Abstract
A typing procedure based on polymorphism of the coagulase gene (coa) was used to discriminate Staphylococcus aureus isolated from Minas Gerais dairy cows with mastitis. Amplification of the gene from the 64 S. aureus isolates produced 27 different polymerase chain reaction (PCR) products; 60 isolates showed only 1 amplicon, and 4 showed 2 amplicons. The isolates were grouped into 49 types by analyzing the restriction fragment length polymorphism (RFLP) of the coa gene; the 10 most common types accounted for 39% of the isolates. The results demonstrate that many variants of the coa gene are present in the studied region, although only a few predominate.
Résumé
Une procédure de typage utilisant le polymorphisme du gène de la coagulase (coa) a été utilisée afin de distinguer les isolats de Staphylococcus aureus provenant de vache laitière avec mammite dans la région de Minas Gerais. L’amplification du gène de 64 isolats de S. aureus a résulté en 27 produits de réaction d’amplification en chaîne (PCR) différents; 60 isolats n’ont donné qu’un seul amplicon et 4 ont donné deux amplicons. Les isolats ont été groupés en 49 types suite à l’analyse du polymorphisme de taille des fragments de restriction du gène coa; les 10 types les plus fréquents représentant 39 % des isolats. Les résultats démontrent que bien que plusieurs variants du gène coa soient présents dans la région étudiée seulement quelques uns prédominent.
(Traduit par Docteur Serge Messier)
Introduction
Bovine mastitis continues to be one of the most significant endemic illnesses in dairy herds in terms of responsibility for economic losses for the producer as well as the dairy industry (1). Although mastitis can be caused by a variety of pathogens, Staphylococcus aureus is considered the most frequent (2).
Natural populations of S. aureus have shown considerable variability in genome content (3,4). This variability has contributed to the emergence of distinct epidemiologic profiles that are dependent on the strains prevalent in a herd, which suggests the need to identify such strains or subtypes before applying specific measures to control mastitis (5).
In the last few years, numerous molecular techniques have been used to identify and compare S. aureus subtypes. Amplification of the coagulase gene (coa) has been considered a simple and accurate method for typing S. aureus isolated from distinct sources (6–11). The results of epidemiologic research based on analysis of the coa gene suggest that few S. aureus subtypes are responsible for most cases of bovine mastitis, and these are widespread (8,11,12).
In Brazil, little information is available about the genetic diversity of S. aureus isolated from cows with mastitis. The purpose of this study was to use coa gene polymorphism to identify S. aureus subtypes isolated from mastitis cases in dairy herds in Minas Gerais state.
Materials and methods
Bacterial strains
We used 64 S. aureus isolates from the milk of cows with mastitis. The milk samples had been obtained between 1994 and 1997 from different dairy herds in Minas Gerais state, identified by Cardoso (13), and kept frozen at −20°C in tryptic soy broth (TSB; Biobrás, São Paulo, Brazil) containing 15% glycerol until molecular tests were carried out.
Gene typing
Extraction and purification of DNA
We extracted and purified bacterial DNA according to previously published methods (14,15). Bacterial cell lysates were prepared from 0.5 mL of overnight TSB cultures. After centrifugation at 12 000 × g for 10 min, the bacterial pellets were washed with 500 μL of Tris-hydrochloride-ethylene diamine tetraacetic acid (EDTA) (TE) buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA) and centrifuged again. The pellets were resuspended in 200 μL of TE buffer, pH 7.5, with 15 U of lysostaphin (2 mg; Sigma-Aldrich Brasil, São Paulo, Brazil) per milliliter, and incubated at 37°C for 1 h. Next, 15 μL of proteinase K, 20 mg/mL (Invitrogen Brasil, São Paulo, Brazil), was added and the suspension incubated at 56°C for 1 h. The suspension was then heated at 95°C for 15 min to inactivate the proteinase K. An equal volume of phenol-chloroform was added and the mixture centrifuged at 12 000 × g for 10 min. The supernatant was extracted with an equal volume of phenol-chloroform and then chloroform. The DNA in the supernatant was mixed with 2 volumes of 95% ethanol and stored overnight at −20°C. The mixture was then centrifuged at 12 000 × g for 5 min. The DNA pellet was washed with ice-cold 70% ethanol, recentrifuged, and dried by tube inversion. The DNA was suspended in 100 μL of sterile TE, pH 7.5, quantified in a spectrophotometer (at 260 nm), and kept frozen at −20°C.
