Abstract
Background
Heparan sulfate (HS) is an ubiquitous component of the extracellular matrix that binds and modulates the activity of growth factors, cytokines and proteases. Animals with defective HS biosynthesis display major developmental abnormalities however the processes that are affected remain to be defined. D-glucuronyl-C5-epimerase (Glce) is a key HS chain modifying enzyme that catalyses the conversion of glucuronic acid into iduronic acid, a biosynthetic step that enhances HS biological activity. In this study the role of Glce during early zebrafish development has been investigated.
Results
Two Glce-like proteins (Glce-A and -B) are expressed in zebrafish at all times. They are the products of two distinct genes that, based on chromosomal mapping, are both orthologues of the same single human gene. Transcripts for both proteins were detected in fertilized zebrafish embryos prior to the onset of zygotic transcription indicating their maternal origin. At later developmental stages the epimerases are expressed widely throughout gastrulation and then become restricted to the hindbrain at 24 h post-fertilization. By monitoring the expression of well characterized marker genes during gastrulation, we have found that misexpression of Glce causes a dose-dependent expansion of the ventral structures, whereas protein knockdown using targeted antisense morpholino oligonucleotides promotes axis dorsalization. The ventralizing activity of Bmp2b is enhanced by Glce overexpression whereas Glce knockdown impairs Bmp2b activity.
Conclusion
Glce activity is an important determinant of of dorso-ventral axis formation and patterning in zebrafish. In particular Glce acts during gastrulation by affecting Bmp-mediated cell specification. The results obtained further corroborate the concept that HS encodes information that affect morphogenesis during early vertebrate development.
Background
Heparan sulfate proteoglycans (HSPG) are macromolecules found in all connective tissues, extracellular matrices and on the surface of cells [1]. Their most prominent feature is the presence of one or more heparan sulfate (HS) chains covalently attached to a core protein [2]. The heterogeneity of the HSPG is due to the variation of the core protein, as well as to the type and size of the HS chains. Configuration variation in the disaccharide bonds and the position of sulfation leads to increased diversity in the chemical and structural properties of these chains. HS is composed of repeating disaccharide units of D-glucuronic acid (GlcA) or L-iduronic acid (IdoA) both of which may be 2O-sulfated, and unsubstituted, N-acetylated, or N-, 3O- or 6O-sulfated glucosamine (Glc). Forty-eight different disaccharides are possible but because of constraints in the biosynthetic process, only 23 have been identified in HS as biosynthetic intermediates [3]. Typical HS chains contain relatively short segments of modified sequences represented by IdoA-GlcNS derivatives of different sulfation content dispersed among large sections of unmodified (GlcA-GlcNAc) units.
The biosynthesis of HS occurs in the Golgi and involves the sequential modification of the nascent polysaccharide chain [4,5]. The conversion of GlcA into IdoA is a critical modification mediated in mammals by a single enzyme: D-glucuronyl-C5-epimerase (GLCE) [4,6]. Epimerization of GlcA increases the flexibility of HS chain thereby enhancing its ability to interact with proteins [2,7-9]. IdoA is the preferential substrate of the HS 2-O-sulfotransferase. Disaccharide units containing IdoA-2-O-S are organized in clusters along the HS chain and are specifically recognized by growth factors and morphogens [5,10]. The essential role played by Glce during development is demonstrated by the fact that transgenic mice that are Glce-null generally express highly abnormal HS structures and die neonatally [11,12]. C. elegans expressing mutated Glce, display abnormal neuronal development characterized by specific axonal and cellular guidance defects [13].
Much of the information concerning the role of HS in development has been obtained from studies in D. melanogaster [14]. An important concept arising from those studies is that the establishment of a morphogen gradient necessary for early patterning requires HSPG. This function is likely to involve the polysaccharide chain since morphogens such as Wingless (Wg) [15], Decapentaplegic (the orthologe of the vertebrate bone morphogenetic protein 4, Bmp4) [16,17], Hedgehog (Hh) [18] and several fibroblast growth factors (FGFs) [19] bind to HS. The specific role of HS in vertebrate development however remains conjectural and the developmental mechanisms that are affected have not been clearly identified. In zebrafish, lack of uridine 5'-diphosphate glucose dehydrogenase [20], an enzyme required for the biosynthesis of extracellular matrix glycosaminoglycans including HS, affects bone and heart morphogenesis. In mice [21] and zebrafish [22] the disruption of HS biosynthesis affects the nervous system development that can be ascribed to the effect HS has on the activity of multiple morphogens. In this paper we report that Glce's activity affects the establishment of the embryonic dorso-ventral (D/V) axis through a mechanism involving the bone morphogenetic proteins (Bmps).
Results
Cloning and chromosomal mapping of zebrafish glce
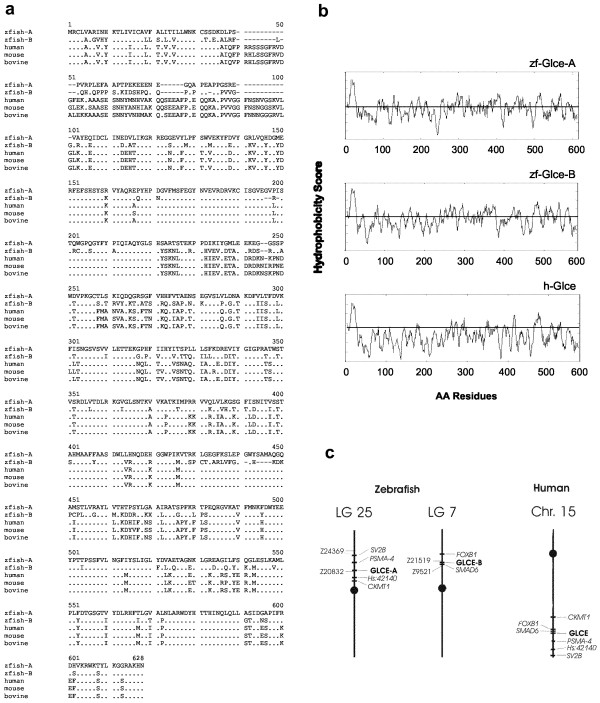
Zebrafish glce-A and glce-B genes both encode proteins of 585 amino acids. The gene products are homologous to the human protein sequence (67% and 73% respectively) (fig. 1a). Compared to the human, mouse and bovine sequences, the zebrafish proteins lack part of the N-terminus. The C-terminal domain is the most conserved region of Glce. Analysis of the hydrophobicity index determined utilizing the SOSUI [23] and the TMPRED [24] algorithms, reveals the presence in both zebrafish proteins of a conserved hydrophobic domain of ~20 amino acids located between residue 10 and 30 at the N-terminus of the proteins (fig. 1b). As the mammalian enzyme, zebrafish epimerase is a "type-two" transmembrane protein with predicted localization in the Golgi apparatus [4]. The glce-A and the glce-B locus mapped to linkage group LN25 between markers Z24369 and Z20832 and to linkage group 7 between markers Z21519 and Z9521, respectively (fig. 1c). Based on the mapping of neighbor genes, both chromosomal regions are synthenic with human chromosome 15q22, i.e. the region harboring the epimerase gene.
