Abstract
Tetrameric hemoglobins are the most widely used systems in studying protein cooperativity. Allosteric effects in hemoglobins arise from the switch between a relaxed (R) state and a tense (T) state occurring upon oxygen release. Here we report the 2.0-Å crystal structure of the main hemoglobin component of the Antarctic fish Trematomus newnesi, in a partial hemichrome form. The two α-subunit iron atoms are bound to a CO molecule, whereas in the β subunits the distal histidine residue is the sixth ligand of the heme iron. This structure, a tetrameric hemoglobin in the hemichrome state, demonstrates that the iron coordination by the distal histidine, usually associated with denaturing states, may be tolerated in a native-like hemoglobin structure. In addition, several features of the tertiary and quaternary organization of this structure are intermediate between the R and T states and agree well with the R → T transition state properties obtained by spectroscopic and kinetic techniques. The analysis of this structure provides a detailed pathway of heme–heme communication and it indicates that the plasticity of the β heme pocket plays a role in the R → T transition of tetrameric hemoglobins.
Keywords: protein denaturation|protein function|x-ray structure|Antarctic
In vertebrate hemoglobins (Hbs) the binding of exogenous ligands is associated with a variation of the heme iron coordination (1, 2). Indeed, in unliganded states high-spin iron is pentacoordinated, with the proximal histidine residue in the fifth coordination position, whereas, upon binding of ligands such as O2 and CO, a low-spin hexacoordination is observed. Notably, low-spin complexes may also be obtained by the replacement of the exogenous ligand with a residue of the globin chain. In fact, under unusual circumstances, iron can coordinate both the proximal and distal histidine residues (3). Depending on the oxidation state of the iron, these bishistidine complexes are called hemichrome [Fe(III)] or hemochrome [Fe(II)]. Over the years hemichromes have been considered as precursors of Hb denaturation processes, such as unfolding, precipitation, and heme dissociation. In fact, hemichrome formation, which is rather slow in human Hb, is accelerated by denaturing agents. Under denaturing conditions, the formation of the bishistidine complex can be favored by the altered structure of the globin, which allows the binding of the distal His to the iron.
More recently, Rifkind and coworkers (3–5) have demonstrated that hemichrome and hemochrome states can be obtained under nondenaturing conditions and that bishistidine complexes may represent a populated conformational substate of native human Hb. It is also worth mentioning that hemichrome states of myoglobin and Hb are associated with heart disease and blood abnormalities, respectively. Nevertheless, structural information on these compounds is still rather limited. So far, three-dimensional structures of Hbs in hemichrome states have been reported only for monomeric Hb extracted from the sea cucumber Caudina arenicola (6) and for dimeric nonsymbiotic rice Hb (7). On the other hand, difficulties in trapping hemichrome states in vertebrate Hbs have hitherto prevented structure determinations of tetrameric Hbs in the hemichrome state.
We have recently shown that, in contrast to human Hb, the major Hb component (HbTn) of the Antarctic fish Trematomus newnesi rapidly forms hemichromes when exposed to air (8). This finding has permitted structural studies on this system. Here we report the 2.0-Å crystal structure of HbTn in a partial hemichrome state. This structure is characterized by a different binding state of the α and β chains. A CO molecule is bound to the α heme iron, whereas a bishistidine complex is observed at the β heme. Surprisingly, the analysis of the quaternary structure and of the critical interface α1β2(α2β1) reveals that this hemichrome has an intermediate structure between the relaxed (R) and tense (T) Hb functional states.
Materials and Methods
Purification, Crystallization, and Data Collection.
The major Hb component of T. newnesi was purified from the hemolysate by ion-exchange chromatography as previously described (9). The carbonmonoxy form of HbTn (HbTnCO), when exposed to air, rapidly evolves toward the hemichrome state (8). To investigate the structural features of this form, several crystallization trials of air-exposed HbTnCO with monomethoxypolyethylene glycol (MPEG) 5000 as precipitant were set up at 20°C. The free interface diffusion technique was used by pouring a solution of HbTnCO, exposed to air for 2 hr, in 60 mM Tris⋅HCl pH 7.6 (10 μl) onto a solution of 10% (wt/vol) MPEG 5000 (10 μl) in 100 mM sodium acetate, pH 5.8, in a capillary (final pH 7.4). The capillaries were sealed in air. Additional details regarding HbTn hemichrome formation and crystallization have been reported elsewhere (8).
Single crystals suitable for x-ray analysis were obtained in 2–3 days. From one crystal, x-ray diffraction data were collected up to 2.0-Å resolution by using a Nonius DIP2030b imaging plate mounted on a Nonius FR591 rotating anode. Statistics of data processing are reported in Table 1.
Table 1.