Polymerase chain reaction (PCR) amplification
The 3′ end region of the coa gene was amplified with use of the internal primer pair previously reported by Aarestrup et al (8): Coag2, 5′-ACCACAAGGTACTGAATCAACG-3′ (bases 1432 to 1453); and Coag3, 5′-TGCTTTCGATTGTTCGATGC-3′ (bases 2399 to 2418). For PCR, each reaction mixture contained 1 to 2 μL of target DNA (approximately 350 ng/μ L), 1 μL of each of the primers (50 pmol), 0.8 μL of a mix of deoxynucleotide triphosphates (200 μM each), 0.2 μL of Taq polymerase (1 U), and 3 μL of PCR 10× buffer (500 mM of KCl; 100 mM of Tris-HCl, pH 8.4; 1% Triton X-100; and 15 mM of MgCl2). The volume of this mix was adjusted to 40 μL with sterile water. Evaporation was prevented by the addition of 50 μL of sterile mineral oil. Amplification was carried out in a thermal cycler (MJ MiniCycler; Bio-Rad Laboratories, Hercules, California, USA) as follows: initial denaturation at 94°C for 2 min, 30 cycles of amplification (denaturation at 95°C for 30 s, annealing at 55°C for 2 min, and extension at 72°C for 4 min), and extension at 72°C for 7 min.
Analysis of restriction fragment length polymorphism (RFLP)
Restriction analysis of the PCR products was performed with AluI (Invitrogen), according to the manufacturer’s instructions. We incubated a mixture of 10 μL of PCR products and 10 U of AluI in a thermal cycler at 37°C for 1 h (8).
Agarose gel electrophoresis
The PCR products and AluI digests were separated in 2% and 5% agarose gel, respectively, with 10 mg/mL of an aqueous solution of ethidium bromide (Pharmacia Brasil, São Paulo, Brazil), and photographed under ultraviolet illumination. We used bacteriophage DNA of Phi29 digested with HindIII as a molecular marker.
Specificity testing
To test the specificity of the primer pair, we analyzed the DNA of S. epidermidis American Type Culture Collection (ATCC) 12228, S. intermedius 08/96PE-FUNED, and S. aureus ATCC 25923.
Reproducibility testing
We tested PCR reproducibility by interassay analysis of 5 randomly chosen isolates, tested for 5 consecutive d. We tested RFLPPCR reproducibility by twice submitting 4 different PCR products to AluI digestion.
Data analysis
The sizes, in base pairs (bp), of the PCR and RFLP products were estimated with the LabImage gel-analysis software program (version 2.7.0; Kapelan Bio-Imaging Solutions, Halle-Saale, Germany). The within-gel standard error (sχ̄) was calculated by estimated-size analysis from the PCR products of 2 isolates run electrophoretically 4 times. The discriminatory power of the typing method was determined according to the numerical index described by Hunter and Gaston (16). The D-value indicates the probability that 2 isolates randomly selected from the test population will be assigned to different typing groups. The following formula was used:
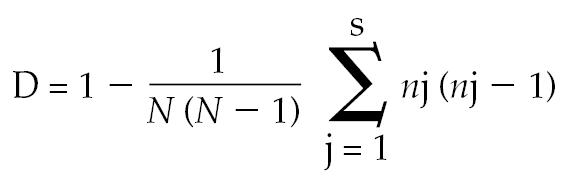 |
with D = discriminatory index, s = total number of different types, nj = number of isolates representing each type, and N = total number of isolates in the sample population.
Results
Electrophoresis sχ̄
The within-gel error was calculated as 8 bp. Thus, PCR products or restriction fragments with this difference were considered the same.
