, et al., personal communication). 3D7C2 was collected from the volunteer, 8 days after the third infection. Parasitemia was estimated to be 0.5/μl at time of collection.
Fig 1.
Lineage of 3D7 P. falciparum samples used in this study.
The blood samples of B1, B2, C1, and C2 were not cultured before freezing.
Cloning of 3D7B1 and 3D7B2 Parasites and Cultivation in Vitro.
3D7B1 and -B2 were thawed and immediately plated onto 96-well plates at theoretical concentrations of 3, 1, and 0.3 infected erythrocytes per well. Culture media containing fresh uninfected erythrocytes were added to the wells to a total volume of 100 μl at 5% hematocrit. Culture media were changed on days 4 and 8. Five microliters of the culture was retrieved on day 9 and used in a PCR to identify culture wells that contained parasites (20). On days 12 and 13, the positive culture wells were resuspended, and 40% of the original contents was transferred to RNase-free tubes and frozen at −80°C. In this experiment, only parasite clones from 0.3 and 1 parasitized cell per well were used (average positive wells ranged from 10 to 30%). A total of 24 3D7B1 clones and 12 3D7-B2 clones were examined.
Isolation of Total RNA from Parasites.
Total RNA was extracted directly from frozen blood samples (0.3–1 ml) and cloned parasite lines (40 μl) by using the RNeasy Mini Kit, QIAshredder, RNase-Free DNase Set, and Carrier RNA (Qiagen, Hilden, Germany), or the NucleoSpin RNAII kit (Macherey–Nagel, Düren, Germany). We followed the manufacturer's instructions and included a second elution step to maximize the yield. Carrier RNA was added to Lysis buffer at a final concentration of 10 μg/ml. RNasin Ribonuclease Inhibitor (Promega, Madison, WI) was added to the RNA and stored at −80°C.
Reverse Transcription–PCR (RT-PCR) to Amplify the Expressed Duffy-Binding-Like Domain α (DBLα) Region.
All samples were tested to ensure they were negative for DNA by using the primers that would be used in the subsequent RT-PCR without reverse transcriptase before proceeding with the RT-PCR. The in vivo samples were tested for DNA with two rounds of PCR (40 + 40 cycles) regardless of whether one or two rounds of PCR was performed to amplify the var transcripts. The in vitro samples were tested with one round of PCR of 40 cycles. We used the One-Step RT-PCR kit (GIBCO/BRL) and universal primers (αAF and αBR) for amplifying region of DBLα (AFBR) (21). These primers were reported to have limited primer bias (21). The first-round cycling conditions for all samples were: 1 cycle of 42°C for 30 min; 94°C, 2 min followed by 40 cycles of 93°C, 30 sec; 42°C, 50 sec; and 72°C, 1 min. For samples 3D7-C1, C2, we performed a second round of PCR using AF2 (5′-AGTTTTGCNGAYATWGG-3′) and BR3 (5′-AGAWAYTGHGGNACRTARTC-3′) primers that were designed to prime all DBLα sequences. AmpliTaq Gold (Roche Molecular Biochemicals) was used for the second round. Cycling conditions were: 1 cycle of 93°C, 10 min; followed by 93°C, 30 sec; 42°C, 50 sec; 72°C, 1 min, for 40 cycles.
Cloning and Sequencing.
The amplified AFBR fragments of 3D7A, B1, B2, C1, and C2 were cloned into pGEM-T (Promega) or PCR-Script (Stratagene), following the manufacturer's protocols. Approximately 40 insert-containing clones were selected from each sample and sequenced by using vector primers. The amplified AFBR fragments from the in vitro clones were sequenced directly without cloning into vectors.
Sequence Analysis.
AFBR sequences were aligned with the previously described AFBR set of DBLα fragments of var genes (21) and assigned an AFBR number. Each AFBR sequence was used to search the complete sequence of P. falciparum 3D7 chromosomes 2 (22) and 3 (23) to assign chromosomal locations of the corresponding var genes.
RT-PCR to Confirm Transcripts Were Full-Length.
