Abstract
We report a microarray of cantilevers to detect multiple unlabeled biomolecules simultaneously at nanomolar concentrations within minutes. Ligand-receptor binding interactions such as DNA hybridization or protein recognition occurring on microfabricated silicon cantilevers generate nanomechanical bending, which is detected optically in situ. Differential measurements including reference cantilevers on an array of eight sensors can sequence-specifically detect unlabeled DNA targets in 80-fold excess of nonmatching DNA as a background and discriminate 3′ and 5′ overhangs. Our experiments suggest that the nanomechanical motion originates from predominantly steric hindrance effects and depends on the concentration of DNA molecules in solution. We show that cantilever arrays can be used to investigate the thermodynamics of biomolecular interactions mechanically, and we have found that the specificity of the reaction on a cantilever is consistent with solution data. Hence cantilever arrays permit multiple binding assays in parallel and can detect femtomoles of DNA on the cantilever at a DNA concentration in solution of 75 nM.
Microarray methods employing the detection of specific biomolecular interactions are now an indispensable tool for disease diagnosis (1, 2), genome research (3, 4), and drug discovery (5). However most current approaches, for example DNA microarrays and enzyme-linked immunosorbent assays (ELISAs), rely on the labeling of samples with a fluorescent or radioactive tag. This highly sensitive procedure is time-consuming and expensive. The chemical modification and global amplification of the nucleic acid samples are achieved by PCR, although this process potentially can introduce artifacts caused by the preferential amplification of certain sequences (6). Alternative label-free methods include surface plasmon resonance (SPR) (7, 8) and quartz crystal microbalance (9), which rely on mass detection. We recently reported a nanomechanical biodetection mechanism by using miniaturized silicon cantilevers (10) that require no external probes or labeling and can operate autonomously. We observed that when biospecific interactions occur between a receptor immobilized on one side of a cantilever and a ligand in solution, the cantilever bends, which is detected optically. The general applicability of this label-free detection method has been shown for DNA hybridization, the detection of single base mismatches (10), protein-antibody recognition of IgG by immobilized protein A (10), and the detection of prostate-specific antigens (11). This nanoactuation mechanism has important advantages, because cantilevers are microfabricated by standard low-cost silicon technology and, by virtue of the size achievable, are extremely sensitive to detect biological interactions. For example, we can measure the hybridization of label-free 12-mer oligonucleotides. Furthermore, these miniaturized sensors are highly suitable for parallelization into arrays (12). We have scaled-up this technology and now report a microarray of cantilevers that can directly detect multiple unlabeled biomolecules in a single-step reaction without any sample manipulation (see Fig. 1A). In addition to the ability to perform multiple independent experiments, an array of cantilevers permits an internal reference sensor, which is essential for biospecific detection. Automatic-injection protocols require only 100 μl of sample and give rapid in situ analysis within minutes. The signal transduction process is repeatable when denaturation or unbinding agents are used, enabling cyclic operation.
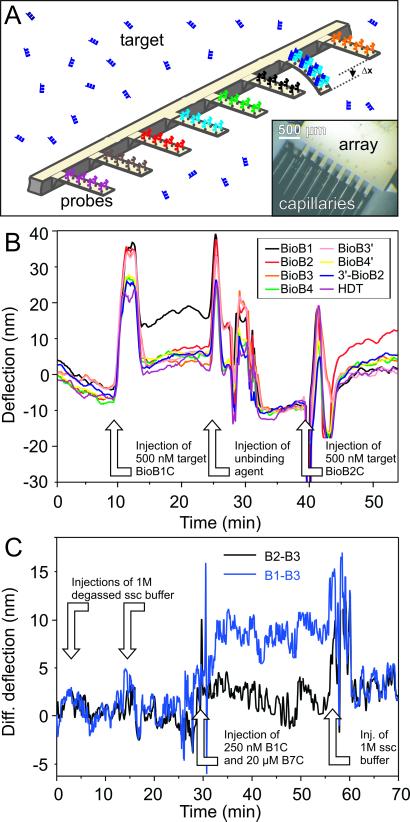
Figure 1.