Gene amplification products
The Coag2 and Coag3 primer pair produced 27 amplicons, which ranged from approximately 579 to approximately 1442 bp. Sizes 790, 759, 725, and 579 bp were the most frequent, accounting for 52% of the studied isolates; 60 isolates had only 1 PCR product, and the other 4 had 2 PCR products, with sizes of approximately 972 and 579 (2 isolates), 972 and 739, and 950 and 800 bp (Figure 1). There was no amplification product of the DNA from the other Staphylococcus species. However, amplicons were observed for all S. aureus isolates investigated (100% typability).
Figure 1.
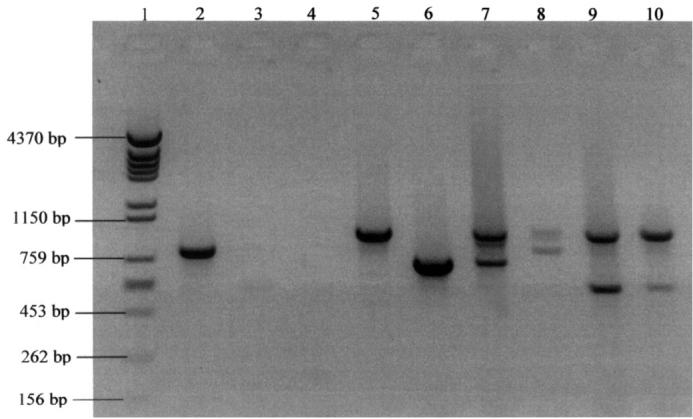
Electrophoretic profile, in 2% agarose gel, of polymerase chain reaction (PCR) products of Staphylococcus aureus coagulase gene isolated from cows with mastitis: lane 1 — molecular marker; lane 2 — positive control, S. aureus American Type Culture Collection 25923; lanes 3 and 4 — negative controls, S. epidermidis and S. intermedius; lanes 5 and 6 — isolates with only 1 amplicon; lanes 7 to 10 — isolates with 2 amplicons.
Reproducibility of the PCR products was demonstrated with 100% of the tested isolates. Although there was some variation in intensity, the bands were always present and their sizes reproducible. The discriminatory index for the PCR-based typing method was 0.92.
Restriction-fragment patterns
The numbers of AluI RFLP patterns, by PCR product and genotype frequency, are shown in Tables I and II, respectively. Most of the PCR products generated a single pattern, but 9 products (those of 1006, 950, 880, 838, 790, 759, 725, 602, and 579 bp) generated 2 or more patterns. The isolates with only 1 PCR amplicon produced 2 to 4 restriction fragments, whereas those with 2 amplicons produced 5 to 6 restriction fragments (Figure 2).
Table I.
Numbers of distinct patterns of restriction fragment length polymorphism (RFLP), determined by AluI testing of polymerase chain reaction (PCR) products of the coagulase gene of Staphylococcus aureus, isolated from cows with mastitis
| PCR products (approximate number of base pairs [bp]) | Number of isolates | Number of distinct RFLP patterns |
|---|---|---|
| 1442 | 1 | 1 |
| 1262 | 1 | 1 |
| 1181 | 1 | 1 |
| 1165 | 1 | 1 |
| 1129 | 1 | 1 |
| 1113 | 1 | 1 |
| 1080 | 1 | 1 |
| 1042 | 1 | 1 |
| 1026 | 2 | 1 |
| 1006 | 2 | 2 |
| 994 | 1 | 1 |
| 972-579 | 2 | 1 |
| 972-739 | 1 | 1 |
| 950-800 | 1 | 1 |
| 950 | 2 | 2 |
| 900 | 1 | 1 |
| 880 | 2 | 2 |
| 850 | 1 | 1 |
| 838 | 3 | 2 |
| 824 | 1 | 1 |
| 800 | 1 | 1 |
| 790 | 10 | 4 |
| 759 | 13 | 11 |
| 725 | 5 | 4 |
| 684 | 1 | 1 |
| 602 | 2 | 2 |
| 579 | 5 | 2 |
| Total | 64 | 49 |
Table II.