Data collection and refinement statistics
| Measurement | Value |
|---|---|
| Crystal data | |
| a, Å | 89.85 (91.17)* |
| b, Å | 88.19 (88.06)* |
| c, Å | 55.14 (55.25)* |
| β, ° | 97.8 (97.7)* |
| Space group | C2 |
| Data processing | |
| Resolution range, Å | 12.0–2.0 |
| No. of observations | 64,025 |
| No. of unique reflections | 27,020 |
| Completeness, % | 92 |
| Rmerge† | 0.077 |
| Refinement | |
| Resolution range, Å | 12.0–2.0 |
| R factor‡ | 0.179 |
| Rfree§ | 0.212 |
| No. of protein atoms (αβ dimer) | 2,136 |
| No. of heme groups | 2 |
| No. of CO molecules | 1 |
| No. of water molecules | 71 |
| Root-mean-square deviations from ideal values | |
| Bond lengths, Å | 0.010 |
| Bond angles, ° | 1.45 |
| Dihedral angles, ° | 19.7 |
| Improper angles, ° | 1.44 |
The values in parenthesis refer to HbTnCO crystals (PBD code 1T1N).
Rmerge = ΣhklΣi|Ii − 〈I〉|/|Ii|.
R factor = Σhkl(∥Fhklobs∥ − k|Fhklcalc∥)/Σhkl|Fhklobs|.
Rfree = Σh(∥Fobs| − k|Fcalc∥)/Σh|Fobs∥, where h is a statistical subset (10%) of data.
The crystals are monoclinic (space group C2), with cell dimension a = 89.85 Å, b = 88.19 Å, c = 55.14 Å, and β = 97.79°. An αβ dimer constitutes the asymmetric unit. Interestingly, these crystals are nearly isomorphous to the crystals of HbTnCO (10) obtained from a 2 M ammonium sulfate medium (pH 8.0) (Table 1).
Refinement.
The structure of HbTnCO [Protein Data Bank (PDB) ID code 1T1N] (10) was used as starting model in the refinement, which was carried out by using the program x-plor (11). The overall position of the αβ dimer was initially refined by rigid body minimization. Subsequently, the α and β chains were refined as two distinct rigid units. The R factor of this model was 29.8% in the resolution shell of 10–2.0 Å.
The rigid body refinement cycles were followed by positional refinements and B factor optimizations. Each refinement run was followed by manual intervention using the molecular graphic program o (12) to correct minor errors in the position of the side chains. The electron density maps corresponding to the heme regions showed that a CO molecule was bound to the heme iron in the α chain, whereas in the β chain there was clear indication of the formation of a bishistidine complex. Following the indications of the electron density, a bishistidine complex was modeled in the heme region of the β chain. To confirm the evidence coming from the electron density maps, several refinement cycles were carried out without imposing any geometrical restraints to the interactions of the iron atoms of both α and β chains with the axial ligands. After these refinements small distortions from the ideal geometry were observed for the ligands of both heme groups. In the subsequent refinement cycles, geometrical restraints were imposed only for the iron/axial ligand interactions of the α chain. In the last steps of the refinement, 71 water molecules were identified by a home-made automated procedure and included in the refining model. A detailed description of the refinement statistics of the structure of air-exposed HbTnCO, hereafter referred to as HbTn[α(CO)β(hemi)]2, is reported in Table 1. The figures were generated by using the programs molscript (13), bobscript, and RASTER3D (14).
Comparative Analyses of Hb Structures.
To analyze the tertiary structure variations associated with hemichrome formation the structure of HbTn[α(CO)β(hemi)]2 was compared with that of HbTnCO (10). Furthermore, the carbonmonoxy (HbTbCO) (PDB ID code 1PBX) (15) and deoxy (HbTb-deoxy) (PDB ID code 1HBH) (16) forms of Trematomus bernacchii Hb were used as reference structures to evaluate the position of HbTn[α(CO)β(hemi)]2 along the R → T transition pathway. It is worth mentioning that T. bernacchii and T. newnesi Hb sequences differ by only 14 residues (9). Given the similarity of the R → T structural transitions in fish and mammalian Hbs (16), nearly identical results were obtained when unliganded and fully liganded forms of human Hb were used as reference structures.
Following the procedure of Baldwin and Chothia (17), the BGH core, constituted by the residues of helices B, G, and H, was used in superimpositions. The relative orientations of α1β1/α2β2 dimers in the tetramer were evaluated by using a stepwise superimposition procedure (17–19). In particular, after the α1β1 dimers for a pair of structure had been optimally fitted, the angle needed to superimpose the dimers α2β2 was evaluated.