(A) The preparation of an active cantilever biosensor array and an illustration of the basic principle of nanomechanical label-free biodetection are shown. (Inset) The incubation of individual gold-coated cantilevers (dimensions: 500 × 100 × 1 μm) in microcapillaries, each containing a different solution of thiolated probe DNA. The schematic illustrates how target DNA injected into solution will hybridize sequence-specifically to its complementary partner immobilized on a particular cantilever. Hybridization generates a compressive surface stress, which causes the cantilever to bend with respect to a reference probe-coated cantilever, giving rise to a differential bending signal, Δx, of 10 nm. (B) Absolute deflection signals from an eight-cantilever array were monitored in real time. By convention, a positive deflection signal corresponds to the downward bending of the cantilever (away from the gold coating) because of the generation of a compressive surface stress. A negative signal corresponds to the upward motion of the cantilever resulting from a tensile surface stress. Changes in environment (index of refraction, temperature, etc.) might result in lever movements, but then all levers react simultaneously to the changes (see discussion in the Fig. 2 legend). Throughout the process, the absolute deflection of individual cantilevers was recorded, and simultaneously we extracted the differential signal (e.g., the deflection of the BioB1-functionalized cantilever minus that of a reference, noncomplementary sequence BioB4-functionalized cantilever). Analogous hybridization experiments were performed by injecting other target sequences and gave similar differential signals between cantilevers functionalized with complementary probes and reference cantilevers modified with noncomplementary sequences [e.g., injection of 500 nM target BioB1C in 1 M NaCl sodium citrate hybridization (SSC) buffer]. The cantilevers are deflected but will regain their equilibrium within minutes. Washing the array with dehybridization agents (injection of unbinding agent) dehybridized the specifically interacting oligos, and then the array is ready for the next cycle (injection of 500 nM target BioB2C). The experiments demonstrate that the differential bending is clearly sequence-specific and provides an unambiguous “yes” or “no” response. (C) Detection of 250 nM BioB1C in the presence of 20 μM BioB7C. B2 and B3 denote reference cantilevers in this experiment, because no BioB2C and BioB3C is injected. The differential response B1–B3 shows a clear signal of ≈7 nm (blue curve), whereas the difference of two reference cantilevers only shows a base line (black).
To quantify the specificity and molecular sensitivity of cantilever-array technology, we used synthetic oligonucleotides, the structure and properties of which can be changed systematically, because we already have shown that it is possible to detect 12-mers, 16-mers, and single base mismatches (10). The specificity is determined by the thermodynamics of the ligand-receptor binding interaction on the cantilever, and we show that it is possible to measure thermodynamic equilibrium constants by using cantilevers. The sensitivity of cantilever arrays corresponds to the number of molecular interactions that gives rise to a nanomechanical response. We have determined this number by using radiolabeled probes. Our systematic experiments also provided new insight into the molecular origin of the nanomechanical signal, which is important because the observed bending signal depends on both the thermodynamic target-probe affinity and the nanomechanical response of the cantilever.
Materials and Methods
Cantilever Preparation.
Microfabricated arrays of eight identical silicon cantilevers with 250-μm pitch and a spring constant of 0.02 N/m were provided by the micro- and nanomechanics group at the IBM Zurich Research Laboratory. Before use, cantilevers were cleaned with piranha and 10% hydrofluoric acid in water. Clean arrays were coated on one side with a 2-nm titanium adhesion layer followed by 20 nm of gold by using an electron double-beam evaporator (BOC Edwards, Sussex, U.K.). The functionalization of each cantilever with a different thiolated probe sequence was performed in parallel and under identical conditions by using microcapillaries (see Fig. 1A). Individual cantilevers were inserted into microcapillaries filled with a 40 μM solution of thiolated probe DNA in triethyl ammonium acetate buffer (TEAA, 50 mM) for 20 min, rinsed, and dried in nitrogen. Functionalized arrays could be stored for several days at 4°C without significant loss in performance. Before use, the arrays were equilibrated for several hours in saline SSC buffer until the differential signal became stable.
Hybridization Experiments.