Frequency of coagulase genotypes in the isolated Staphylococcus aureus
| Genotype
|
|||
|---|---|---|---|
| Type code | PCR products (bp) | RFLP pattern (bp) | Frequency (%) |
| 1 | 1442 | 453-301-162-80 | 1.6 |
| 2 | 1262 | 453-301-162 | 1.6 |
| 3 | 1181 | 453-301-162-80 | 1.6 |
| 4 | 1165 | 453-301-162-80 | 1.6 |
| 5 | 1129 | 453-301-162-80 | 1.6 |
| 6 | 1113 | 552-229-80 | 1.6 |
| 7 | 1080 | 453-301-162-80 | 1.6 |
| 8 | 1042 | 453-301-162-80 | 1.6 |
| 9 | 1026 | 453-301-162 | 3.1 |
| 10 | 1006 | 453-301-162-80 | 1.6 |
| 11 | 1006 | 453-301-162 | 1.6 |
| 12 | 994 | 453-301-162-80 | 1.6 |
| 13 | 972-579 | 453-377-301-215-162-80 | 3.1 |
| 14 | 972-739 | 453-301-215-162-80 | 1.6 |
| 15 | 950-800 | 453-301-184-162-80 | 1.6 |
| 16 | 950 | 453-301-162-80 | 1.6 |
| 17 | 950 | 453-229-80 | 1.6 |
| 18 | 900 | 453-301-162-80 | 1.6 |
| 19 | 880 | 453-301-162-80 | 1.6 |
| 20 | 880 | 715-80 | 1.6 |
| 21 | 850 | 301-184-162-80 | 1.6 |
| 22 | 838 | 667-80 | 1.6 |
| 23 | 838 | 602-80 | 3.1 |
| 24 | 824 | 552-244 | 1.6 |
| 25 | 800 | 301-244-184-80 | 1.6 |
| 26 | 790 | 508-229-80 | 1.6 |
| 27 | 790 | 715-80 | 3.1 |
| 28 | 790 | 636-80 | 6.3 |
| 29 | 790 | 602-80 | 4.7 |
| 30 | 759 | 301-184-162-80 | 1.6 |
| 31 | 759 | 229-184-162-80 | 3.1 |
| 32 | 759 | 453-215-80 | 1.6 |
| 33 | 759 | 453-229-80 | 3.1 |
| 34 | 759 | 453-184-80 | 1.6 |
| 35 | 759 | 730-80 | 1.6 |
| 36 | 759 | 667-80 | 1.6 |
| 37 | 759 | 636-100 | 1.6 |
| 38 | 759 | 579-229 | 1.6 |
| 39 | 759 | 579-100 | 1.6 |
| 40 | 759 | 579-80 | 1.6 |
| 41 | 725 | 301-244-184-80 | 1.6 |
| 42 | 725 | 636-80 | 3.1 |
| 43 | 725 | 602-80 | 1.6 |
| 44 | 725 | 552-244 | 1.6 |
| 45 | 684 | 334-229 | 1.6 |
| 46 | 602 | 334-229 | 1.6 |
| 47 | 602a | 1.6 | |
| 48 | 579 | 334-229 | 6.3 |
| 49 | 579a | 1.6 | |
Not digested by AluI
PCR — polymerase chain reaction; RFLP — restriction fragment length polymorphism; bp — base pairs
Figure 2.