Results
The structure of HbTn[α(CO)β(hemi)]2 was refined to an R factor of 17.9% (Rfree 21.2%) by using diffraction data in the resolution range 12–2.0 Å. The final model includes 71 water molecules. The electron density is well defined for both the main chain and the side chains of most of the residues. As frequently reported in R state Hbs, the regions corresponding to the CD loop (residues 43–53) and at the C terminus (residues 143–146) of the β subunit are completely disordered. Therefore, these regions were not included in the final model. The stereochemical parameters of the refined structure (Table 1) are in close agreement with those obtained for well-refined protein structures at 2.0-Å resolution. In particular, 93.7% of the residues are located in the most favored regions of the Ramachandran plot. It is worth mentioning that no residue is located in either generously allowed or disallowed regions. Although HbTnCO and HbTn[α(CO)β(hemi)]2 structures were derived from isomorphous crystals, relevant variations of tertiary and quaternary structure of the protein could be identified.
Structure of the Heme Regions.
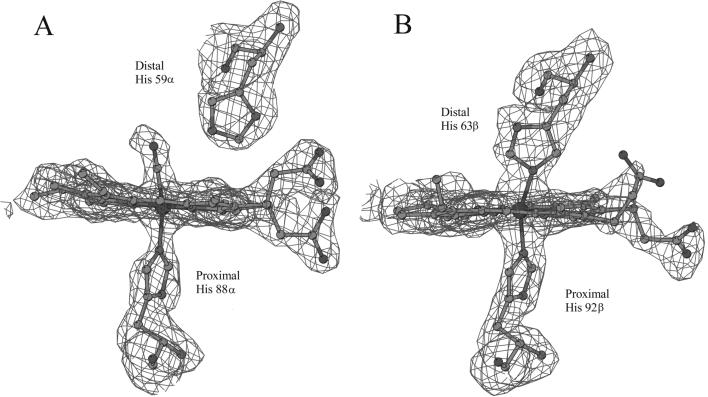
Differences in the heme structures of the α and β subunits of HbTn[α(CO)β(hemi)]2 were clear since the inspection of the first electron density maps. In particular, the overall conformation of the heme regions in the α subunit is virtually identical to that reported for HbTnCO (10). As shown in Fig. 1A, the heme iron is hexacoordinated. A CO molecule fits the electron density of the sixth coordination position very well. The analysis of the geometry of iron coordination reveals that CO is slightly bent, with a Fe–C–O angle of 162°. This value agrees well with those found in crystal structures of carbonmonoxy Hbs determined at similar resolutions. Recent high-resolution crystallographic studies on myoglobin have demonstrated that the bending of the Fe–C–O angle may be associated with the presence in the crystal of a fraction of oxidized aquomet form (20). The moderate bending of this angle indicates that in HbTn[α(CO)β(hemi)]2 crystals only a limited fraction of α hemes might have been oxidized to the aquomet state, despite the air exposure of the protein during crystallization.
Figure 1.
Electron density maps (contoured at 2.5 σ) corresponding to the α (A) and β (B) heme regions.
A completely different picture emerges from the analysis of the electron density maps corresponding to the β heme (Fig. 1B). The iron atom coordinates, in fact, both the proximal (92β) and the distal (63β) histidine residues. This bishistidine complex, in principle indicative of either a hemochrome [Fe(II)] or a hemichrome [Fe(III)] state, may be identified as a hemichrome. Indeed, UV–visible spectroscopic analyses of air-exposed solutions of HbTn clearly indicate that the protein evolves toward the formation of the hemichrome. The tendency of HbTn to form hemichrome states also occurs in the presence of monomethoxypolyethylene glycol, which was used as a precipitant in the crystallization. The geometry of this bishistidine form displays distinctive features compared with other functional bishistidine complexes (6, 7). In particular, the Nɛ2–Fe–Nɛ2 angle, usually close to 180° in the latter structures, deviates significantly from linearity (160°) in HbTn[α(CO)β(hemi)]2. The nonlinearity of the Nɛ2–Fe–Nɛ2 angle may be ascribed to the strain of the protein matrix, which prevents the formation of a hemichrome with ideal geometry. Nevertheless, the structure demonstrates that formation of a hemichrome, usually associated with the Hb degradation pathway, is compatible with a well-defined native-like three-dimensional structure of the protein.