Different concentrations of target DNA were injected automatically at various flow rates. All measurements were taken at 22°C. After binding, hybridized oligonucleotides were denatured chemically by purging the cell with dehybridization agents, e.g., 30% urea salts in buffer. The dehybridization efficiency depends on injection speed and injection volume.
Instrument.
The cantilever array was mounted at an angle of 11° toward the incoming laser beam. The beam is redirected to the detector with an adjustable mirror. Time-multiplexed vertical-cavity surface-emitting lasers with regulated power supply (operated at 1 Hz, wavelength 760 nm, Avalon Photonics, Zurich, Switzerland) were used in combination with adjustable optics to yield a pitch of 250 μm. A linear position-sensitive detector (SiTek Electro Optics, Partille, Sweden) was used for beam-deflection readout of each cantilever with an accuracy of 0.1 nm, preamplified, and the data were stored by using a National Instruments (Austin, TX) PCMCIA 16XE50 (16 bit, 200 kS/s) data-acquisition card. The instrument is driven by LABVIEW software to control liquid exchange via a syringe pump (GENIE, Kent Scientific, Torrington, CT), and a 10-position valve selector (Rheodyne, Rohnert Park, CA), data acquisition, and data processing.
Radiolabeling Experiments.
The amount of chemisorbed probes and the kinetics of oligonucleotide packaging were monitored by using 32P-labeled oligonucleotides. The quantification of the samples was done by digital imaging of the radiolabeled oligonucleotide surfaces produced with a PhosphorImager scanner and quantified by using internal standards and IMAGEQUANT software (Molecular Dynamics).
The 20-mer oligonucleotide with the sequence 5′-CCGTTGCGATGTCAGTGGTA-SH-3′ with a (CH2)6-SH modification on the 3′ end was labeled on the 5′ end with Redivue [γ-32P]ATP (Amersham Biosciences) and by using the T4 polynucleotide kinase (Amersham Biosciences). The integration efficiency of 32P was ≈10% and has been checked by scintillation counting. Residual [γ-32P]ATP was removed by passing the labeled oligonucleotides through ProbeQuant spin columns (Amersham Biosciences). DTT was removed by extracting five times into ethyl acetate. The final concentration of the radioactively labeled and thiol-modified oligos was adjusted with cold thiol-modified oligos to a concentration of 3 μM. The labeled oligonucleotides then were incubated on 1-cm2 square-shaped gold-coated silicon substrates in various buffers for various lengths of incubation time. After incubation the samples were rinsed thoroughly with incubation buffer.
Results and Discussion
By using microcapillaries, individual cantilevers were modified in parallel with a different sequence of thiolated DNA (probe). These 12-mer sequences (see Table 1) are derived from the Escherichia coli biotin operon gene B by using a PCR primer program (OMIGA, Oxford Molecules, Oxford), and termed BioB1, BioB2, BioB3, and BioB4 (EMBL accession no. J04423). All hybridization experiments were performed in a temperature-controlled liquid cell containing a modified cantilever array immersed in SSC hybridization buffer at 22°C. Different solutions of complementary DNA (targets: BioB1C, BioB2C, etc.) were injected into the liquid cell, and the mechanical bending of individual cantilevers was tracked by using time-multiplexed vertical-cavity surface-emitting lasers.
Table 1.
Oligonucleotides purified twice by HPLC and PAGE gel purification and used as obtained from Microsynth (Balgach, Switzerland)
| Type of DNA | Name | Base sequence 5′–3′ |
|---|---|---|
| Thiolated probes | BioB1 | ACA TTG TCG CAA |
| BioB2 | TGC TGT TTG AAG | |
| BioB3 | CCG GAA GAT TGC | |
| BioB4 | GGA AGC CGA GCG | |
| Complementary targets | BioB1C | TTG CGA CAA TGT |
| BioB2C | CTT CAA ACA GCA | |
| BioB3C | GCA ATC TTC CGG | |
| BioB4C | CGC TCG GCT TCC | |
| BioB7C | AGA TCG CGC CGG | |
| Targets with extensions | BioB2C-(A)12-3′ | CTT CAA ACA GCA AAA AAA AAA AAA |
| 5′-(A)6-BioB2C-(A)6)-3′ | AAA AAA CTT CAA ACA GCA AAA AAA | |
| 5′-(A)12-BioB2C | AAA AAA AAA AAA CTT CAA ACA GCA |
The thiol modification with a 5′ HS(CH2)6 linker enables covalent binding to gold-coated cantilever surfaces.