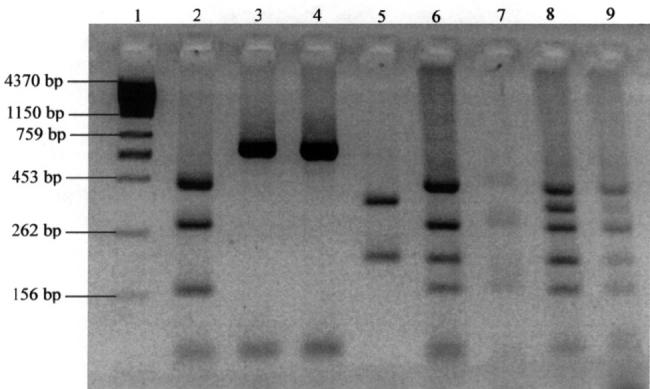
Electrophoretic profile, in 5% agarose gel, of AluI restriction fragments of polymerase chain reaction (PCR) products: lane 1 — molecular marker; lanes 2 to 5 —isolates with only 1 amplicon; lanes 6 to 9 — isolates with 2 amplicons.
The agarose gel analysis of the AluI RFLP patterns showed 49 different types. Types 9, 13, 23, 27, 28, 29, 31, 33, 42, and 48 were the most common and accounted for 39% of the isolates. The remaining 39 types accounted for only 1 isolate each (1.6% of the total).
Reproducibility of the AluI RFLP was observed with all repeatedly tested PCR products. The PCR products of 2 isolates were not digested by the AluI; therefore, this method had 97% typability. The discriminatory index was 0.99.
Discussion
Production of coagulase is an important phenotypic feature, used worldwide to identify S. aureus. Although the role of this protein in S. aureus infection is not completely understood, the variability of the 3′ end region of the coa gene is the basis for a typing method used for isolates from infected humans (6,7,17) and animals (8,18). By this method, we detected many different genotypes among the studied isolates, which suggests that S. aureus has considerable heterogeneity in the sampled region. Although many genotypes were detected, only a few predominated. In an earlier study, performed in the south of Brazil (18), 7 coa PCR types were observed, and 2 accounted for more than 50% of the isolates. Thus, in some Brazilian regions most cases of mastitis may be caused by S. aureus strains with the same coa genotype.
According to Su et al (12), the presence of few types may permit greater efficiency in measures to control S. aureus mastitis, since the important virulence factors could be specifically targetted. Furthermore, these researchers demonstrated that organisms with the predominant coagulase genotypes were more resistant to neutrophil activities than those with the rare genotypes, which indicated that specific features in the former may help them overcome host defence mechanisms.
Calculation of the discriminatory power of the typing method yielded high values: 0.92 for PCR amplification and 0.99 for RFLP analysis. According to Hunter and Gaston (16), an index greater than 0.90 can be interpreted with confidence and is thus desirable. Although the number of analyzed strains was low, the high number of AluI RFLP patterns and the absence of an epidemiologic relation between the isolates may have contributed to the high RFLP index.
The primer pair amplified more than 1 PCR product in 4 isolates, which suggests the presence of different allelic forms of the coa gene. Some S. aureus isolates express more than 1 immunologic form of the coagulase protein (6), but this appears to be a very uncommon finding: only Goh et al (6) and Schwarzkopf and Karch (7) had reported it.
With the PCR method, an amplification product was not observed for the DNA of another coagulase-positive species of Staphylococcus. According to Aarestrup et al (8), this could indicate that the S. aureus coa gene differs from the coa gene of other Staphylococcus species that have the ability to coagulate mammalian plasma. These authors suggested that specific coa gene primers could be used to identify and discriminate between coagulase-positive Staphylococcus species.
In conclusion, our results demonstrate that although mastitis in the studied region is caused by S. aureus strains that have many variants of the coa gene, only a few coa gene variants predominate. Further studies are needed to determine the common characteristics of the predominant strains. The information gathered could be used to develop control measures for mastitis caused by S. aureus.
Acknowledgments
The authors are grateful to the Veterinary School of the Federal University of Minas Gerais and the National Research Council of Brazil for financial support.
Footnotes
This project was reported as a portion of Dr. Rodrigues da Silva’s doctoral thesis Genotipagem e avaliação do potencial enterotoxigênico de amostras de Staphylococcus aureus isoladas de mastite caprina e bovina. Belo Horizonte, Minas Gerais, Brazil: Escola de Veterinária, Universidade Federal de Minas Gerais, 2004.