Figure 1.
Cloning and structural analysis of zebrafish glce. (a) Alignment of Glce from zebrafish, human, mouse, and bovine. Conserved AAs are dotted. (b) Hydrophobicity plot of zebrafish and human Glce. Values above the line represent positive hydrophobicity scores. (c) Chromosomal mapping of zebrafish glce-A and glce-B and homology to the human Glce locus.
Glce is maternally and zygotically derived
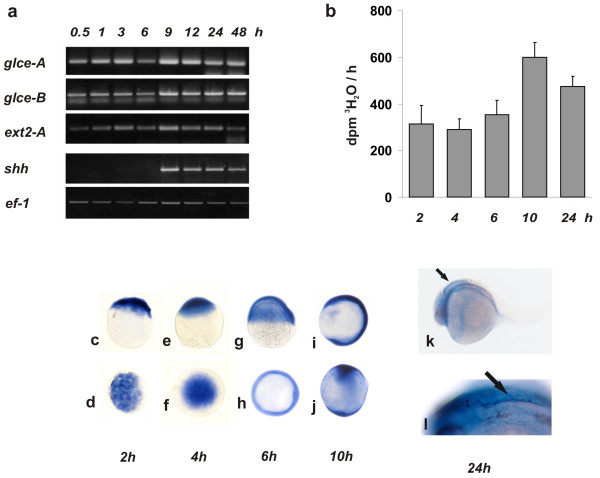
glce transcript level was examined at different developmental stages and compared to that of ext2-A, an HS polymerase acting upstream the epimerization step [3]. The expression of shh which is activated during gastrulation, and that of ef-1 which is expressed at similar level throughout development, were also monitored. glce-A, glce-B and ext2-A transcripts were present in fertilized embryos at developmental stages prior to the onset of zygotic transcription, indicating that these messages are maternally derived. The expression of HS biosynthesis enzymes reached a peak at the onset of gastrulation following midblastula transition (fig. 2a). Assay of epimerase activity in embryonic extracts at different developmental stages, was consistent with the level of mRNA encoding these proteins. In particular epimerase activity at 10 hpf was twice that observed at the 64-cells stage (fig. 2b).
Figure 2.
glce mRNA expression pattern in developing embryos. (a) RT-PCR analysis of the transcript level. Thirty embryos were collected in Tri-Reagent at different developmental stages as indicated. cDNA was generated from total RNA (1 μg) using Sensiscript reverse transcriptase primed with oligo-dT at 37°C for 2 h. PCR reactions (25 cycles) were performed in duplicate and analyzed on 1% agarose gel. (b) Glce enzymatic activity in embryos at different developmental stage. At each time point 20 embryos were dechorionated and homogenized. For the enzymatic assay, a cell lysate was incubated (2 h at 28°C) with labeled bacterial K5 heparosan substrate and the 3H2O liberated as result of the epimerization of GlcA into IdoA, measured. The bars represent the mean ± SD of the values from three independent determinations. (c-l) whole-mount in-situ hybridization of glce-a in embryos at different stages of development. (c-j) Top row: lateral views. Bottom row: animal pole views. (c,d) blastoderm at 64 cells stage; (e,f) dome stage; (g,h) shield stage; (i,j) 3 somite stage. (k) 24 hpf embryo showing showing intense glce staining at the perimeter of the forth ventricle as indicated by the arrow-heads. (l) enlargment of the embryo brain forth ventricle area.
Temporal and spatial expression of Glce
glce transcripts were localized in embryos at different developmental stages by in-situ hybridization using gene-specific antisense riboprobes (fig. 2c–l). During the early stages a diffuse staining was observed throughout the blastoderm. At the beginning of segmentation, staining was detected along the entire dorsal axis (fig. 2i,j). At 24 hpf, however, glce expression was higher in the newly forming brain (fig. 2k). At this site epimerase transcripts were mostly detected at the perimeter of the forth ventricle (fig. 2l). The staining specificitity was confirmed through the use of sense control riboprobes in which case only background staining was seen. No significant differences were detected in the expression pattern of the two glce genes.
Overexpression of Glce causes ventralization and potentiates Bmp activity
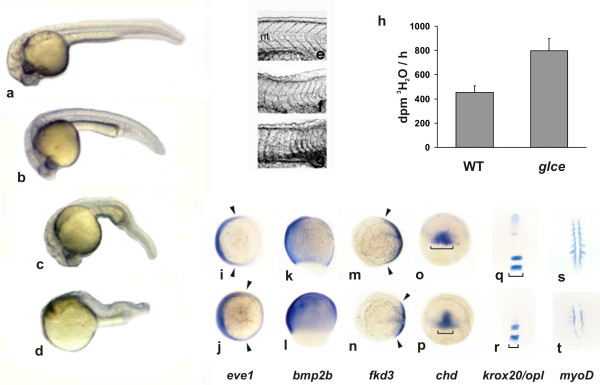
To investigate the functional significance of the epimerase during zebrafish development the protein was ectopically expressed by injecting embryos (1–2 cell stage) with capped in vitro-transcribed mRNA. The majority of injected embryos displayed a ventralizing phenotype whose severity correlated with the dose of mRNA injected (250 to 1000 pg) (table 1). The affected embryos had smaller head size, expanded blood islands, and abnormal tail somites (fig. 3b,c,f). More strongly ventralized embryos also lacked a notochord and developed somites that were not chevron-shaped and were fused in the midline below the neural tube (fig. 3g). Overexpression of glce-B produced an identical spectrum of phenotypes as the overexpression of glce-A. However, the highest frequency of severely affected embryos was observed when 250 or 500 pg of glce-A and glce-B mRNA were administered together in which case most of the animals failed to form an anterior axis (fig. 3d). In this treatment group, epimerase enzymatic activity at 10 hpf was 73% higher the level detected in uninjected embryos (fig 3h).
Table 1.