From var gene sequences in the PlasmoDB database, primers were synthesized to amplify across the 3′ intron region in 3D7AFBR13 (forward 5′-AGCACCAGAGGAACTTCCACCTGG-3′ and reverse 5′-TGGTATATCACTAGGTGTATTTGG-3′) and 3D7AFBR41(forward 5′-TTTTGTGGTCTTAATGGCGATGAG-3′ and reverse 5′-GTGGAGATGGATGTTCCTTTGTGC-3′). As the 3′ end of the var gene corresponding to 3D7AFBR16 was not in the PlasmoDB database at the time (Ver. 24, March 2001), for this gene a primer pair corresponding to the 3′ end of exon 1 (131 bp upstream from the intron) was synthesized (forward 5′-AGACACCAAATCCAAAGACATCCA-3′ and reverse 5′′ATTTTGTGTATCACTAGGTGTGTC-3′. The 3′ ends of the corresponding var gene transcripts were amplified by using conditions described above for RT-PCR amplification of the DBLα region.
Comparison of var Transcripts in 3D7B2 Sample (Experiment 2) at Ring and Trophozoite Stages.
One vial of 3D7B2 sample was thawed and the contents divided into three aliquots. One aliquot was immediately processed to isolate total RNA (0 h), and the remaining two aliquots were put into cultures to allow parasites to develop to mature stages. These parasites were harvested at 12 and 24 h. Total RNA was isolated from these samples. Thick films were made at each time point and examined by microscopy. The proportion of different parasite developmental stages was derived by counting 60 parasites at each time point.
Data Analysis.
To estimate the true proportion of each DBLα type in the sample, values (t1 and t2) corresponding to the two tails of the binomial distribution (with n being the number of sequences sampled) were generated for proportions between 0 and 1 such that P(Y ≤ t1) = α1, P(Y > t2) = α2, and α1 + α2 = 0.05 (where Y is a binomial random variable). The number of times each DBLα type was seen in an individual sample was then compared with t1 and t2, with the estimated confidence limits (hereafter referred to as the 95% confidence limits) being the minimum and maximum population proportions for which the sample frequency is not greater than t2 and not less than or equal to t1, respectively.
Results
Composition of var Transcripts in the Parent Parasite 3D7A Before Mosquito Infection.
In the parent 3D7 clone on which the mosquitoes were fed, we found 13 different transcribed DBLα sequences (Table 1). AFBRNT1, a previously undescribed DBLα sequence, was the dominant transcript (23/50). AFBR8 (8/50), -21 (5/50), and -23 (3/50) were observed more than twice, although the remaining nine types were observed only once or twice.
Table 1.
Type of DBLα sequences and their frequencies in 3D7 lineage
| 3D7 DBLα types
|
No. of sequences seen and frequencies | ||||||
|---|---|---|---|---|---|---|---|
| In vitro samples | In vivo samples | In vitro samples | |||||
| Premosquito 3D7A (50 seq.) | Postmosquito 3D7B1 (39 seq.) | Postmosquito 3D7B2 (41 seq.) | Post-blood passage 3D7C1 (32 seq.) | Post-blood passage 3D7C2 (49 seq.) | 3D7B1 clones (25 seq.) | 3D7B2 clones (10 seq.) | |
| AFBR3 | 1 (2%) | 3 (8%) | — | — | — | 1 (4%) | — |
| AFBR4 | 1 (2%) | — | — | 1 (3%) | — | — | — |
| AFBR7 | — | — | 2 (5%) | — | 9 (18%) | 3 (12%) | — |
| AFBR8 | 8 (16%) | — | — | — | — | — | — |
| AFBR11 | 1 (2%) | 3 (8%) | — | — | — | — | — |
| AFBR12 | — | 2 (5%) | — | — | — | — | — |
| AFBR13 | — | 4 (10%) | 4 (10%) | 15 (47%) | 13 (27%) | 2 (8%) | — |
| AFBR14 | — | — | 1 (2%) | — | — | 2 (8%) | — |
| AFBR16 | 1 (2%) | 19 (49%) | 21 (51%) | — | — | 2 (8%) | — |
| AFBR18 | — | — | — | — | — | — | 2 (20%) |
| AFBR19 | 2 (4%) | — | — | — | — | — | — |
| AFBR20 | — | — | — | — | — | — | 1 (10%) |
| AFBR21 | 5 (10%) | — | 1 (2%) | — | — | — | — |
| AFBR23 | 3 (6%) | — | — | — | — | — | — |
| AFBR24 | — | 1 (3%) | 1 (2%) | — | — | — | — |
| AFBR27 | — | — | — | — | — | 2 (8%) | — |