The data in Fig. 1B show that when a 500 nM solution of the BioB1C target sequence is injected into the liquid cell, it finds its complementary BioB1 sequence immobilized on the surface of one particular cantilever on the array within minutes and causes the cantilever to bend downward. The bending occurs because the hybridization reaction produces a difference in surface stress between the probe-functionalized gold side of the cantilever and the lower silicon surface, which produces a compressive surface-stress signal. Hybridization generated an average differential deflection signal of 9.8 nm, which can be converted into a compressive surface stress of 2.7 × 10−3 N/m, by using Stoney's equation (13). This signal amplitude is consistent with our previous work to detect single base mismatches (10). The differential signals were observed to be reproducible within ±1 nm for a given concentration. The arrays can be regenerated after every hybridization experiment with dehybridization agents, e.g., urea salts in buffer, which are known to break the hydrogen bonds between complementary base pairs. A single array could be recycled more than 10 times without significant loss in performance, corresponding to more than 100 independent experiments per cantilever array.
To prove sequence specificity, a mixture of 250 nM BioB1C and 20 μM BioB7C was injected (i.e., 80-fold excess of nonmatching DNA as a background). The differential signals B1–B3 (probe–reference) and B2–B3 (reference–reference) are shown in Fig. 1C.
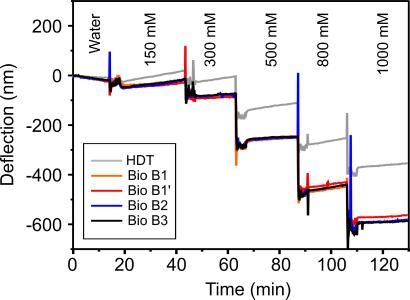
To illustrate the importance of differential measurements, an array was exposed to various buffer NaCl concentrations (range, 150–2,000 mM) where four cantilevers were coated with probe DNA sequences and one reference cantilever coated with hexadecanethiol, which has no ionizable groups (see Fig. 2). The experiment demonstrates the sensitivity of probe-coated sensors to small changes in buffer salt concentration and the importance of differential measurements for accurate and reliable biodetection on a cantilever array. Our data show that the effect of salt is independent of the probe base sequence, and therefore the reference cantilever in all hybridization studies is coated with noncomplementary DNA.
Figure 2.
The absolute response of an array of probe-coated cantilevers to different SSC buffer NaCl concentrations. Four cantilevers were coated with different DNA probe sequences, namely BioB1 (BioB1′ denotes another cantilever coated with BioB1), BioB2, and BioB3, and one reference cantilever was modified with hexadecane thiol, which has no ionizable or polar groups. The large response of the reference hexadecane thiol cantilever, 380 nm, reflects the significant nonspecific contribution and changes in refractive index of the buffer solutions to the absolute signal of probe-coated cantilevers. However, a differential measurement deconvoluted the signal to reveal the component that was specific to only the probe DNA, namely a differential deflection of 200 nm arising from the electrostatic repulsion between phosphate groups on neighboring probe molecules and charge screening by Na+ in solution. The stepwise appearance of the signal after buffer exchange reflects both the efficient exchange of solvent in the liquid cell and the stability of the instrument.