Effect of the ectopic expression of Glce on Bmp2b ventralizing activity. Capped glce and bmp2b mRNAs were generated by reverse transcription from full length cDNA clones. After extraction in phenol/chloroform and precipitation in isopropanol, mRNA was dissolved in Danieau's buffer and the concentration assessed by UV reading (260 nm). Embryos at one or two cell stage were injected with 1–3 nl of mRNAs to achieve the indicated dose. Each injection session consisted of 2–3 treatment groups of 30 embryos each and several experiments were performed to reach the sample number indicated. Embryos with increasing degree of ventralization were ranked according to previously established criterias [40,41]. Ventralized V1 embryos show reduced eye size and shorter body. In addition to these abnormalities V2 embryos display abnormal notocord, reduced anterior somites, and enlarged blood islands. Ventralized V3 embryos have little or no head structures and no notochord. V4 embryos display gross body abnormalities and lack of anterior structures.
| Injected mRNA | Strength of Ventralization | |||||
| No. | Wild Type | V1 | V2 | V3 | V4 | |
| (%) | (%) | (%) | (%) | (%) | ||
| Uninjected | 120 | 100 | 0 | 0 | 0 | 0 |
| glce-A (250 pg) | 60 | 65 | 23 | 10 | 2 | 0 |
| glce-B (250 pg) | 60 | 54 | 16 | 28 | 2 | 0 |
|
glce-A (125 pg) glce-B (125 pg) |
30 | 60 | 7 | 23 | 10 | 0 |
|
glce-A (250 pg) glce-B (250 pg) |
30 | 37 | 6 | 10 | 47 | 0 |
|
glce-A (500 pg) glce-B (500 pg) |
60 | 26 | 0 | 38 | 35 | 0 |
| bmp2b (20 pg) | 50 | 0 | 26 | 37 | 26 | 11 |
|
bmp2b (20 pg) glce-A (250 pg) |
60 | 0 | 0 | 15 | 45 | 40 |
|
bmp2b (20 pg) glce-B (250 pg) |
30 | 0 | 0 | 26 | 44 | 30 |
Figure 3.
Effect of Glce overexpression on embryo morphogenesis and HS composition. Embryos at 1–2 cells stage were injected with glce-A and/or glce-B mRNA and observed at 24 hpf. (a) wild type embryo. (b,c) mild and moderately ventralized embryos showing enlarged blood sac (indicated by an arrow). (d) Severely ventralized embryos displaying dramatically reduced head and trunk. (e-g) High-contrast images of the somites and the notocord (nt) structures in control (e), and in mildly and moderately ventralized embryos (f,g). Note the loss of chevron-like structure of the somites and the narrowing or disappearance of the notocord in embryos overexpressing glce (f,g). (h) Epimerase enzymatic activity at 10 hpf in controls (WT) and in embryos injected with glce-A plus glce-B mRNA (250 pg each). The enzymatic assay was performed as described in Fig. 2b. (i-t) Whole mount in-situ hybridization with D/V markers in embryos during gastrulation and at 5 somite stage. Top row: wild type embryos. Bottom row: embryos injected with 250 pg each of glce-A and glce-B mRNA. (i,j) eve1 staining viewed from the animal pole at shield stage. The arrowheads point to the edges of the expression range of the marker; (k,l) bmp2b, lateral view with the dorsal side to the right at 70% epiboly; (m,n) fkd3, animal pole view at 70% epiboly; (o,p) chordino, dorsal view at 50% epiboly; (q,r) krox-20/opl double staining (head view) and (s,t) myo-D (dorsal view) in embryos at 5 somite stage. Note in (r) the narrow expression domain of krox20 in embryos injected with glce mRNA whereas opl transcript is undetectable.
In order to better characterize the phenotype of embryos overexpressing Glce, the expression of dorsal and ventral markers were analyzed by in situ hybridization [25,26]. The expression domain of eve1, a marker of ventral and lateral regions at early gastrula stages, was expanded at the shield stage (fig. 3i,j). Likewise the range of expression of bmp2b was greatly enlarged in embryos at the 70% epiboly stage (fig. 3k,l). In contrast expression of fkd3, a marker of the presumptive neuroectoderm, was reduced by Glce overexpression (fig. 3m,n). Similarly the expression domain of chordin, a marker of the dorsal organizer, was reduced (fig. 3o,p). The fact that overexpression of the epimerase alters the pattern but does not prevent the expression of dorsal determinants, is consistent with the idea that Glce acts on D/V axis formation downstream the Wnt/β-catenin pathway that regulates chordin gene expression [27]. glce is also a target of the Wnt/β-catenin transactivation pathway [28] raising the possibility that zygotic glce expression is coordinated with that of other D/V determinants.
Because head size is affected following ectopic expression of glce (fig 3b–d) the expression pattern of the neuroectoderm-specific markers krox 20 [29] and opl (zic1) [30] was determined during somitogenesis. The expression of myoD, a transcript specifically localized to somitic mesoderm was also examined [31]. During somitogenesis, krox-20 is normally expressed in hindbrain rhombomeres R3 and R5 both of which are dorsal ecdoderm derivatives. A reduced area of krox-20 expression was detected at the 5 somite stage in most of the embryos injected with 250 pg each of glce-A and glce-B mRNA whereas opl expression was undetectable (fig. 3q,r). myoD expression in the cells adjacent to the axial mesoderm and in the somitic furrows was also reduced implying a role of Glce in establishing mesodermal cell fate (fig 3s,t).
Since both the phenotype and marker gene expression pattern following ectopic expression of Glce is reminiscent of that of chordino [30,32-34], ogon [33,35,36] and radar [37,38] mutants or of embryos misexpressing alk 8 [39] in which the ventralizing activity of Bmps is enhanced, we examined whether Bmp2b activity is potentiated by Glce. For this purpose we titrated the dose of injected bmp2b mRNA to achieve a preponderance of partially ventralized embryos displaying V2 and V3 phenotype severity [40,41] (table 1). This same dose (20 pg) was then administered together with glce-A and/or glce-B mRNA (250 pg). Following the treatment, the majority of embryos exhibited the most severe V4 class phenotype consistent with Glce activity potentiating the effect of Bmp2b.
Glce availability affects Bmp-mediated ventralization
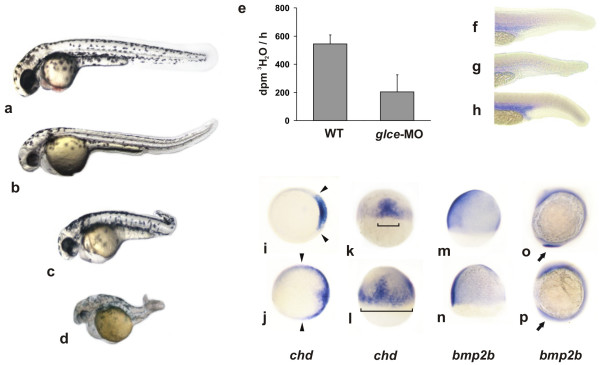
The effect of Glce protein knockdown on D/V axis formation was next examined by administering antisense morpholino oligonucleotides (MO) targeting glce-A and glce-B transcripts. Most of the embryos after injection of 4 ng of antisense MO, displayed a mild dorsalized phenotype with reduced ventral tail fin (fig. 4b and table 2). At higher dose (8 ng) about half of the morphants showed kinked tail, enlarged heart cavity and in some animals the atrioventricular boundaries failed to form (fig. 4c). A dramatic shortening and reduction of the body mass with tail coiling similar to the phenotype associated with mutation of Bmp2b, and Bmp7 [34,41] was observed in the majority of the embryos receiving the highest dose (16 ng) of MO (fig. 4d). In this group of morphants the epimerase enzymatic activity was significantly decreased (34% of the control at 8 hpf) (fig. 4e). The inability of the MOs to completely abolish the Glce activity suggests that at this time residual maternally derived enzyme is still active. In spite of this, the effect of Glce knockdown on ventral cell fate was already detectable in the mild morphants as revealed by the faint staining of gata1 expressing blood islets (fig. 4g), a ventral tissue derivative [42]. In stronger morphants, a broadening of the chordin expression domain wasobserved (fig. 4i–l). In addition, consistent with the axis dorsalization, the expression of bmp2b at shield stage was severely reduced as also evidenced by its undetectable expression in the tail during somitogenesis (fig. 4m–p). The administration of human or zebrafish glce mRNAs to embryos injected with MO rescued the enzymatic activity and prevented the development of the most severe dorsalized phenotypes (table 2).