| AFBR28 | — | 2 (5%) | — | — | 1 (2%) | 2 (8%) | 3 (30%) |
| AFBR29 | — | — | — | 7 (22%) | — | — | — |
| AFBR30 | — | — | — | — | — | — | 2 (20%) |
| AFBR32 | — | — | — | — | — | 1 (4%) | — |
| AFBR34 | — | — | — | — | — | 1 (4%) | — |
| AFBR37 | 1 (2%) | — | — | — | — | — | — |
| AFBR39 | — | — | — | — | — | 2 (8%) | — |
| AFBR40 | — | 1 (3%) | — | — | — | 1 (4%) | — |
| AFBR41 | — | — | 6 (15%) | — | 21 (43%) | 6 (24%) | 2 (20%) |
| AFBR42 | 2 (4%) | 1 (3%) | 1 (2%) | — | — | — | — |
| AFBR46 | 1 (2%) | 3 (8%) | — | — | — | — | — |
| AFBR48 | — | — | 4 (10%) | 6 (19%) | 5 (10%) | — | — |
| AFBRNT1 | 23 (46%) | — | — | — | — | — | — |
| AFBRNT2 | 1 (2%) | — | — | — | — | — | — |
| AFBRNT3 | — | — | — | 3 (9%) | — | — | — |
| No. of var types | 13 | 10 | 9 | 5 | 5 | 12 | 5 |
seq., sequences.
Frequency is the no. of sequences of each type of DBLα as a percentage of the total no. sequenced.
“—”: none observed.
Nos. in bold indicate the dominant types.
Composition of var Transcripts After the Initial 8 Days of Asexual Stage Infection in Mosquito-Infected Volunteers.
Samples 3D7B1 and 3D7B2 were obtained from volunteers 1 and 2, who were infected by 25 and 30 mosquito bites, respectively, of which 8 and 9 were sporozoite positive, respectively. Parasites passed through the liver stage and were first detected in the bloodstream of volunteers 1 and 2 on days 5.5 and 6.5, respectively (20). On the basis of the number of ring-infected cells detected on day 5 or 6 in these volunteers, which was approximately 10 parasites per milliliter of blood, each volunteer had a parasite load of approximately 50,000 in the first asexual cycle. It is likely that these parasites originated from one or two infected and ruptured hepatocytes, i.e., the infection was initiated from very few sporozoites (20), hence the starting heterogeneity is likely to be limited. A further three or four asexual cycles (6 and 8 days) were completed in volunteers 1 and 2, respectively. We observed 10 types of transcribed DBLα sequences in 3D7B1 and 9 types in 3D7B2 (Experiment 1) (Table 1). AFBR16, seen only once in the parent clone, was the dominant transcript in both 3D7B1 and -B2 with comparable frequencies of approximately 50%. AFBRNT1, the dominant transcript in the parent clone (46% in 3D7A) and AFBR8, the next most abundant transcript (16% in 3D7A), were not seen in either postmosquito sample. Because the 95% confidence limits for the frequency of these two types were 34–58% and 10–28%, respectively, the lack of parasites transcribing these types in the postmosquito samples is significant (P < 0.05). The two samples shared three other types of transcripts, AFBR13, -24, and -42, at similar low frequencies. Although the remaining observed types were different between the 3D7B1 and -B2 samples, a closer examination revealed that there was considerable overlap in the 95% confidence limits, suggesting that the frequencies of var transcripts were not different between 3D7B1 and -B2.
Infections in volunteers 1 and 2 were terminated on days 13 and 14 (days 6 and 8 of asexual stage infection), respectively. No agglutinating antibodies were detected in the volunteers during infection and 2 wk after infection (data not shown).
var Transcripts Detected Are Full-Length.
Correctly spliced RT-PCR products spanning the intron close to the 3′ end of var genes were obtained for AFBR13 and -41. Because the genomic sequence of the short 3′ exon of AFBR16 was not known, it was not possible to test for correctly spliced full-length transcripts for this type. However, an RT-PCR product of the 3′ end of the long exon 1 obtained by using specific AFBR16 primers suggested that full-length transcripts from this gene were present.
var Transcripts Detected in the Clinical Samples Were Comparable to Those Detected in Mature Stages.