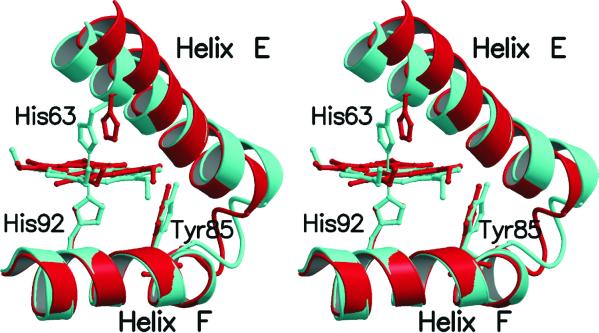
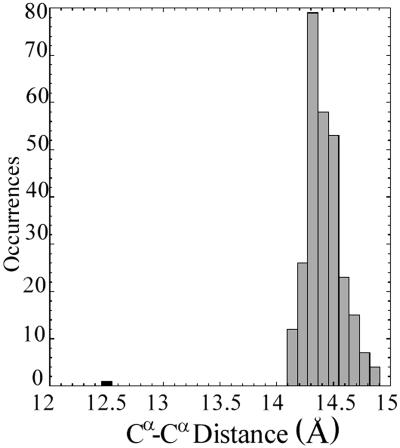
The hemichrome formation in the β subunit of HbTn is associated with a significant modification of the pocket in which the heme group is located. Indeed, a concerted scissors-like motion of helices E and F and a significant displacement of the heme group (about 1 Å) are essential for the bishistidine coordination of the iron (Fig. 2). A quantitative evaluation of the entity of this scissors-like motion of the EF segment may be provided by the analysis of the distance between the Cα atoms of the proximal and the distal histidine residues, which belong to helix F and E, respectively. This distance, which is almost invariantly close to 14.5 Å (±0.2) in α and β chains of Hb structures extracted from different organisms and in a variety of liganded states, is as low as 12.5 Å in the β subunit in HbTn[α(CO)β(hemi)]2 (Fig. 3). Therefore, this scissoring-like motion of the EF moiety, produced by the simultaneous iron coordination of both the proximal and distal histidine residues, represents a significant alteration of the tertiary structure of the protein.
Figure 2.
Stereo drawing showing the scissors-like motion of the β chain EF corner occurring upon hemichrome formation. The EF regions of HbTn[α(CO)β(hemi)]2 (cyan) and HbTnCO (red) are shown after superimposition of the F helix. For clarity the propionate groups of heme have been omitted.
Figure 3.
Distribution of the Cα–Cα distances between the proximal and the distal histidine residues in 276 α and β chains belonging to 80 tetrameric Hbs reported in the PDB (38). The value corresponding to the β chain of HbTn[α(CO)β(hemi)]2 is colored in black.
Hemichrome Formation and Structural Modifications of β Subunits.
The closure movement of the EF fragment associated with hemichrome formation propagates to the other structural elements of the tertiary structure of the β subunit. Hemichrome formation produces relevant modifications of the FG corner. In particular, the N-terminal region of HbTn[α(CO)β(hemi)]2 FG corner assumes a conformation that is more similar to that found in HbTb-deoxy (16) than in HbTnCO (10) and HbTbCO (15).
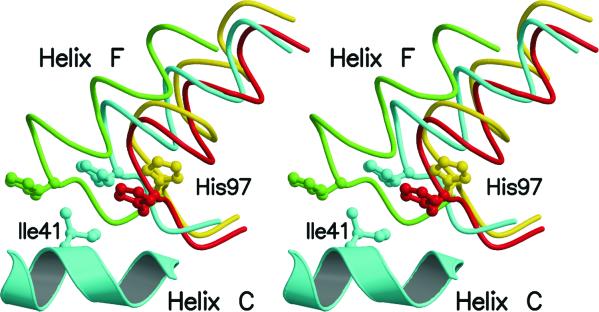
It is worth noting that the βFG corner is one of the key segments of the α1β2(α2β1) interface, which is a critical region for the allosteric regulation of tetrameric Hb. In fact, this region is directly connected to the αC helix and therefore any movement of the βFG region is easily transmitted to the other chains of the tetramer (17). As shown in Fig. 4, the displacement of the FG corner produces variations in the position of His-97β (see below) whose location is crucial in stabilizing the quaternary structure of liganded and unliganded forms of tetrameric Hbs.
Figure 4.
Stereo drawing showing the relative orientation of His-97β side chain and helix C of the α chain at the α1β2 of liganded and unliganded Hbs. HbTnCO, HbTn[α(CO)β(hemi)]2, HbTb-deoxy and the R2 carbonmonoxy form of human Hb are colored in red, cyan, green, and yellow, respectively. The residues of helix C of the α chains have been used in the superimposition. For clarity only helix C of HbTn[α(CO)β(hemi)]2 is shown.
Modification of the α1β2 Interface.