Dr. Rodrigues da Silva’s current address is Universidade Federal Rural de Pernambuco, Unidade Acadêmica de Garanhuns, Rua Ernesto Dourado, 82, Centro-55290-000, Garanhuns, Pernambuco, Brazil.
References
- 1.Philpot WN. Milk quality and mastitis control: past, present and future. PanAmerican Congress on Milk Quality and Mastitis Control. 2002:39–53. [Google Scholar]
- 2.Brito JRF, Brito MAVP. Programas de controle das mastites causadas por microrganismos contagiosos e do ambiente. Juiz de Fora, Brazil: Embrapa CNPGL, 1998.
- 3.Phonimdaeng P, O’Reilly M, Nowlan P, Bramley AJ, Foster TJ. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol. 1990;4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald JR, Reid SD, Ruotsalainen E, et al. Genome diversification in Staphylococcus aureus: molecular evolution of a highly variable chromosomal region encoding the staphylococcal exotoxin-like family of proteins. Infect Immun. 2003;71:2827–2838. doi: 10.1128/IAI.71.5.2827-2838.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zecconi A, Piccinini R. Teoria e prática de controle de mastite por Staphylococcus aureus. Napgama. 1999;5:4–11. [Google Scholar]
- 6.Goh S-H, Byrne SK, Zhang JL, Chow AW. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32:2407–2412. doi: 10.1128/jcm.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aarestrup FM, Dangler CA, Sordillo LM. Prevalence of coagulase gene polymorphism in Staphylococcus aureus isolates causing bovine mastitis. Can J Vet Res. 1995;59:124–128. [PMC free article] [PubMed] [Google Scholar]
- 9.Hookey J, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiou C-S, Wei H-L, Yang L-C. Comparison of pulsed-field gel electrophoresis and coagulase gene restriction profile analysis techniques in the molecular typing of Staphylococcus aureus. J Clin Microbiol. 2000;38:2186–2190. doi: 10.1128/jcm.38.6.2186-2190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlegelová J, Dendis M, Benedík J, Babák V, Rysánek D. Staphylococcus aureus isolates from dairy cows and humans on a farm differ in coagulase genotype. Vet Microbiol. 2003;92:327–334. doi: 10.1016/s0378-1135(02)00409-1. [DOI] [PubMed] [Google Scholar]
- 12.Su C, Herbelin C, Frieze N, Skardova O, Sordillo LM. Coagulase gene polymorphism of Staphylococcus aureus isolates from dairy cattle in different geographical areas. Epidemiol Infect. 1999;122:329–336. doi: 10.1017/s0950268899002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso HFT. Identificação de fatores de virulência e susceptibilidade a antimicrobianos de Staphylococcus aureus isolados de amostras de leite bovino em Minas Gerais [dissertação mestrado]. Belo Horizonte, Minas Gerais, Brazil: Escola de Veterinária, Universidade Federal de Minas Gerais, 1999.
- 14.Chen T-R, Hsiao M-H, Chiou C-S, Tsen H-Y. Development and use of PCR primers for the investigation of C1, C2 and C3 enterotoxin types of Staphylococcus aureus strains isolated from foodborne outbreaks. Int J Food Microbiol. 2001;71:63–70. doi: 10.1016/s0168-1605(01)00564-5. [DOI] [PubMed] [Google Scholar]
- 15.Rosec JP, Gigaud O. Staphylococcal enterotoxin genes of classical and new types detected by PCR in France. Int J Food Microbiol. 2002;77:61–70. doi: 10.1016/s0168-1605(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 16.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montesinos I, Salido E, Delgado T, Cuervo M, Sierra A. Epidemiologic genotyping of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis at a university hospital and comparison with antibiotyping and protein A and coagulase gene polymorphisms. J Clin Microbiol. 2002;40:2119–2125. doi: 10.1128/JCM.40.6.2119-2125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange C, Cardoso M, Senczek D, Schwarz S. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet Microbiol. 1999;67:127–141. doi: 10.1016/s0378-1135(99)00031-0. [DOI] [PubMed] [Google Scholar]