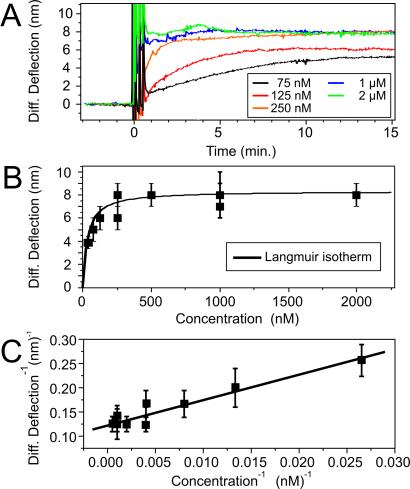
Thermodynamic measurements focused on the specific binding of the BioB2C target in solution to the surface-bound probe BioB2. The cantilever differential bending signal was found to vary with the concentration of the injected target BioB2C in solution (range, 75–2,000 nM). The data in Fig. 3A show the differential deflection signals for various target concentrations as a function of time. At low concentrations (75 nM), the differential signal slowly increased, reflecting the diffusion-limited approach of the target toward the surface-bound probe. After 12 min, equilibrium is achieved, and the signal stabilizes to reach a differential equilibrium value of 5.3 nm. At concentrations above 250 nM, the signal rapidly reached equilibrium within 5 min to give a maximum 8.1 ± 0.8 nm differential deflection. Subsequent target injections of increasingly higher concentrations did not change the magnitude of the equilibrium saturation signal (8.1 nm). Hybridization experiments on different cantilever arrays showed similar concentration-dependent signals with a standard deviation of ≈10%. Fig. 3B shows the equilibrium differential deflection signal as a function of target concentration.
Figure 3.
(A) The target concentration-dependent studies show the time scale for the cantilever to equilibrate to different concentrations of oligonucleotides. Systematic hybridization measurements were performed on a single cantilever, and sequence-specific differential signals were extracted by using a reference cantilever. The spikes in the differential signal result from the injection of the solutions into the fluid chamber, but rapid equilibration is reached within a few minutes. Depending on injection speed and volume, the dehybridization of the double helix was achieved by a urea injection. (B) Concentration-versus-deflection plot of many levers shows that the hybridization of the oligonucleotides follows the Langmuir isotherm model (15). (C) Langmuir plot (15) of the data in B.
These concentration-dependent data were fitted to the Langmuir adsorption isotherm model (14) to derive the thermodynamic surface-solution equilibrium constant, K−1. The theory was developed originally to describe the reaction of gas-phase molecules on a metal surface but also can model solution-phase data. Assuming the target-probe binding events are independent and unaffected by surface coverage, the Langmuir adsorption isotherm states
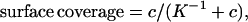 |
where c is the concentration of target molecules in solution. The surface coverage (occupancy) is a ratio of the number of bound molecules to the total number of available binding sites. If we assume the bending signal is proportional to the surface coverage,
 |
where a is the proportionality constant. At full occupancy (saturation) a is a measure of the nanomechanical response of the cantilever, independent of target concentration. A plot of deflection−1 versus concentration−1 is shown in Fig. 3C. In 750 mM NaCl buffer, K−1 = 41 × 10−9 M and a = 8.1 ± 0.4 nm. The Gibbs free energy for the hybridization of DNA on a cantilever was calculated to be ΔG(BioB2 cantilever) = 41.4 kJ/mol. This value was smaller but comparable to theoretical solution-phase data, ΔG(BioB2 solution) = 50.5 kJ/mol, calculated from the Watson–Crick base sequence (15). It is highly agreeable with the most favorable binding measurements taken on other supported phase methods. For example, our nanomechanical thermodynamic data agree very favorably with recent SPR imaging data [Kd = 1.8 × 108 M−1 for thiolated 18-mer probes (8)] by using a highly optimized SPR configuration. Clearly tethering one end of an oligonucleotide to a surface is expected to affect the formation of duplex with target molecules in solution. Surface-bound probes are constrained and no longer able to diffuse and behave as they would in free solution, and thus hybridization efficiencies are reduced. However, supported phase-detection methods have important advantages, because very small quantities of material are required, chips can be reused, and multiple oligonucleotides can be screened in parallel. The nature of the underlying surface, coverage (packing density), and the linker chemistry are important factors, and oligonucleotide probes modified with short alkanethiol linkers on gold surfaces have been studied extensively by radiolabeling, SPR (7, 8), and quartz crystal microbalance (9).
The influence of charge screening on the thermodynamic equilibrium constant was investigated at different NaCl concentrations (150, 750, and 1,000 mM). The concentration-dependent deflection measurements were modeled to Langmuir adsorption isotherms (see Table 2). As expected, K−1 decreased with increasing NaCl concentration because of enhanced charge screening. Interestingly, a remained relatively independent of charge screening.