As mentioned above, the contacts between βFG and αC at α1β2 are essential for the heme–heme communication in tetrameric Hbs. It is worth mentioning that the R and T forms of vertebrate Hbs are characterized by different relative positions of the His-97β side chain and of the C helix of the α chain. In unliganded states, the imidazole ring of His-97β is positioned between residues 41α and 44α. The interface in the T state is further stabilized by the hydrogen bond between Asp-99β and Tyr-42α side chains. On the other hand, in the R state His-97β side chain is located between residues 38α and 41α of helix C. In the R state the distance between Asp-99β and Tyr-42α side chains is as large as 8.0 Å, and the α2β1 interface is characterized by the hydrogen bond between Asp-95α and Asn-102β side chains. According to Baldwin and Chothia (17), the βFG and αC region “has the characteristic of a switch, in that there are two distinct stable packing arrangements and any intermediate position would be unstable … [since] His-97β has to move past the side chain of 41α.” Interestingly, as shown in Fig. 4, in HbTn[α(CO)β(hemi)]2 the His-97β side chain takes the intermediate position between those that the residue has in the R and T states. Therefore, this residue is located in an apparently disallowed position. The displacement of the βFG corner in HbTn[α(CO)β(hemi)]2 (Fig. 4), as a consequence of hemichrome formation, makes this location of His-97β accessible. It should be pointed out that the location of the side chain of this histidine in HbTn[α(CO)β(hemi)]2 is also very different from the position taken by this residue in the fully liganded R2 state of human Hb (18) (Fig. 4).
Even more intriguing is the observation that interactions that are considered typical of either the R or the T state (2) are present at α1β2. In particular, one of the two alternative conformations of the Asp-99β side chain, observed in the crystal structure of HbTn[α(CO)β(hemi)]2, interacts with Tyr-42α side chain, as in Hb deoxy structures. Similarly, the hydrogen bond between Asp-α95 and Asn-β102, typically found in liganded R structures, is also observed in HbTn[α(CO)β(hemi)]2.
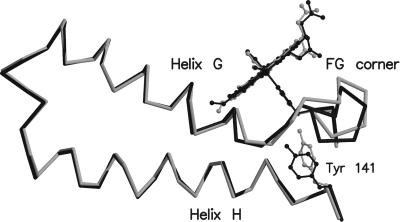
The structural variations induced by hemichrome formation at the β heme cause significant changes of the FG region of the α subunits. Structural comparisons between HbTnCO and HbTn[α(CO)β(hemi)]2 show that modification of the helix αC position causes a displacement of the αFG region that is also accompanied by conformational rearrangement of the Tyr-141 side chain (Fig. 5). Indeed, the deoxy-like displacement of the αFG segment in HbTn[α(CO)β(hemi)]2 leaves enough room for the insertion of the tyrosine side chain, which assumes a conformation typically found in deoxy-Hbs (1, 21).
Figure 5.
Structural modifications of the α chain produced by hemichrome formation in the β chain. HbTn[α(CO)β(hemi)]2 and HbTnCO are colored in gray and black, respectively.
A Comparative Analysis of the Quaternary Structure of HbTn[α(CO)β(hemi)]2 with Other Tetrameric Hbs.
The analysis of the overall quaternary structure of HbTn[α(CO)β(hemi)]2 confirms that this is an intermediate structure between the thoroughly characterized R and T states. As shown in Table 2, in which the root-mean-square deviations between several Hb structures is reported, HbTn[α(CO)β(hemi)]2 tetramer, although more similar to the R states, lies between the end points of the R → T transition. This is also evident from Fig. 6, in which the overall quaternary structures of Antarctic fish Hbs are displayed.
Table 2.
Comparison of fish Hb quaternary structures
| Structures | Root-mean-square deviations, Å*
|
α1β1/α2β2 rotation angle, °† | ||
|---|---|---|---|---|
| α1β1 dimer | α2β2 dimer‡ | Tetramer | ||
| HbTbCO vs. HbTb-deoxy | 0.37 | 4.80 (0.40)§ | 1.59 | 11.5 |
| HbTbCO vs. HbTn[α(CO)β(hemi)]2 | 0.39 | 1.97 (0.37)§ | 0.68 | 4.6 |
| HbTn[α(CO)β(hemi)]2 vs. HbTb-deoxy | 0.32 | 3.06 (0.36)§ | 1.08 | 7.5 |
| HbTnCO vs. HbTn[α(CO)β(hemi)]2 | 0.37 | 1.98 (0.37)§ | 0.71 | 4.8 |
| HbTnCO vs. HbTbCO | 0.21 | 0.47 (0.21)§ | 0.25 | 1.6 |
| HbTn[α(CO)β(hemi)]2 vs. R2Hb¶ | 0.77 | 5.54 (0.74)§ | 1.75 | 17.2 |
The N, Cα, C atoms of the BGH core were used for the superimpositions.
The α1β1/α2β2 rotation angle is the angle required to superimpose the α2β2 dimer after the fitting of the α1β1 dimer.
These values correspond to the root-mean-square deviations of the α2β2 dimers after the fitting of the α1β1 dimers.