Figure 4.
Effect of Glce knockdown on embryo morphogenesis. (a-d) Embryos at 1–2 cells stage were injected with a mix (8 ng each) of glce-A and glceB MO and the phenotype scored at 48 hpf. (a) wild type embryo. (b,c) mild phenotypes displaying reduced head volume and ventral fin extension. (d) severe phenotype with shortened A/P axis and loss of ventral structures. (e) Epimerase enzymatic activity in 10 hpf embryos and effect of Glce knockdown. The enzymatic assay was performed as described in Fig. 2b. (f-p) Whole mount in-situ hybridization with D/V markers of embryos at different developmental stages. (f) gata1 expression in wild-type embryos, (g) glce morphants, and (h) in embryos overexpressing glce at 24 hpf. (i-l) chordino at 50% epiboly in wild type (top) and morphants (bottom). (i,j) animal pole view, (k,l) dorsal view. (m,n) bmp2b at 70% epiboly and (o,p) at 3 somite stage in wild type (top) and morphants (bottom). Note the absence of bmp2b expression in the presumptive tail of the morphants as indicated by the arrows.
Table 2.
Effect on D/V axis formation of antisense targeting of glce. Capped mRNA and glce-MOs were dissolved in Danieau's buffer and injected as 1–3 nl bolus into the yolk of one- to two-cell embryos as described in Table 1. Capped human GLCE-mRNA was generated from a full length cDNA clone. The embryos were classified according to the severity of the observed phenotype at 48 hpf based on a classification of the dorsalization severity observed in embryos that had only received MOs. Mildly dorsalized embryos show reduced ventral tail fin extension. Moderately dorsalized embryos display kinked tail, enlarged heart cavity and in some case the absence of atrioventricular boundary. Severely affected morphants show dramatic shortening and reduction of the body mass with tail coiling. For the rescue experiments, embryos were first injected with MO and after randomization half received the indicated amount of mRNA before reaching the 4-cell stage.
| Treatment | Phenotype Severity | ||||
| No. | Wild | Mild | Moderate | Severe | |
| (%) | (%) | (%) | (%) | ||
| Uninjected | 42 | 100 | 0 | 0 | 0 |
| glce-A MO (4 ng) | 62 | 35 | 46 | 19 | 0 |
| glce-A MO (8 ng) | 40 | 20 | 25 | 39 | 16 |
| glce-A MO (16 ng) | 28 | 12 | 19 | 31 | 38 |
| glce-B MO (16 ng) | 30 | 25 | 19 | 30 | 26 |
|
glce-A MO (8 ng) glce-B MO (8 ng) |
30 | 15 | 10 | 30 | 45 |
|
glce-B MO (16 ng) glce-A mRNA (200 pg) |
68 | 28 | 38 | 22 | 12 |
|
glce-B MO (16 ng) glce-B mRNA (200 pg) |
30 | 32 | 45 | 15 | 8 |
|
glce-A MO (16 ng) Human GLCE mRNA (200 pg) |
35 | 21 | 41 | 29 | 9 |
In order to assess the dependency of Bmp activity on Glce level, embryos were injected with either 50 pg of bmp2b mRNA or 100 pg of bmp4 to generate a preponderance of V3-V4 ventralized embryos (table 3). Following randomization, half of the injected embryos received a mix of MOs targeting both glce-A and glce-B transcripts. About two-third of the embryos receiving the MOs displayed a normal-to-mild (V1-V2) ventralized phenotype whereas few developed the most severe V4 class phenotype. These results are in stark contrast to the embryos that had only received bmp2b and bmp4 mRNA supporting the concept that Glce is required for Bmp-mediated ventralization.
Table 3.
Effect of antisense MO on the ventralizing activity of Bmps. Capped mRNA and glce MOs were injected into the yolk of one- to two-cell embryos as in the experiments of table 1. The resulting phenotypes were scored at 48 hpf. For the rescue experiments, embryos were first injected with bmp2b or bmp4 mRNA and after randomization half received the indicated amount of MO before reaching the 4-cell stage. Phenotype severity was scored as in Table 1.
| Treatments | Strength of Ventralization | |||||
| No. | Wild | V1 | V2 | V3 | V4 | |
| (%) | (%) | (%) | (%) | (%) | ||
| Uninjected | 40 | 100 | 0 | 0 | 0 | 0 |
| bmp2b mRNA (50 pg) | 62 | 0 | 0 | 6 | 18 | 76 |
|
bmp2b mRNA (50 pg) glce-A MO (8 ng) glce-B MO (8 ng) |
60 | 15 | 15 | 31 | 25 | 16 |
| bmp4 mRNA (100 pg) | 60 | 0 | 0 | 0 | 15 | 85 |
|
bmp4 mRNA (100 pg) glce-A MO (8 ng) glce-B MO (8 ng) |
60 | 21 | 18 | 23 | 23 | 15 |
Discussion
In mammals, HS plays a crucial role in a variety of important biological processes including the regulation of blood coagulation, cell adhesion and differentiation, angiogenesis, and virus infection [1,3,43]. Most of the information concerning the role of HSPG in development has been obtained in the invertebrate model organism D. melanogaster and support the idea of a major functional role for HS in the morphogen's gradient establishment [3,14,44,45]. The fly mutants Sugarless [46], fringe connection [47], sulfateless [48], and tout-velu [49,50], display cuticle abnormalities that are reminiscent of the phenotypes exhibited by the mutations in Wg, Hh, or FGF and suggest an involvement of HS in the activity of these morphogens. In Drosophila the lack of HS also affects the body axis formation, but this effect is evident only at later stages of development [14]. Compared to the wealth of data generated in invertebrate species, the functional role of HSPG in vertebrate development is still poorly investigated. Transgenic mice carrying null-mutations in genes coding for enzymes implicated in HS chain polymerization [51], glucosamine N- or IdoA 2O-sulfation [52,53], and GlcA epimerization [11] are not viable leading to the conclusion that HS encoding critical structural epitopes is required for normal embryonic development [54]. The developmental mechanisms affected by the lack or by structurally aberrant HS, however remain to be assessed.