To investigate whether the var transcripts observed in clinical peripheral samples, where the majority of parasites are at ring stage, represent those in trophozoites, we compared var transcripts in 3D7B2 without culturing (0 h) and with 12- and 24-h in vitro cultivation, respectively. At 0 h, 97% of the parasites were at ring stage and 3% at early trophozoite stage. The proportion of ring-stage parasites decreased to 56.7 and 15% after 12- and 24-h culture, respectively, and the proportion of trophozoites increased to 42% (24% late trophozoites) and 75% (69% late trophozoites), respectively. Early schizonts (two to eight nuclei) appeared at 12 h (1.7%), and the proportion increased to 10% at 24 h. The total number of var transcripts detected at these time points were 10, 12, and 10, respectively. There is no difference in the number of var transcripts detected at the three developmental-stage time points and between this (Experiment 2) and the first experiment (Experiment 1) (Table 2). AFBR16 was the dominant DBLα sequence in all samples taken at different time points. The 95% confidence limits on the frequencies of other var transcripts overlapped between different time points and between this and the first experiment (Table 2).
Table 2.
Comparison of DBLα transcripts observed in 3D7-B2 between experiments 1 and 2 and between different stages of parasite life cycle
| DBLα types
|
No. (%) of sequences seen and estimated proportion in population | |||||||
|---|---|---|---|---|---|---|---|---|
| 3D7-B2 (Exp. 1) (41 seq.) | 3D7-B2 (Exp. 2–0 h) (32 seq.) | 3D7-B2 (Exp. 2–12 h) (32 seq.) | 3D7-B2 (Exp. 2–24 h) (32 seq.) | |||||
| No. of seq. seen | Confidence limits on frequency (%) | No. of seq. seen | Confidence limits on frequency (%) | No. of seq. seen | Confidence limits on frequency (%) | No. of seq. seen | Confidence limits on frequency (%) | |
| AFBR 1 | — | 0–8 | 1 (3%) | 1–15 | — | 0–10 | — | 0–10 |
| AFBR 4 | — | 0–8 | — | 0–10 | 1 (3%) | 1–15 | — | 0–10 |
| AFBR 7 | 2 (5%) | 2–16 | 1 (3%) | 1–15 | 2 (6%) | 2–20 | — | 0–10 |
| AFBR 12 | — | 0–8 | 2 (6%) | 2–20 | 1 (3%) | 1–15 | 1 (3%) | 1–15 |
| AFBR 13 | 4 (10%) | 4–23 | — | 0–10 | 2 (6%) | 2–20 | 1 (3%) | 1–15 |
| AFBR 14 | 1 (2%) | 1–12 | — | 0–10 | — | 0–10 | — | 0–10 |
| AFBR 16 | 21 (51%) | 37–65 | 11 (34%) | 21–51 | 8 (25%) | 14–42 | 8 (25%) | 14–42 |
| AFBR 18 | — | 0–8 | 2 (6%) | 2–20 | 1 (3%) | 1–15 | — | 0–10 |
| AFBR 20 | — | 0–8 | 1 (3%) | 1–15 | — | 0–10 | — | 0–10 |
| AFBR 21 | 1 (2%) | 1–12 | — | 0–10 | — | 0–10 | — | 0–10 |
| AFBR 22 | — | 0–8 | — | 0–10 | — | 0–10 | 1 (3%) | 1–15 |
| AFBR 24 | 1 (2%) | 1–12 | — | 0–10 | — | 0–10 | — | 0–10 |
| AFBR 25 | — | 0–8 | — | 0–10 | — | 0–10 | 2 (6%) | 2–20 |
| AFBR 27 | — | 0–8 | — | 0–10 | — | 0–10 | 1 (3%) | 1–15 |
| AFBR 28 | — | 0–8 | 4 (12%) | 5–28 | 2 (6%) | 2–20 | 2 (6%) | 2–20 |
| AFBR 34 | — | 0–8 | — | 0–10 | 1 (3%) | 1–15 | — | 0–10 |
| AFBR 37 | — | 0–8 | — | 0–10 | 1 (3%) | 1–15 | — | 0–10 |
| AFBR 41 | 6 (15%) | 7–28 | 8 (25%) | 14–42 | 5 (16%) | 7–31 | 3 (9%) | 4–24 |
| AFBR 42 | 1 (2%) | 1–12 | 1 (3%) | 1–15 | — | 0–10 | 6 (19%) | 9–35 |
| AFBR 48 | 4 (10%) | 4–23 | 1 (3%) | 1–15 | 7 (22%) | 12–38 | 7 (22%) | 12–38 |
| AFBR-NT2 | — | 0–8 | — | 0–10 | 1 (3%) | 1–15 | — | 0–10 |
| No. of var types | 9 | 10 | 12 | 10 | ||||
seq., sequences.