The value given in parenthesis corresponds to the root-mean-square deviations of the α2β2 dimers after their superimposition.
This notation identifies the fully liganded R2 state of human Hb.
Figure 6.
Cα trace of HbTn[α(CO)β(hemi)]2 (cyan), HbTbCO (purple), and HbTb-deoxy (green). The residues of the α1β1 dimers have been superimposed. Residues belonging to the α1β1 and α2β2 dimers are mostly located in the lower and in the upper part of the figure, respectively. The labeled residues Asp-5, together with Lys-95 and Pro-47, belong to β2 and α2 chains, respectively. The axis required to superimpose the α2β2 dimers of HbTbCO and HbTb-deoxy is normal to the figure and its position is marked by a black circle. The gray circle represents the intersection of the axis required to superimpose the α2β2 dimers of HbTn[α(CO)β(hemi)]2 and HbTb-deoxy with a plane parallel to the figure and passing through the center of mass of the tetramer. The two axes form an angle of 15°.
These findings are corroborated by a more detailed analysis of the quaternary structure of HbTn[α(CO)β(hemi)]2, which was carried out by using the stepwise superimposition of the αβ dimers frequently adopted in Hb structure comparisons (see Materials and Methods). To optimally superimpose the α2β2 dimers of HbTbCO (15) and HbTb-deoxy (16) after the fitting of the α2β2 dimers a further rotation of 11.5° is required (Table 2). Similar values (≈14°) have been reported in comparisons of human R and T Hb states (17). When the same procedure is applied to HbTn[α(CO)β(hemi)]2 and HbTb-deoxy the rotation required to superimpose the α2β2 dimers is 7.5°. These findings demonstrate that HbTn[α(CO)β(hemi)]2 structure is closer to HbTb-deoxy than to HbTbCO. Interestingly, the rotation axis required to superimpose the α2β2 dimers of HbTb-deoxy and HbTn[α(CO)β(hemi)]2 is nearly coincident with the axis required to superimpose HbTb-deoxy and HbTbCO (Fig. 6). A similar rotation, of about 4.7°, is required to superimpose the α2β2 dimer of HbTn[α(CO)β(hemi)]2 with the dimers of both HbTbCO and HbTnCO. This overall picture is confirmed when the human R and T Hbs are taken as reference structures (data not shown) (22, 23). Finally, the overall quaternary structure of HbTn[α(CO)β(hemi)]2 is very different from the structure of the R2 human Hb state (18) (Table 2), which has been demonstrated to lie outside the pathway from R to T (24).
Discussion
Vertebrate Hbs have been extensively studied both functionally and structurally because they are the first and the most important molecular model for protein cooperativity. Allosteric effects in Hb arise from the switch between an R and a T state occurring upon oxygen binding and release (1, 2). Although the end points of this transition have been thoroughly characterized, structural information on intermediate species is still rather limited, despite the extensive use of a variety of experimental techniques (19, 25–34).
In this context, the mixed structure of HbTn[α(CO)β(hemi)]2 herewith reported displays a number of interesting features. In fact, in this structure the α and β subunits show different binding states. In particular, the α irons are bound to the exogenous CO ligand; the β irons are also hexacoordinated, but the sixth ligand is the distal histidine residue, thus producing a hemichrome structure. Hemichromes were thought to be one of the first steps along the degradation pathway of Hbs, but they have also been shown to be a detectable substate of Hb (3). The mixed form of HbTn[α(CO)β(hemi)]2, to our knowledge the first example of a tetrameric Hb in the hemichrome state, demonstrates that coordination, at least in the β subunits, of the iron by distal His may be tolerated in a native-like Hb structure. Furthermore, the identification of a different binding state of the α and β iron in HbTn[α(CO)β(hemi)]2 is an unusual finding among structural studies on tetrameric Hbs. In fact, most of Hbs structures with α and β chains in different binding states have been obtained by ad hoc chemical modifications of the protein. This study provides a strong indication that the EF corner of the α and β subunits in the tetramer have a remarkably different intrinsic flexibility. It is worth mentioning, however, that investigations carried out on isolated chains of human Hb demonstrate that α chains are more prone to form hemichrome than β chains (35). Altogether these data, although derived from Hb of different species, seem to indicate that the quaternary structure may play a role in modulating the propensity of α and β chains to form hemichromes in the tetramer.
