Abstract
Introduction
Nosocomial pathogens are responsible for severe infections and are typically acquired in healthcare settings within days of patient admission. One of the primary contributors to the spread of healthcare-associated infections (HAIs) is the transfer of bacteria from inanimate surfaces to patients.
Methods
A cross-sectional study was done at Zewditu Memorial Hospital from June to November 2024. For this study, 204 inanimate surfaces from operating rooms and intensive care units were swabbed and cultured on MacConkey and blood agar. Bacteria were identified based on their colony morphology, gram staining, and conventional biochemical tests. Antimicrobial susceptibility testing was done using disk diffusion. Extended-spectrum beta-lactamase (ESBL) producing gram-negative bacteria were confirmed phenotypically using the double disc synergy and combination disc methods, while carbapenemase producers were identified using the Modified Carbapenem inactivation method. Methicillin-resistant Staphylococcus aureus (MRSA) was detected via the Cefoxitin Disk Diffusion Test. A P-value of less than 0.05 was considered statistically significant. Data were analyzed using SPSS version 20.
Results
Of the 204 swabbed samples, 77.45% (n = 158/204) showed bacterial growth, yielding a total of 171 bacterial isolates. Among these, Gram-positive bacteria comprised 71.3% (n = 122/171), while Gram-negative bacteria accounted for 28.7% (n = 49/171). The most prevalent isolates were Coagulase-negative Staphylococcus (CoNS), making up 46.1% (n = 79/171), followed by Bacillus spp. at 21.6% (n = 37/171). Out of the total isolates, 55 were identified as pathogenic bacteria based on their potential to cause disease and selected for antibiotic resistance testing. P. aeruginosa 36.7% (n = 18/49), Acinetobacter spp. 16.3% (n = 8/49), E. coli 14.2% (n = 7/49) and S. aureus 4.9% (n = 6/122) were the commonest pathogenic bacteria identified. Gram-negative bacteria showed high resistance to ampicillin (67.3%), amoxicillin and clavulanic acid (61.2%), ciprofloxacin (63.2%), sulfamethoxazole-trimethoprim (63.2%), cefepime (57.1%), and piperacillin-tazobactam (55.1%). Similarly gram-positive bacteria showed high resistance to azithromycin (100%), penicillin (100%), clindamycin (100%), and erythromycin (100%). Multidrug resistance was observed in 61.8% (n = 34/55) of the tested gram-negative and gram-positive isolates. The incidence of ESBL-producing and carbapenemase-producing bacteria among the suspected gram-negative bacteria was 26.5% (n=13/49) and 12.2% (n=6/49), respectively. The prevalence of MRSA was 50% (n=3/6).
Conclusion
The study identified a significant presence of multidrug-resistant bacteria in the hospital environment and on inanimate surfaces, emphasizing the urgent need for effective infection prevention and control measures.
Introduction
Nosocomial pathogens are microorganisms that cause hospital-acquired infections (HAIs), which are contracted in healthcare settings [1]. Healthcare-associated infections pose significant risks to both patient and public health, leading to higher morbidity rates, prolonged hospital stays, and increased mortality [2]. Factors such as inadequate infection prevention and hygiene practices, improper equipment sterilization, and the emergence of resistant bacterial strains contribute to the spread of HAIs [3]. Contaminants from inanimate objects and surfaces in intensive care units (ICUs) and operating rooms (ORs) exacerbate patient mortality and morbidity, as patients in these areas are particularly vulnerable to infections [2].
The ability of bacteria to spread from inanimate objects to humans depends on several factors, including the type of microorganism, the size of the inoculum, the environment’s temperature and humidity, the porosity of the object, the presence of organic matter, the microorganism’s ability to form biofilms, and the effectiveness of infection control measures [4]. Both gram-positive and gram-negative bacteria have been found on inanimate surfaces. These bacteria can survive for several months on dry surfaces, but their survival rate increases in humid and cooler environments [2]. Bacteria such as Klebsiella pneumoniae, Escherichia coli, Staphylococcus aureus, Proteus spp., Pseudomonas aeruginosa, and Enterococcus spp. are major contributors to healthcare-acquired infections and are widely recognized as potentially deadly pathogens in hospital settings [5,6].