95% confidence limit on the true frequency at which the DBL type occurs, estimated from the observed frequency and the no. sequenced.
“—”: none observed.
Nos. in bold indicate the dominant type.
Composition of var Transcripts After 8 Days Asexual Infection in Volunteers Who Received Blood Passage.
3D7C1 and -C2 parasite samples were collected from volunteers 3 and 4, respectively, 8 days after receiving i.v. injection of 3D7B2-infected erythrocytes. A rapid changeover of var transcript types was observed in both samples. Five transcribed DBLα sequences were detected in 3D7C1 and in -C2. The diversity of var transcripts in these volunteers was considerably less than that observed in 3D7B1 and -B2. The dominant transcript in 3D7B2, AFBR16, was not seen at all in these samples. Instead, AFBR13 and -41 had become the dominant var transcripts in 3D7C1 and -C2, respectively. Not only did the major types in C1 and C2 differ from B1, they also differed from each other. AFBR7 (18%, 95% confidence limits 10–31%) and AFBR41 (43%, 95% confidence limits 30–55%) in C2 were not observed in C1; AFBR29 (22%, 95% confidence limits 12–37%) in C1 was not observed in C2 (Table 1).
Composition of var Transcripts in 3D7B1 and -B2 Clones Grown in Vitro.
We examined var transcripts for 24 of the 3D7B1 clones that were harvested after 12 and 13 days of culturing. Nine of the 24 clones gave sequences on both days, of which seven clones showed identical DBLα sequences on both days. Two clones gave different sequences between days 12 and 13. The remaining 15 clones gave a sequence on one of the days. PfEXP1 was used as a control for the quantity of RNA. When a sample failed to give a var sequence, PfEXP1 signal was usually also low (data not shown). Because these transcripts were directly sequenced from the RT-PCR products, clones in which there had been a substantial switching of expressed var gene would give a mixture of sequences. However, sequences in 22 of 24 clones were single unambiguous DBLα sequences, with only 2 of the 24 clones showing a mixture of 2 sequences each. A much higher diversity of DBLα types was found in these clones than in either the premosquito culture or the four in vivo samples (B1, B2, C1, and C2), with 12 different DBLα observed of 25 sequences (Table 1). The frequency of AFBR16, the most frequent DBLα type in the blood taken before culturing, had decreased substantially in the cultured clones from 49 to 8% (P < 0.01). AFBR41, the most frequent DBLα type in 3D7C2 sample, was also the most frequent type in the cultured clones.
For 3D7B2 clones, only day 12 samples and only 12 clones were examined. However, the results were consistent with those from the B1 cultured clones. Of 12 3D7B2 clones, 9 gave single DBLα sequences and 3 mixed. The diversity was similarly high (5 DBLα types of 10 readable sequences), and AFBR16 (51% of transcripts in the blood taken for culturing) was not detected in the clones (Table 1).
Location of Transcribed var Genes on Chromosomes 2 and 3.
A total of 34 var genes were observed to actively transcribe in this study (Tables 1 and 2). Four of these var genes were found in the subtelomeric regions of chromosomes 2 and 3: AFBR42 and -30 on the left and right arms of the chromosome 2, respectively, and AFBR13 and -19 on the left and right arms of the chromosome 3, respectively. On these two chromosomes, the four var genes were the genes closest to the telomeres and were transcribed in the direction from telomere to centromere.
Discussion
This study is the first, to our knowledge, to report the composition and variation of transcribed var genes during acute-stage P. falciparum infections in humans. As the samples were taken from peripheral vessels of volunteers, parasite RNA obtained from these samples largely represents transcriptions of ring-stage parasites, which was confirmed by microscopic examination.