Table 2.
Summary of the effect the buffer NaCl concentration has on the thermodynamic equilibrium dissociation constant (K−1), the proportionality constant (a), and the Gibbs free energy for hybridization interaction occurring on the cantilever surface (ΔG)
| [NaCl], mM | K−1, nM | a, nm | ΔG, kJ/mol |
|---|---|---|---|
| 150 | 120 ± 30 | 8.3 ± 0.8 | 39.5 |
| 750 | 33 ± 8 | 8.3 ± 0.8 | 42.3 |
| 1,000 | 31 ± 5 | 9.8 ± 1.0 | 42.8 |
The sensitivity of the cantilever-array technology is determined by the number of binding events that give rise to the nanomechanical surface-stress signal. In all experiments, the areal density of probe DNA was kept constant, and the number of molecules on a single cantilever was determined by using 32P-radiolabeled probes. Instead of directly performing radiolabeling experiments on the small cantilevers, measurements were performed on square-shaped pieces of silicon wafer (1 cm2) coated with titanium and gold similarly to the cantilevers, which have been used. The surfaces were exposed to 3 μM radioactively labeled and thiolated DNA by using identical buffer conditions as in the cantilever preparation. To check the influence of ionic conditions, three different incubation buffers were used [75% TEAA buffer (50 mM)/25% EtOH, 75% TEAA/100 mM NaCl, and 75% TEAA/500 mM NaCl]. A maximum signal, or packing density of 1.3 × 1013 probes per cm2, was obtained after only 5 min of incubation. The density of probe DNA was unaffected by the salt concentration of the buffer or dilutions with ethanol. This value is in agreement with other reports for 12-mers (16, 17) and corresponds to 1.5 × 1010 probes per cantilever, where a single-stranded DNA molecule occupies an area of 3.2 nm2, which approaches the theoretical maximum packing density (ref. 16; this estimate does not take into account the surface roughness of the gold film). Radiolabeling experiments by Tarlov and coworkers have shown that less than 10% of these immobilized probes are free to hybridize because of steric crowding, electrostatic repulsion between neighboring surface species, and nonspecific adsorption of thiolated probes via polar side groups (16–18). Therefore we postulate that a differential signal of 8.1 nm corresponds to the formation of 109 double helices or femtomoles of DNA binding on the cantilever. To our knowledge, this is one of the highest levels of sensitivity achieved by a label-free oligonucleotide-detection method by virtue of the tiny reaction area on the cantilever (cantilever dimensions, 500 × 100 μm2) compared with SPR imaging (500 × 500 μm), conventional SPR (mm2), and quartz crystal microbalance (cm2). If we assume that the probe-target binding events are independent, then a single duplex molecule exerts a compressive stress of 10−12 N/m.
The question that remains unanswered is what causes the cantilever to bend downward after target-probe hybridization? Understanding the molecular origin of the surface-stress signal is critical, because the observed bending depends on both the thermodynamic target-probe affinity and the nanomechanical response of the cantilever. Moreover, these two factors may be convoluted such that a strong binding affinity will not necessarily produce a large surface stress. The Langmuir adsorption isotherm makes it possible to separate and study the relative contributions of the two factors independently. Two possible mechanisms for the generation of the compressive surface stress after hybridization are increased electrostatic repulsion between charged phosphate groups on duplex DNA and an increase in chain-packing density on the cantilever surface. Others have suggested that the signal is governed by the change in configurational entropy after hybridization and observed a tensile surface stress for 20-mers and longer (19). After hybridization, electrostatic as well as steric effects are repulsive, which could bend the cantilever downward. The data in Table 2 show that the nanomechanical saturation signal was relatively independent of buffer NaCl concentration and suggests that electrostatic repulsive interactions after hybridization are small. The high density of probes immobilized on the surface of the cantilever suggests that physical steric crowding is the dominant component of the nanomechanical signal. This hypothesis was tested by optimizing the accessibility of probe DNA in a mixed monolayer with 6-mercapto-1-hexanol by using the protocol developed by Herne and Tarlov (16, 17). The differential hybridization signal dropped to 1 nm, which is close to the detection limit of our instrument, and confirms that surface preparations and the spatial arrangement of probe DNA are critical.