The analysis of the tertiary and quaternary structure of HbTn[α(CO)β(hemi)]2 provides interesting information on the structural features of the intermediate state along the pathway of the R → T transition identified by the pioneering work of Perutz (21). Indeed, the coordination of distal His to the β iron produces a scissoring motion of helices E and F, which, in turn, triggers (i) several deoxy-like modifications of the tertiary structure of the two chains, mainly localized at the α1β2(α2β1) interface, and (ii) a significant change in the quaternary structure, which is displaced toward the T state. HbTn[α(CO)β(hemi)]2 structure reveals that the α1β2 interface is characterized by interactions that are believed to be a signature of either the R or the T state. These structural features of the tertiary and quaternary organization of HbTn[α(CO)β(hemi)]2 agree well with the transition state properties obtained by Raman spectroscopy and kinetic techniques. In particular, Raman spectroscopy studies reported by Spiro and coworkers (28, 29) have demonstrated that a scissors-like motion of helices E and F is an early important event in the transition from the R to the T state. Furthermore, kinetic studies carried out by Eaton et al. (26) have shown that the transition state has an R-like overall structure. Notably, these authors roughly estimated that the αβ dimers in the transition state rotated by 3° with respect to the native R structure. This value is remarkably similar to the rotation required to superimpose the α2β2 dimers of HbTnCO and HbTn[α(CO)β(hemi)]2. Therefore, the data reported here indicate the scissoring-like motion of the EF pocket may initiate the cascade of events that favor the switch from the R to the T state. The existence of reversible bishistidine complexes, such as hemochrome, as a populated substate of native Hbs (3) suggests that this form may play a functional role in tetrameric Hbs, although this hypothesis is not supported by mutational studies, which show that the removal of β chain distal histidine marginally affects the properties of human Hb (36). More simply, transient scissoring-like motions of the β chain EF corner may occur upon substrate release. It is worth noting that the association and dissociation rate constants of CO and O2 in R state hemoglobin are much larger for the β chain than for the α chain, the β distal pocket being less hindered (37).
In conclusion, the HbTn[α(CO)β(hemi)]2 structure simultaneously provides information on the oxidation and the functional pathways of tetrameric Hbs. The analogies of the structural alterations occurring during these distinct events indicate that there is a limited number of ways to modify Hb structure.
Acknowledgments
This paper is dedicated to the memory of Prof. Alfonso Maria Liquori. We thank Mr. Giosue' Sorrentino for his skilful technical assistance and Mr. Luca De Luca for his help in the picture lay-out. This work was financially supported by grants from the Italian Program for Antarctic Research to L.M. and G.d.P.
Abbreviations
- Hb
hemoglobin
- R
relaxed
- T
tense
- HbTnCO
carbonmonoxy form of the major Hb component of Trematomus newnesi
- HbTn[α(CO)β(hemi)]2
structure of the major Hb component of T. newnesi in a partial hemichrome state
- HbTbCO
carbonmonoxy form of Trematomus bernacchii Hb
- HbTb-deoxy
T. bernacchii Hb in the deoxy state
- PDB
Protein Data Bank
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates of HbTn[α(CO)β(hemi)]2 have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1LA6).
References
- 1.Perutz M F. Nature (London) 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 2.Perutz M F, Wilkinson A J, Paoli M, Dodson G G. Annu Rev Biophys Biomol Struct. 1998;27:1–34. doi: 10.1146/annurev.biophys.27.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Rifkind J M, Abugo O, Levy A, Heim J. Methods Enzymol. 1994;231:449–480. doi: 10.1016/0076-6879(94)31030-0. [DOI] [PubMed] [Google Scholar]
- 4.Levy A, Rifkind J M. Biochemistry. 1985;24:6050–6054. doi: 10.1021/bi00343a005. [DOI] [PubMed] [Google Scholar]
- 5.Levy A, Kuppusamy P, Rifkind J M. Biochemistry. 1990;29:9311–9316. doi: 10.1021/bi00492a002. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell D T, Kitto G B, Hackert M L. J Mol Biol. 1995;251:421–431. doi: 10.1006/jmbi.1995.0445. [DOI] [PubMed] [Google Scholar]