Several papers have reported that in vitro, ring-stage parasites in long-term culture P. falciparum lines (3D7 and FCR3) transcribe many copies of var genes, whereas trophozoites express only one var gene, which is responsible for cytoadherence (7, 9, 21). Additionally, it appears that in 3D7, some of the multiple transcripts detected at ring stage were truncated (21). To investigate whether the 3D7 parasite transcribes var genes in the same manner in vivo, we compared var transcripts in samples without culturing and cultured for 12 or 24 h to mature trophozoites. Our results showed that there was no difference in the total number of var genes transcribed or in the proportion of each transcribed var gene between these time points in this parasite line. In addition, the three major var transcripts were found to be full length, not truncated messages as previously reported (21). These results suggest that the var gene transcripts detected in the in vivo samples are likely to be representative of the proteins expressed on the surface of the infected erythrocytes, although direct evidence of protein expression was not obtained because of limited parasite material. It is also highly unlikely that the detected transcripts resulted from leaky transcription or primer bias, because the types of var gene transcripts detected in the first two volunteers were different from those in subsequent volunteers and in cultured parasites. Therefore, our results indicate that in vivo, transcription of var genes in ring-stage parasites is representative of that in later stages and suggest that, at least in this line of parasite, the control of var gene expression in vivo is different from that observed in vitro. In fact, it has been well documented that as parasites adapt to in vitro cultivation, genomic changes occur frequently (24). These observations suggest that the in vitro adaptation process may alter the control mechanism of var gene expression.
The first two volunteers were infected via mosquito bites. We detected 10 and 9 different var transcripts in the parasite samples taken 12.5 and 13.5 days postinoculation. The observed transcripts represent approximately 20% of the ≈50 var genes in the 3D7 genome. The composition and frequency of the var transcripts detected in these samples were dramatically different from the parent parasite used to infect the mosquitoes. These changes included the disappearance of the var transcript that was dominant in the parent parasite population (AFBRNT1) and the dominance of a different transcript, AFBR16, in both volunteers. Significantly, the same profile was observed in two separate samples from one volunteer (3D7B2) and in a sample of a second volunteer (3D7B1), both of whom were infected with sporozoites from different mosquitoes, hence from different fertilized zygotes. Thus, the propensity for early expression of AFBR16 appears to be an intrinsic property of the set of var genes in 3D7 and not because of some chance event.
These observations would be consistent with a single var type, AFBR16, transcribed in all parasites in the first generation post liver, with a switching rate averaging 16% per generation up to the day of harvest on the fourth cycle post liver. This switching rate is much higher than the 2% switching rate observed for a different line of P. falciparum in vitro (25) and orders of magnitude higher than the average switching rate predicted to sustain a chronic infection by mathematical modeling (14, 15).
Examination of the var transcript frequencies in the next passage of these parasites in vivo and in vitro sheds further light on subsequent switching events. Surprisingly, we could not detect any AFBR16 transcripts after passaging infected blood (3D7B2) from one of the volunteers infected by mosquito bites into a further two volunteers and allowing the parasites to grow for only four more generations. The switch rate required to produce this result would need to be much greater than the average rate of 16% per generation estimated above, because at a rate of 16%, approximately 25% of the parasites would be expected to still express AFBR16 when samples C1 and C2 were taken.
After AFBR16, the rank order of the frequency for the remaining var transcripts in the 3D7B2 sample was similar to the rank order in one of the blood recipient volunteers (3D7C2) (Spearman rank correlation 0.78, P = 0.01). However, in the sample from the other volunteer (3D7C1), AFBR41, the second most abundant var transcript in the donor (3D7B2) and the most dominant type in 3D7C2, was not detected. Instead, AFBR13, one of the next two most dominant transcripts in 3D7B2, dominated in 3D7C1. The results seen in vivo parallel the observations made in vitro after the blood from the mosquito-infected donors was cloned and cultured for a further 12 and 13 days. Similar to 3D7C1 and -C2, AFBR16 was either not detected (cultured 3D7B2) or became a minor component (cultured 3D7B1), again indicating that transcription of this var gene was rapidly turned off.
In contrast to the fast switching of AFBR16, the lower complexity of var transcripts in the 3D7C1 and -C2 blood samples compared with that of the mosquito-infected volunteers suggests that the switch rate for successive var genes after AFBR16 is much slower. Only five var types were detected in the 3D7C1 and -C2 samples compared with 9 and 10 types detected in the 3D7B2 and -B1 samples, respectively. Had successive switching continued at the same rate as the initial switch from AFBR16, a much more complex mixture of transcripts would have been expected in 3D7C1 and -C2.