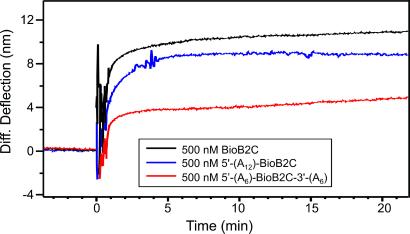
The ability of microarrays to detect and discriminate specific recognition sites or coding regions on a long sequence of target DNA is important for the analysis of genomic DNA (1). The influence of 5′- and/or 3′-overhanging extensions on the nanomechanical signal was investigated with 24-mer targets. Three different constructs were studied, each designed to contain a section complementary to the surface-bound probe BioB2 and a nonspecific polyadenine tail: BioB2C-(A)12-3′ had a 3′ (adenine)12 tail, 5′-(A)6-BioB2C-(A)6-3′ had (adenine)6 tails at both 5′ and 3′ positions, and 5′-(A)12-BioB2C had an (adenine)12 tail at the 5′ end only (see Table 1). Hybridization between all three 24-mer targets and the immobilized BioB2 probe produced a compressive surface stress; the magnitude of the differential signal depended on the position of the tail (see Fig. 4). An injection of 500 nM BioB2C gave a differential signal of 10 nm (the data were corrected for 3 nm/h thermal drift); 5′-(A)12-BioB2C 8 nm, 5′-(A)6-BioB2C-(A)6-3′ 6 nm, and BioB2C-(A)12-3′ always gave the smallest signal (1 nm). The relative bending signals reflect the steric hindrance of the overhanging extensions of the long target DNA and their position with respect to the surface-immobilized probe. Concentration-dependent experiments were performed on 5′-(A)12-BioB2C, and the data were modeled to the Langmuir adsorption isotherm. We found K−1 (cantilever, 5′-(A)12-BioB2C) = 222 × 10−9 M and a = 12.3 nm and ΔG (cantilever, 5′-(A)12-BioB2C) = 38 kJ/mol in 1 M NaCl buffer. As expected the hybridization efficiency of a target with a 5′-overhang extension is significantly lower than that of the complementary oligonucleotide (20). Notably, the nanomechanical response was also larger than for 5′-(A)6-BioB2C-3′-(A)6, suggesting that a 5′ overhang, dangling into solution (i.e., away from the gold-coated cantilever surface), enhances the compressive surface-stress signal (data not shown).
Figure 4.
The nanomechanical response after hybridization to targets with different overhanging extensions. Sequential experiments were performed on a single cantilever followed by dehybridization washing steps, and differential measurements were extracted. The oligonucleotide B2C is the perfect matching complement, whereas 5′-(A)12-BioB2C (Table 1) has an overhanging single-stranded DNA end toward the lumen dangling away from the cantilever surface, and 5′-(A)6-BioB2C-(A)6-3′ has overhanging ends on both sides of the immobilized probe on the cantilever surface.
To conclude, we have developed a nanomechanical cantilever array for multiple quantitative biomolecular detection. This technology complements and extends current DNA and protein microarray methods, because nanomechanical detection requires no labels, optical excitation, or external probes and is rapid, highly specific, sensitive, and portable. We can simultaneously detect nanomolar concentrations of different unlabeled DNA sequences within minutes and discriminate overhangs through differential measurements. The nanomechanical response is sensitive to the concentration of oligonucleotides in solution, and thus we can determine how much of a given biomolecule is present and active. We show that it is possible to investigate the thermodynamics of the biomolecular interaction mechanically and find that the specificity of the reaction on a cantilever is consistent with the most favorable surface-supported binding-affinity measurements. Hence arrays permit multiple binding assays in parallel. Our experiments suggest that mainly steric hindrance effects cause the compressive surface-stress bending signal, a mechanism that has the sensitivity to detect femtomoles of oligonucleotides binding on a cantilever. We have shown already that it is possible to detect single base mismatches, and in principle cantilever arrays also could quantify gene-expression levels of mRNA, protein–protein, drug-binding interactions, and other molecular recognition events in which physical steric factors are important. Furthermore, the ability to sequence-specifically detect short oligonucleotide 12-mers suggests that nanomechanical sensors will be useful for analyzing small molecule-binding interactions. Fabricating thinner cantilevers will enhance the molecular sensitivity further, and integrating arrays into microfluidic channels will reduce the amount of sample required significantly (21). In contrast to SPR, cantilevers are not limited to metallic films, and other materials will be explored, e.g., cantilevers made from polymers. In addition to surface-stress measurements, operating cantilevers in the dynamic mode will provide information on mass changes, and current investigations will determine the sensitivity of this approach. Currently it is possible to monitor more than 1,000 cantilevers simultaneously with integrated piezoresistive readout (12), which in principle will allow high-throughput nanomechanical genomic analysis, proteomics, biodiagnostics, and combinatorial drug discovery.