- 7.Hargrove M S, Brucker E A, Stec B, Sarath G, Arredondo-Peter R, Klucas R V, Olson J S, Phillips G N., Jr Structure Fold Des. 2000;8:1005–1014. doi: 10.1016/s0969-2126(00)00194-5. [DOI] [PubMed] [Google Scholar]
- 8.Riccio A, Vitagliano L, di Prisco G, Zagari A, Mazzarella L. Acta Crystallogr D. 2001;57:1144–1146. doi: 10.1107/s0907444901007739. [DOI] [PubMed] [Google Scholar]
- 9.D'Avino R, Caruso C, Tamburrini M, Romano M, Rutigliano B, Polverino de Laureto P, Camardella L, Carratore V, di Prisco G. J Biol Chem. 1994;269:9675–9681. [PubMed] [Google Scholar]
- 10.Mazzarella L, D'Avino R, di Prisco G, Savino C, Vitagliano L, Moody P C, Zagari A. J Mol Biol. 1999;287:897–906. doi: 10.1006/jmbi.1999.2632. [DOI] [PubMed] [Google Scholar]
- 11.Brunger A. x-plor, User's Guide. A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 12.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 15.Camardella L, Caruso C, D'Avino R, di Prisco G, Rutigliano B, Tamburrini M, Fermi G, Perutz M F. J Mol Biol. 1992;224:449–460. doi: 10.1016/0022-2836(92)91007-c. [DOI] [PubMed] [Google Scholar]
- 16.Ito N, Komiyama N H, Fermi G. J Mol Biol. 1995;250:648–658. doi: 10.1006/jmbi.1995.0405. [DOI] [PubMed] [Google Scholar]
- 17.Baldwin J, Chothia C. J Mol Biol. 1979;129:175–220. doi: 10.1016/0022-2836(79)90277-8. [DOI] [PubMed] [Google Scholar]
- 18.Silva M M, Rogers P H, Arnone A. J Biol Chem. 1992;267:17248–17256. [PubMed] [Google Scholar]
- 19.Mueser T C, Rogers P H, Arnone A. Biochemistry. 2000;39:15353–15364. doi: 10.1021/bi0012944. [DOI] [PubMed] [Google Scholar]
- 20.Stec B, Phillips G N., Jr Acta Crystallogr D. 2001;57:751–754. doi: 10.1107/s0907444901001731. [DOI] [PubMed] [Google Scholar]
- 21.Perutz M F, Fermi G, Luisi B, Shaanan B, Liddigton R C. Acc Chem Res. 1987;20:309–321. [Google Scholar]
- 22.Fermi G, Perutz M F, Shaanan B, Fourme R. J Mol Biol. 1984;175:159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- 23.Derewenda Z, Dodson G, Emsley P, Harris D, Nagai K, Perutz M, Renaud J P, Reynaud J P. J Mol Biol. 1990;211:515–519. doi: 10.1016/0022-2836(90)90262-k. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan R, Rose G D. Proc Natl Acad Sci USA. 1994;91:11113–11117. doi: 10.1073/pnas.91.23.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzozowski A, Derewenda Z, Dodson E, Dodson G, Grabowski M, Liddington R, Skarzynski T, Vallely D. Nature (London) 1984;307:74–76. doi: 10.1038/307074a0. [DOI] [PubMed] [Google Scholar]
- 26.Eaton W A, Henry E R, Hofrichter J. Proc Natl Acad Sci USA. 1991;88:4472–4475. doi: 10.1073/pnas.88.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho C. Adv Protein Chem. 1992;43:153–312. doi: 10.1016/s0065-3233(08)60555-0. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers K R, Spiro T G. Science. 1994;265:1697–1699. doi: 10.1126/science.8085153. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman V, Rodgers K R, Mukerji I, Spiro T G. Science. 1995;269:1843–1848. doi: 10.1126/science.7569921. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher M A, Dixon M M, Kluger R, Jones R T, Brennan R G. Nature (London) 1995;375:84–87. doi: 10.1038/375084a0. [DOI] [PubMed] [Google Scholar]
- 31.Paoli M, Dodson G, Liddington R C, Wilkinson A J. J Mol Biol. 1997;271:161–167. doi: 10.1006/jmbi.1997.1180. [DOI] [PubMed] [Google Scholar]
- 32.Englander J J, Rumbley J N, Englander S W. J Mol Biol. 1998;284:1707–1716. doi: 10.1006/jmbi.1998.2279. [DOI] [PubMed] [Google Scholar]
- 33.Gibson Q H. Biochemistry. 1999;38:5191–5199. doi: 10.1021/bi982970t. [DOI] [PubMed] [Google Scholar]
- 34.Eaton W A, Henry E R, Hofrichter J, Mozzarelli A. Nat Struct Biol. 1999;6:351–358. doi: 10.1038/7586. [DOI] [PubMed] [Google Scholar]
- 35.Rachmilewitz E A, Peisach J, Blumberg W E. J Biol Chem. 1971;246:3356–3366. [PubMed] [Google Scholar]
- 36.Nagai K, Luisi B, Shih D, Miyazaki G, Imai K, Poyart C, De Young A, Kwiatkowsky L, Noble R W, Lin S H, Yu N T. Nature (London) 1987;329:858–860. doi: 10.1038/329858a0. [DOI] [PubMed] [Google Scholar]
- 37.Mathews A J, Rohlfs R J, Olson J S, Tame J, Renaud J P, Nagai K. J Biol Chem. 1989;264:16573–16583. [PubMed] [Google Scholar]
- 38.Berman H M, Bhat T N, Bourne P E, Feng Z, Gilliland G, Weissig H, Westbrook J. Nat Struct Biol. 2000;7:957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]