A total of 34 var transcripts were detected in this study. Four of these transcribed var genes were located at the subtelomeric regions of the published chromosomes 2 and 3, as the last gene to the telomeres. The results suggest that, unlike the trypanosome vsg genes, var genes may be transcribed from multiple sites. For these four copies of var genes, the locations are similar to the transcribed trypanosome vsg genes, except that these var genes are in the opposite orientation with 5′ end of the gene toward the telomeres. The detailed location for the remaining 30 var transcripts can be published when the genome sequencing of 3D7 is itself published. We expect that the transcribed var genes are located at both telomeric and internal regions of the chromosomes, as has been previously reported for a different P. falciparum line (26).
From these data, we hypothesize that the var gene switching process is reset as parasites go through the sexual cycle in the mosquito and the hepatic stage. Similar reinitiation of the antigenic variation process was also observed in the Plasmodium chabaudi model (27), suggesting that a reinitiation mechanism may be universal in malaria parasites. We also propose that in the blood stage, parasites switch on the expression of the first var gene and then switch away from this type at a very high rate. The subsequent switching from the later types occurs at a slower rate. We assume that this rate is so slow that some selection pressure, either through antibody in vivo or through changing accessibility of adherence ligands in vivo or in vitro, is needed to allow ready observation of new var types (14, 15).
The observation that there is a preferred var gene transcript after the mosquito stage extends the similarities between var gene switching in P. falciparum and vsg gene switching in trypanosomes. However, despite this similarity and the similarity in chromosome location, it appears that the mechanism of antigenic variation will be quite different.
Acknowledgments
We thank Dr. Greg Lawrence (Queensland Institute of Medical Research) for coordinating informed consent and the volunteers for allowing us to use their stored samples. We thank Dr. Graham Brown and his colleagues (Walter and Eliza Hall Institute) for performing agglutination antibody tests and Drs. Richard Carter and Lisa Ranford-Cartwright (Institute of Cell, Animal and Population Biology, University of Edinburgh) for providing the 3D7A parasites and information of the parasite culture history. We also thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P. falciparum (3D7) public before publication of the completed sequence. This project is funded by the National Institutes of Health Grant AI47500-02. The experimental work was performed at the Australian Army Malaria Institute. The Sanger Centre (United Kingdom) provided sequence for chromosomes 1, 3–9, and 13, with financial support from the Wellcome Trust. A consortium composed of the Institute for Genome Research, along with the Naval Medical Research Center (U.S.), sequenced chromosomes 2, 10, 11, and 14, with support from the National Institute of Allergy and Infectious Diseases/National Institutes of Health, the Burroughs Wellcome Fund, and the Department of Defense. The Stanford Genome Technology Center sequenced chromosome 12, with support from the Burroughs Wellcome Fund. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania and Monash University (Melbourne, Australia), supported by the Burroughs Wellcome Fund.
Abbreviations
DBLα, Duffy-binding like domain α
AFBRs, 3D7 αAF/αBR PCR products of the DBLα
RT-PCR, reverse transcription–PCR
References
- 1.Su X. Z., Heatwole, V. M., Wertheimer, S. P., Guinet, F., Herrfeldt, J. A., Peterson, D. S., Ravetch, J. A. & Wellems, T. E. (1995) Cell 82, 89-100. [DOI] [PubMed] [Google Scholar]
- 2.Baruch D. I., Pasloske, B. L., Singh, H. B., Bi, X., Ma, X. C., Feldman, M., Taraschi, T. F. & Howard, R. J. (1995) Cell 82, 77-87. [DOI] [PubMed] [Google Scholar]