In this study the specific role of Glce has been investigated. GlcA epimerization endows the nascent polysaccharide HS with increased biological activity and is necessary to direct further chain sulfation at specific sites [3,6]. In mammals the enzyme is a single gene product whereas in zebrafish two genes have been identified likely arising from gene duplication. Transcripts for the two epimerases are already detected during the early cell divisions indicating a maternal contribution to the zygotic pool. The temporal expression pattern of glce closely resemble that of ext2-A suggesting that HS chain elongation and GlcA epimerization may be activated at the same time. Up to 12 hpf glce expression is detected along the entire axis. At 24 hpf however, the epimerase is highly expressed in the hindbrain, most notably along the perimeter of the fourth ventricle. In the hindbrain, at this developmental stage, Glce may play a specialized role involving axonal guidance as postulated based on observations made in C. elegans with mutated glce [13]. It will be of interest in future studies to compare the pattern of expression of glce and ext2 with that of the other enzymes involved in polysaccharide chain formation and sulfation to test the hypothesis that HS structure is developmentally regulated. For example zebrafish glucosamine 6O-sulfotransferase which act downstream to the biosynthetic step catalyzed by Glce, is not maternally derived [55] suggesting that GlcA epimerization and glucosamine sulfation represents two distinct pathways regulating HS structure during development.
The fact that the expression of Glce is rather ubiquitous throughout gastulation, has given us the opportunity to investigate the role of this enzyme by globally perturbing its level either by injecting capped mRNA or antisense oligonucleotides. When misexpressed the protein produced a ventralized phenotype similar to that observed in null-mutants for genes ogon [33,35,36], radar [37,38] and chordino [25,36] that directly modulate the function of the ventralizing agents Bmps albeit through different mechanisms. This phenotypic similarity lead us to focus on the role of Glce with respect to the activity of Bmps. During development the cell fate in zebrafish depends on the position within the embryo during blastula and gastrula stages. Positional information to cells are provided through the establishment of an activity gradient of Bmp proteins that promotes ventral specification in a dose dependent manner [40,56,57]. The idea that the specification of cells fate along the D/V axis is mediated through Bmp acting as terminal effectors, is supported by the fact that activation of the Bmp signaling pathway is a rather late event during embryogenesis and by the observation that functional inactivation of the zebrafish genes Bmp2b (swirl) [40] and Bmp7 (snailhouse) [34] both result in dramatic suppression of ventral fates and expansion of dorsal structures. Bmp2b and Bmp4 interact with HS through a cluster of positively charged aminoacids located at the N-terminus outside the receptor-binding domain of the protein [17]. Additional studies indicate that the interaction of Bmps with HS has important functional significance in that mutations in the HS/heparin binding domain results in an increase in the long-range activity of the morphogens [58]. HS also potentiates the biological activity of Bmps by serving the ligand to their receptor and/or by stabilizing the biological activity of the morphogen by preventing its proteolytic degradation [59]. Changes in IdoA content affecting HS binding to Bmp can thus change Bmp activity through different mechanisms. A spectrum of D/V mutants ranging from strongly ventralized to dorsalized embryos are generated when Glce activity is modulated suggesting that correct axial patterning requires that the activity of the epimerase be maintained within a critical range. An analysis of the structural domain in HS responsible for binding to Bmps can further elucidate what specific role IdoA residues play in this context.
Because HS ability to interact with proteins generally correlates positively with the IdoA content [2], Glce may be involved in the regulation of the activity of other heparin-binding morphogens involved in D/V axis formation. Fgf-8 has been demonstrated to play a key role is the establishment of D/V axis by acting upstream of Bmp2 and Bmp4 [60] and interacts with IdoA-rich regions in HS [61]. In zebrafish Fgf-8 inhibits the expression of Bmps in the ventral part of the embryo leading to the expansion of dorso-lateral derivatives at the expenses of ventral and posterior domains [60,62]. Based on our results an activation of Fgf-8 mediated pathway following ectopic Glce expression seems unlikely since an expansion rather than a reduction of ventral structures has been observed in embryos overexpressing the enzyme. It is possible that Glce overexpression inhibits Fgf-8 mediated signaling. This would occur if Fgf-8 is sequestered in the extracellular matrix by HS enriched in FGF binding regions or if the FGF receptor dimerization is negatively affected by HS [19]. However Fgf-8 null mutants display rather mild D/V abnormalities and similarly to fgf-8 morphants or mutant embryos with disrupted Ras/MAPK-mediated FGF signaling, do not form a midbrain-hindbrain boundary and do not develop the cerebellum [62]. Both these brain structures are present in the glce morphants and in embryos overexpressing glce unless, as a consequence of marked dorsalization or ventralization, the entire body plan is grossly altered. This finding is consistent with the observation that Glce-null mice have normal brain morphology [11] as it would be expected if Fgf-8 function is not affected [63]. Conceivably Fgf-8 mediated D/V patterning is little influenced by perturbation in Glce activity pointing to a downstream mediator of axis formation as sensitive to changes in HS IdoA content. A similar conclusion can be reached with regard to the D/V patterning effect of Wnt which acts upstream to Fgf-8, since activation of this pathway would result in posteriorization of the neural ectoderm affecting the eyes and the midbrain-hindbrain boundary development [64,65].
Taken together our results identify the stage of D/V patterning controlled by Bmp as sensitive to changes in HS structure. Previously it has been shown that mutants of the HSPG Dally have altered Dpp gradient formation resulting in abnormal patterning of the wing imaginal disc [16]. It was hypothesized that the HS chains of Dally bind Dpp and promote the interaction of the morphogen with the cognate cell surface receptor. Decreased interaction with HS, as may occurs when Glce activity is lowered, may reduce the concentration of Bmp available for interaction with the cognate receptors or the receptor-ligand binding is affected. Conversely, as a result of enhanced GlcA epimerization, HS affinity for Bmp may increase enhancing the concentration of the ligand at the receptor site and prolonging the morphogen activity [58,59,66]. The fact that Bmp is antagonized by proteins such as Chordin, Noggin, and Follistatin that require HS for diffusion and activation [67-69], represents an additional potential mechanism of regulation of Bmp activity that is dependent on HS. Chordin is required to dorsalize surrounding neuroectoderm and mesoderm and its expression pattern is affected when Glce activity is altered. A specific class of HSPG strongly potentiates the antagonism of Bmp signaling by Chordin and is necessary for the retention of Chordin by cells [69]. Likewise the interaction of Noggin with HSPG in vivo regulate its diffusion [67]. Conceivably in tissues rich of HS that binds with high affinity to Chordin and Noggin, the range of action of these Bmp antagonists is reduced and the ventralizing effect of Bmps may prevail. Our results support the hypothesis that correct D/V patterning depends upon the regulated expression of specific structural elements in HS and provide the basis for the interpretation of the functional role of Glce in vivo.