Acknowledgments
We thank H. Schmid, U. Drechsler, H. E. Rothuizen, P. Vettiger, R. Allenspach, and P. F Seidler (IBM Zurich Research Laboratory) as well as F. M. Battiston and J.-P. Ramseyer (Institute of Physics, University of Basel, Basel) for helpful discussions. This project was funded partially by the National Center of Competence in Research in Nanoscale Science (Basel), the Swiss National Science Foundation, the Commission for Technology and Innovation (Berne, Switzerland), and the Royal Society (London). R.M. is a Dorothy Hodgkin Research Fellow.
Abbreviation
- SPR
surface plasmon resonance
References
- 1.Lindblad-Toh K, Tanenbaum D M, Daly M J, Winchester E, Lui W O, Villapakkam A, Stanton S E, Larsson C, Hudson T J, Johnson B E, et al. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 2.Duggan D, Bittner M, Chen Y, Meltzer P, Trent J. Nat Genet Suppl. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 3.MacBeath G, Schrieber S L. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 4.Wang D G, Fan J, Siao C, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 5.Southern E, Mir K, Shchepinov M. Nat Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 6.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 7.Georgiadis R, Peterlinz K P, Peterson A N. J Am Chem Soc. 2000;122:3166–3173. [Google Scholar]
- 8.Nelson B P, Grimsrud T E, Liles M R, Goodman R M, Corn R M. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 9.Caruso F, Rodda E, Furlong D N. Anal Chem. 1997;69:2043–2049. doi: 10.1021/ac961220r. [DOI] [PubMed] [Google Scholar]
- 10.Fritz J, Baller M K, Lang H P, Rothuizen H, Vettiger P, Meyer E, Güntherodt H J, Gerber C, Gimzewski J K. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 11.Wu G H, Datar R H, Hansen K M, Thundat T, Cote R J, Majumdar A. Nat Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 12.Lutwyche M I, Despont M, Drechsler U, Dürig U, Häberle W, Rothuizen H, Stutz R, Widmer R, Binnig G K, Vettiger P. Appl Phys Lett. 2000;77:3299–3301. [Google Scholar]
- 13.Stoney G G. Proc R Soc London Ser A. 1909;82:172–175. [Google Scholar]
- 14.Hudson J B. Surface Science: An Introduction. Boston: Butterworth–Heinmann; 1992. pp. 181–183. [Google Scholar]
- 15.Breslauer K J, Frank R, Blocker H, Marky L A. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herne T M, Tarlov M J. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 17.Steel A B, Levicky R L, Herne T M, Tarlov M J. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavicky R, Herne T M, Tarlov M J, Satija S K. J Am Chem Soc. 1998;120:9787–9792. [Google Scholar]
- 19.Wu G, Ji H, Hansen K, Thundat T, Datar R, Cote R, Hagan M F, Chakraborty A K, Majumdar A. Proc Natl Acad Sci USA. 2001;98:1560–1564. doi: 10.1073/pnas.031362498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southern E, Mir K, Shchepinov M. Nat Genet Suppl. 2001;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 21.Thaysen J, Marie R, Boisen A. IEEE International Conference on Micro Electro Mechanical Systems, Technical Digest. New York: IEEE; 2001. pp. 401–404. [Google Scholar]