- 3.Smith J. D., Chitnis, C. E., Craig, A. G., Roberts, D. J., Hudson, T. D., Peterson, D. S., Pinches, R., Newbold, C. I. & Miller, L. H. (1995) Cell 82, 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeder J. C., Cowman, A. F., Davern, K. M., Beeson, J. G., Thompson, J. K., Rogerson, S. J. & Brown, G. V. (1999) Proc. Natl. Acad. Sci. USA 96, 5198-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fried M. & Duffy, P. E. (1996) Science 272, 1502-1504. [DOI] [PubMed] [Google Scholar]
- 6.Rowe J. A., Moulds, J. M., Newbold, C. I. & Miller, L. H. (1997) Nature (London) 388, 292-295. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q., Fernandez, V., Sundström, A., Schlichtherle, M., Datta, S., Hagblom, P. & Wahlgren, M. (1998) Nature (London) 394, 392-395. [DOI] [PubMed] [Google Scholar]
- 8.Gardner J. P., Pinches, R. A., Roberts, D. J. & Newbold, C. I. (1996) Proc. Natl. Acad. Sci. USA 93, 3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherf A., Hernandez-Rivas, R., Buffet, P., Bottius, E., Benatar, C., Pouvelle, B., Gysin, J. & Lanzer, M. (1998) EMBO J. 17, 5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner G. D. H., Morrison, H., Jones, M., Davis, T. M. E., Looareesuwan, S., Buley, I. D., Gatter, K. C., Newbold, C. I., Pukritayakamee, S., Nagachinta, B., et al. (1994) Am. J. Pathol. 145, 1057-1069. [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh K. & Howard, R. J. (1986) Science 231, 150-153. [DOI] [PubMed] [Google Scholar]
- 12.Bull P. C., Lowe, B. S., Kortok, M., Molyneux, C. S., Newbold, C. I. & Marsh, K. (1998) Nat. Med. 4, 358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saul A. (1999) Parasitol. Today 15, 455-457. [DOI] [PubMed] [Google Scholar]
- 14.Molineaux L., Diebner, H. H., Eichner, M., Collins, W. E., Jeffery, G. M. & Dietz, K. (2001) Parasitology 122, 379-391. [DOI] [PubMed] [Google Scholar]
- 15.Paget-McNicol S., Gatton, M., Hastings, I. & Saul, A. (2002) Parasitology 124, 225-235. [DOI] [PubMed] [Google Scholar]
- 16.Vanhamme L., Pays, E., McCulloch, R. & Barry, J. D. (2001) Trends Parasitol. 17, 338-343. [DOI] [PubMed] [Google Scholar]
- 17.Borst P. & Ulbert, S. (2001) Mol. Biochem. Parasitol. 114, 17-27. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J. K., Rubio, J. P., Caruana, S., Brockman, A., Wickham, M. E. & Cowman, A. F. (1997) Mol. Biochem. Parasitol. 87, 49-60. [DOI] [PubMed] [Google Scholar]
- 19.Deitsch K. W., Calderwood, M. S. & Wellems, T. E. (2001) Nature (London) 412, 875-876. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Q., Lawrence, G., Reed, C., Stowers, A., Ranford, C. L., Creasey, A., Carter, R. & Saul, A. (1997) Am. J. Trop. Med. Hyg. 57, 495-500. [DOI] [PubMed] [Google Scholar]
- 21.Taylor H. M., Kyes, S. A., Harris, D., Kriek, N. & Newbold, C. I. (2000) Mol. Biochem. Parasitol. 105, 13-23. [DOI] [PubMed] [Google Scholar]
- 22.Gardner M. J., Tettelin, H., Carucci, D. J., Cummings, L. M., Aravind, L., Koonin, E. V., Shallom, S., Mason, T., Yu, K., Fujii, C., et al. (1998) Science 282, 1126-1132. [DOI] [PubMed] [Google Scholar]
- 23.Bowman S., Lawson, D., Basham, D., Brown, D., Chillingworth, T., Churcher, C. M., Craig, A., Davies, R. M., Devlin, K., Feltwell, T., et al. (1999) Nature (London) 400, 532-538. [DOI] [PubMed] [Google Scholar]
- 24.Biggs B. A., Kemp, D. J. & Brown, G. V. (1989) Proc. Natl. Acad. Sci. USA 86, 2428-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts D. J., Craig, A. G., Berendt, A. R., Pinches, R., Nash, G., Marsh, K. & Newbold, C. I. (1992) Nature (London) 357, 689-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer K., Horrocks, P., Preuss, M., Wiesner, J., Wunsch, S., Camargo, A. A. & Lanzer, M. (1997) Mol. Cell. Biol. 17, 3679-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brannan L. R., Turnar, C. M. R. & Phillips, R. S. (1994) Proc. R. Soc. London Ser. B 256, 71-75. [DOI] [PubMed] [Google Scholar]