Conclusion
The results obtained corroborate the concept that HS encodes information that directs morphogenesis during early vertebrate development. In particular Glce emerges from this study as an important modulator of vertebrate morphogenesis that acts in a dose-dependent fashion on D/V axis formation. Bmp-dependent cell fate specification is the main target of Glce activity. Glce effect may be mediated by potentiating the effect of Bmps or by restricting the range of action of other HS-binding proteins such as Chordin and Noggin that by antagonizing Bmps act as indispensable dorsalizing agents.
Methods
Zebrafish breeding and phenotype scoring
Embryos were obtained from natural mating of wild-type (Oregon, AB) fish and breed, raised and staged according to established criterias [70].
Cloning of zebrafish Glce cDNA
A query of the zebrafish dEST database identified a number of putative clones whose translated sequence matched the N- and C-terminus of the human protein. Analysis of the predicted protein sequence of these clones indicated that zebrafish have two highly homologous Glce proteins. Cloning of the putative glce cDNAs was performed by reverse transcription of adult male zebrafish mRNA (Qiagen Sensiscript) primed with oligo-dT. cDNAs were amplified by PCR by combining primers matching the different possible cDNA terminal sequences. Forward primers were 5'-ATGCGCTGTCTGGTGGCTCGAATCAATC ACAAGACT-3' and 5'-ATGCGTTGTCTGGCAGCCGGTGTTCACTACAAGACC-3'. Reverse primers were 5'-CTAGTTGTGCTTAGCCCGACCTCCTTTCAGGTAAGT-3' and 5'-TTAATTGTGCTTAGCCCTCCCTCCTTTCAGGTAGCT-3'. This strategy resulted in two products of the expected size (~1.8 kb) from two of the four possible primer combinations. The amplified products were named glce-A (GenBank AY388516) and glce-B (AY388517), cloned into pcDNA3.1-TOPO (Invitrogen) and sequenced.
The 5'-end translated region of the zebrafish homologue of human exostosin 2 (EXT2) gene, was cloned using a similar strategy. The existence of two zebrafish ext2 genes were predicted from alignments of published EST sequences. The 5'-end of the ext2-A coding sequence including part of the UT region was cloned by RT-PCR using forward and reverse primers of sequence 5'-CATTCAACTTAAATATTCACCATA-3' and 5'-GGCGCTCAGCAGGTCATTGTATTC-3' respectively. Sequencing of an expected 528 bp fragment confirmed the identity of the amplified cDNA.
Chromosomal mapping of zebrafish glce
In the human, the glce translational start site is located in a 602 bp exon. Assuming that zebrafish glce genes maintain the same genomic organization as the human, PCR primers were designed to amplify the exon containing the translation initiation site for each each zebrafish orthologue. PCR amplification with primers 5'-ATGCGCTGTCTGGTGGCTCGAATC-3' and 5'-AGATGAAGGGCAGATACACCTCGC-3' for glce-A and 5'-ATGCGTTGTCTGGCAGCCGGTGTTCACTACAAG-3' and 5'-GACCTTTAATGGTGGCATCGTCATTGATCAGGC-3' for glce-B using genomic male zebrafish DNA as template, gave products of the expected size (420 bp and 261 bp for glce-A and glce-B respectively) that were cloned in pGEM-T vector and sequenced to confirm their identity. These sets of primers were then used to determine the chromosomal location of each gene by radiation hybrid panel (LN54) mapping.
Antisense targeting of the transcripts
Antisense Morpholino oligonucleotides (MOs) (Gene Tools LLC) were designed to target the 5'-UT region of the genes of interest. glce-A and glce-B MOs had sequence 5'-AGCCATGAGGAACACGTCAGCAAAC-3' and 5'-TCCCTGCTTACCTGCAATGCAAACA-3', respectively. MOs were dissolved in water at a concentration of 4 mM and diluted in Danieu's buffer before injection.
Generation of capped mRNA
Full-length glce cDNAs were subcloned in pT3TS vector at the BglII and EcoRV sites and in vitro-transcribed capped mRNAs were synthesized (T3 mMESSAGE mMACHINE kit, Ambion). mRNAs (1 μg) were tested prior to injection for protein expression in vitro using a rabbit reticulocyte lysate assay kit and 35S-methionine. Labeled proteins were separated on 9% SDS-PAGE gel followed by autoradiography. Human glce mRNA was generated from a full length cDNA clone in pcDNA 3.1 vector using T7 RNA polymerase. The human clone (AY635582) was obtained by RT-PCR using primers of sequence 5'-CTGCATATGCTGTGCTTGGCA-3' and 5'-CTAGTTGTGCTTTGCCCTGCTGCCTTT-3' based on published coding sequences and on cDNA 5'-end extension experiments we have performed [28]. bmp2b and bmp4 capped mRNA were kindly provided by Dr M. Mullins (U.Penn).
Microinjection
MOs and mRNAs were injected (1–3 nl) into the yolk of 1–2 cell embryos [71]. Post-injection (6 h) embryos were sorted, the unfertile/damaged removed and the rest allowed to grow at 28°C for further observation.
RNA in situ hybridization of zebrafish embryos
Antisense digoxigenin-labeled riboprobes were generated using SP6 or T7 RNA polymerase-based labeling kit (Roche). cDNA clones for glce-a, glce-b, chordino, krox-20, opl, myoD, were generated by RT-PCR [see Additional file 1]. bmp2b, fkd3, and eve1 antisense riboprobes were generated by reverse transcription from cDNA clones (cb670, cb114, and cb872 respectively) obtained from the Zebrafish International Resource Center (University of Oregon). Whole embryo in situ hybridization was performed as previously described [72].
Semi-quantitative RT-PCR
RNA was extracted and cDNA generated by reverse transcriptase using oligo-dT primers. An aliquot of the reaction was used as template for PCR amplification using gene-specific primers (Appendix I). Reactions were performed in duplicate and the product generated after 20 and 30 cycles analyzed by agarose electrophoresis to ensure that the products were quantitated during the exponential phase of the chain reaction.
Epimerase enzymatic activity assay
Embryos were collected at the indicated times, dechorionated and washed in ice-cold 25 mM HEPES, pH 7.0 buffer containing 0.1% Triton X-100. Embryos were then homogenized in 200 μl of the same buffer with a pestle that fits tightly into an Eppendorf tube and stored at -70°C. The substrate for the epimerase enzymatic assay consisted of radiolabeled modified bacterial N-acetylheparosan prepared as described previously [28]. The final N-sulfated heparosan product was purified by ion-exchange chromatography and eluted at higher ionic strength than the starting bacterial polysaccharide (0.66 M vs. 0.40 M NaCl). The epimerase enzymatic assay was performed as described by Crawford et al. [4]. Briefly, reactions were set up by combining the homogenates from 20 embryos with labeled substrate (1 nmole ~30,000 dpm) followed by 2 h incubation at 28°C. Reactions were stopped by addition of DEAE-Sepharose (1 ml) equilibrated in 50 mM Na-acetate, 50 mM NaCl pH 4.0 buffer (1:1 volume) followed by 15 min incubation at 4°C. Tritiated water generated as a result of the epimerization of GlcA into IdoA was recovered in the supernatant following centrifugation at 10,000 rpm. The sample and a 800-μl rinse of the DEAE slurry with buffer, were counted for the associated radioactivity in a Wallac 1219 Rackbeta liquid scintillation counter. Values were corrected for a reaction blank obtained by adding the substrate to the embryo homogenate just prior to the addition of DEAE-Sepharose.
Authors' contributions
GG cloned the zebrafish glce gene and the cDNAs used in the study, generated the capped mRNAs, performed the Glce enzymatic assay, and drafted the manuscript. SAF supervised the zebrafish injection experiments, provided training regarding the whole-mount in situ hybridization and performed some of the microscope analysis.
Supplementary Material
Oligonucleotide primers Primers used for semiquantitative RT-PCR analysis and for the generation of riboprobes
Acknowledgments
Acknowledgements
The Authors wish to thank Dr. Mary Mullins for the help provided with the evaluation of the morphants' phenotype, for providing the Bmp capped mRNAs, and for the insightful discussion of the results. Mr Amit Agrawal and Chirag Patel have provided excellent technical support with many of the molecular biology techniques utilized. This work was supported by grant RO1-CA82290 to GG, by a Pew Scholar Award to S.F., and in part by RO1-GM063904 to S.F.
Contributor Information
Giancarlo Ghiselli, Email: giancarlo.ghiselli@jefferson.edu.
Steven A Farber, Email: farber@ciwemb.edu.
References
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Crawford BE, Olson SK, Esko JD, Pinhal MA. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. J Biol Chem. 2001;276:21538–21543. doi: 10.1074/jbc.M103414200. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- Li JP, Gong F, El Darwish K, Jalkanen M, Lindahl U. Characterization of the D-glucuronyl C5-epimerase involved in the biosynthesis of heparin and heparan sulfate. J Biol Chem. 2001;276:20069–20077. doi: 10.1074/jbc.M011783200. [DOI] [PubMed] [Google Scholar]
- Mulloy B, Forster MJ. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- Ferro DR, Provasoli A, Ragazzi M, Casu B, Torri G, Bossennec V, Perly B, Sinay P, Petitou M, Choay J. Conformer populations of L-iduronic acid residues in glycosaminoglycan sequences. Carbohydr Res. 1990;195:157–167. doi: 10.1016/0008-6215(90)84164-P. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Inouye Y, Nagasawa K. Conformational equilibria of the L-iduronate residue in non-sulphated di-, tetra- and hexa-saccharides and their alditols derived from dermatan sulphate. Biochem J. 1990;265:533–538. doi: 10.1042/bj2650533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarana M, Casu B, Lindahl U. Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J Biol Chem. 1994;269:3903. [PubMed] [Google Scholar]
- Li JP, Gong F, Hagner-Mcwhirter A, Forsberg E, Abrink M, Kisilevsky R, Zhang X, Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem. 2004;279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- Bulow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/S0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- Reichsman F, Smith L, Cumberledge S. Glycosaminoglycans can modulate extracellular localization of the wingless protein and promote signal transduction. J Cell Biol. 1996;135:819–827. doi: 10.1083/jcb.135.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, Golden C, Selleck SB. dally, a Drosophila glypican, controls cellular responses to the TGF- beta-related morphogen, Dpp. Development. 1997;124:4113–4120. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem. 1996;237:295–302. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Choi Y, Segal RA. Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development. 2002;129:2223–2232. doi: 10.1242/dev.129.9.2223. [DOI] [PubMed] [Google Scholar]
- Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- Lee JS, von der HS, Rusch MA, Stringer SE, Stickney HL, Talbot WS, Geisler R, Nusslein-Volhard C, Selleck SB, Chien CB, Roehl H. Axon sorting in the optic tract requires HSPG synthesis by ext2 (dackel) and extl3 (boxer) Neuron. 2004;44:947–960. doi: 10.1016/j.neuron.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI. 1998. http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0E.html [DOI] [PubMed]
- Hofmann K, Stoffel W. TMPERD. 1993. http://ch.embnet.org/software/TMPRED_form.html
- Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Nusslein-Volhard C. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Serbedzija GN, McMahon AP. Genetic analysis of dorsoventral pattern formation in the zebrafish: requirement of a BMP-like ventralizing activity and its dorsal repressor. Genes Dev. 1996;10:2452–2461. doi: 10.1101/gad.10.19.2452. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Yamanaka Y, Ryu SL, Hashimoto H, Yabe T, Hirata T, Bae YK, Hibi M, Hirano T. Cooperative roles of Bozozok/Dharma and Nodal-related proteins in the formation of the dorsal organizer in zebrafish. Mech Dev. 2000;91:293–303. doi: 10.1016/S0925-4773(99)00319-6. [DOI] [PubMed] [Google Scholar]
- Ghiselli G, Agrawal A. The human D-glucuronyl C5-epimerase gene is transcriptionally activated through the beta-catenin/TCF4 pathway. Biochem J. 2005 doi: 10.1042/BJ20050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr KB, Schulte-Merker S, Tautz D. Zebrafish zic1 expression in brain and somites is affected by BMP and hedgehog signalling. Mech Dev. 1999;85:147–159. doi: 10.1016/S0925-4773(99)00044-1. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Gonzalez EM, Fekany-Lee K, Carmany-Rampey A, Erter C, Topczewski J, Wright CV, Solnica-Krezel L. Head and trunk in zebrafish arise via coinhibition of BMP signaling by bozozok and chordino. Genes Dev. 2000;14:3087–3092. doi: 10.1101/gad.852400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Bertoglio V, Carmany-Rampey A, Furthauer M, Gonzalez EM, Thisse C, Thisse B, Halpern ME, Solnica-Krezel L. Maternal and zygotic activity of the zebrafish ogon locus antagonizes BMP signaling. Dev Biol. 1999;214:72–86. doi: 10.1006/dbio.1999.9384. [DOI] [PubMed] [Google Scholar]
- Schmid B, Furthauer M, Connors SA, Trout J, Thisse B, Thisse C, Mullins MC. Equivalent genetic roles for bmp7/snailhouse and bmp2b/swirl in dorsoventral pattern formation. Development. 2000;127:957–967. doi: 10.1242/dev.127.5.957. [DOI] [PubMed] [Google Scholar]
- Yabe T, Shimizu T, Muraoka O, Bae YK, Hirata T, Nojima H, Kawakami A, Hirano T, Hibi M. Ogon/Secreted Frizzled functions as a negative feedback regulator of Bmp signaling. Development. 2003;130:2705–2716. doi: 10.1242/dev.00506. [DOI] [PubMed] [Google Scholar]
- Wagner DS, Mullins MC. Modulation of BMP activity in dorsal-ventral pattern formation by the chordin and ogon antagonists. Dev Biol. 2002;245:109–123. doi: 10.1006/dbio.2002.0614. [DOI] [PubMed] [Google Scholar]
- Sidi S, Goutel C, Peyrieras N, Rosa FM. Maternal induction of ventral fate by zebrafish radar. Proc Natl Acad Sci U S A. 2003;100:3315–3320. doi: 10.1073/pnas.0530115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutel C, Kishimoto Y, Schulte-Merker S, Rosa F. The ventralizing activity of Radar, a maternally expressed bone morphogenetic protein, reveals complex bone morphogenetic protein interactions controlling dorso-ventral patterning in zebrafish. Mech Dev. 2000;99:15–27. doi: 10.1016/S0925-4773(00)00470-6. [DOI] [PubMed] [Google Scholar]
- Payne TL, Postlethwait JH, Yelick PC. Functional characterization and genetic mapping of alk8. Mech Dev. 2001;100:275–289. doi: 10.1016/S0925-4773(00)00541-4. [DOI] [PubMed] [Google Scholar]
- Nguyen VH, Schmid B, Trout J, Connors SA, Ekker M, Mullins MC. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev Biol. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- Detrich HWIII, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Perrimon N. Functional binding of secreted molecules to heparan sulfate proteoglycans in Drosophila. Curr Opin Cell Biol. 2000;12:575–580. doi: 10.1016/S0955-0674(00)00134-4. [DOI] [PubMed] [Google Scholar]
- De Cat B, David G. Developmental roles of the glypicans. Semin Cell Dev Biol. 2001;12:117–125. doi: 10.1006/scdb.2000.0240. [DOI] [PubMed] [Google Scholar]
- Hacker U, Lin X, Perrimon N. The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development. 1997;124:3565–3573. doi: 10.1242/dev.124.18.3565. [DOI] [PubMed] [Google Scholar]
- Selva EM, Hong K, Baeg GH, Beverley SM, Turco SJ, Perrimon N, Hacker U. Dual role of the fringe connection gene in both heparan sulphate and fringe-dependent signalling events. Nat Cell Biol. 2001;3:809–815. doi: 10.1038/ncb0901-809. [DOI] [PubMed] [Google Scholar]
- Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/S1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Selleck SB. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem. 2000;275:2269–2275. doi: 10.1074/jbc.275.4.2269. [DOI] [PubMed] [Google Scholar]
- Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- Fan G, Xiao L, Cheng L, Wang X, Sun B, Hu G. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 2000;467:7–11. doi: 10.1016/S0014-5793(00)01111-X. [DOI] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. The molecular phenotype of heparan sulfate in the Hs2st-/- mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Habuchi O, Kimata K. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj J. 2004;21:47–52. doi: 10.1023/B:GLYC.0000043747.87325.5e. [DOI] [PubMed] [Google Scholar]
- Bink RJ, Habuchi H, Lele Z, Dolk E, Joore J, Rauch GJ, Geisler R, Wilson SW, Den Hertog J, Kimata K, Zivkovic D. Heparan sulfate 6-O-sulfotransferase is essential for muscle development in Zebrafish. J Biol Chem. 2003 doi: 10.1074/jbc.M213124200. [DOI] [PubMed] [Google Scholar]
- Mullins MC. Embryonic axis formation in the zebrafish. Methods Cell Biol. 1999;59:159–178. doi: 10.1016/s0091-679x(08)61825-7. [DOI] [PubMed] [Google Scholar]
- Myers DC, Sepich DS, Solnica-Krezel L. Bmp activity gradient regulates convergent extension during zebrafish gastrulation. Dev Biol. 2002;243:81–98. doi: 10.1006/dbio.2001.0523. [DOI] [PubMed] [Google Scholar]
- Ohkawara B, Iemura S, ten DP, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr Biol. 2002;12:205–209. doi: 10.1016/S0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- Takada T, Katagiri T, Ifuku M, Morimura N, Kobayashi M, Hasegawa K, Ogamo A, Kamijo R. Sulfated polysaccharides enhance the biological activities of bone morphogenetic proteins. J Biol Chem. 2003;278:43229–43235. doi: 10.1074/jbc.M300937200. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Thisse C, Thisse B. A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development. 1997;124:4253–4264. doi: 10.1242/dev.124.21.4253. [DOI] [PubMed] [Google Scholar]
- Loo BM, Salmivirta M. Heparin/Heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. J Biol Chem. 2002;277:32616–32623. doi: 10.1074/jbc.M204961200. [DOI] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Kelly GM, Greenstein P, Erezyilmaz DF, Moon RT. Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development. 1995;121:1787–1799. doi: 10.1242/dev.121.6.1787. [DOI] [PubMed] [Google Scholar]
- Saksela O, Moscatelli D, Sommer A, Rifkin DB. Endothelial cell-derived heparan sulfate binds basic fibroblast growth factor and protects it from proteolytic degradation. J Cell Biol. 1988;107:743–751. doi: 10.1083/jcb.107.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine-Saunders S, Viviano BL, Economides AN, Saunders S. Heparan sulfate proteoglycans retain Noggin at the cell surface: a potential mechanism for shaping bone morphogenetic protein gradients. J Biol Chem. 2002;277:2089–2096. doi: 10.1074/jbc.M109151200. [DOI] [PubMed] [Google Scholar]
- Sugino K, Kurosawa N, Nakamura T, Takio K, Shimasaki S, Ling N, Titani K, Sugino H. Molecular heterogeneity of follistatin, an activin-binding protein. Higher affinity of the carboxyl-terminal truncated forms for heparan sulfate proteoglycans on the ovarian granulosa cell. J Biol Chem. 1993;268:15579–15587. [PubMed] [Google Scholar]
- Jasuja R, Allen BL, Pappano WN, Rapraeger AC, Greenspan DS. Cell-surface heparan sulfate proteoglycans potentiate Chordin antagonism of bone morphogenetic protein signaling and are necessary for cellular uptake of Chordin. J Biol Chem. 2004;279:51289–51297. doi: 10.1074/jbc.M408129200. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR, University of Oregon; 1995. [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers Primers used for semiquantitative RT-PCR analysis and for the generation of riboprobes