Gram-negative bacteria (GNB) pose a significant global health threat due to their ability to produce beta-lactamase and carbapenemase, which contribute to antimicrobial resistance [7]. Bacteria producing extended-spectrum beta-lactamase (ESBL) enzymes are responsible for over 19% of nosocomial infections [8]. These enzymes can hydrolyze penicillin, monobactams, and third-generation cephalosporins, while carbapenemase and metallo-beta-lactamase enzymes can break down carbapenem medications [9]. Common ESBL- and carbapenemase-producing bacteria in nosocomial infections include Escherichia coli, Klebsiella pneumoniae, P. aeruginosa, and Enterobacter cloacae [10]. The emergence of novel beta-lactamases in gram-negative bacteria, capable of breaking down cephalosporins and carbapenems, is an alarming development. Infections caused by these organisms increase treatment costs, hospital stays, morbidity, and mortality rates [11].
Penicillin-resistant pneumococci and methicillin- and vancomycin-resistant Staphylococcus aureus are the most common drug-resistant gram-positive bacteria found in healthcare settings [5]. Another bacterium associated with healthcare-associated infections and environmental contamination is Acinetobacter baumannii [12].
The most frequent hospital-acquired infections from ICUs and ORs include bloodstream, urinary tract, surgical site, and respiratory infections [3]. The risk of these infections is heightened by inadequate cleanliness, lack of routine infection prevention measures, insufficient control procedures, and unsafe medical environments [13]. Particularly in developing countries, there is a lack of information on the extent and types of contamination, as well as the microbiological profiles of commonly used medical instruments and inanimate surfaces in hospitals. Therefore, ongoing research in this area is necessary to address and mitigate the problem.
Methods and materials
From June to November 2024, a hospital-based cross-sectional study was conducted at Zewditu Memorial Hospital, located in the Kirkos sub-city, Addis Ababa, Ethiopia. The hospital has a capacity of 231 beds, 11 wards, and 3 emergency wards and it provides medical care to approximately 115,102 patients annually. Swab specimens were collected from adult and neonatal intensive care units (ICUs), the gynecology and pediatric neurology (CDC operation rooms), and the cesarean section units (delivery units). A total of 204 swabs were collected from selected equipment and environments of those operating rooms and intensive care units. All swab samples were collected during the morning after cleaning had been performed and before staff began their daily activities. Sterile swabs, moistened with 85% normal saline, were used to sample high-touch surfaces after careful observation. Swabbing was performed in parallel spaced stripes with slight rotation, followed by perpendicular stripes, in accordance with ISO/DIS 14698−1 guidelines [14]. Each sample was then transferred into a leak-proof container containing tryptone soya broth as a transport medium.
Bacterial culture, identification, and interpretation of culture results
All swab samples were inoculated onto MacConkey agar and Blood agar and incubated aerobically at 37°C for 18–24 hours. Each culture-positive bacteria were identified based on their colony morphology, Gram staining, and biochemical characteristics. Biochemical tests used to identify Gram-negative bacteria were the indole, triple sugar iron agar, urea utilization, citrate utilization, mannitol, lysine iron agar decarboxylation, and oxidase. On the other hand, Gram-positive bacteria were identified using Gram staining, catalase, and coagulase tests, with results interpreted according to laboratory standard operating procedures.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted using disc diffusion following the Clinical and Laboratory Standards Institute (CLSI) guidelines. After placing selected antibiotics on a Muller Hinton Agar (MHA) plate inoculated with the test bacteria, the plate was incubated for 16–18 hours. All Gram-negative bacteria were tested against amikacin, amoxicillin-clavulanic acid (AMC), gentamicin, ampicillin, cefotaxime, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, cefepime, ertapenem, imipenem/meropenem, trimethoprim-sulfamethoxazole (SXT), and piperacillin-tazobactam. Gram-positive bacteria were tested against gentamicin, ciprofloxacin, cefoxitin, trimethoprim-sulfamethoxazole (SXT), penicillin, azithromycin, clindamycin, erythromycin, and tetracycline. Zones of inhibition were measured and interpreted according to the CLSI 34th edition (2024) [15].
Extended-spectrum β-lactamase (ESBL) detection and confirmation
Gram-negative bacteria that were suspected for Extended-spectrum β-lactamase (ESBL) production were confirmed using the double-disk synergy test and combination disc test. Using the double-disk synergy test, a disc containing amoxicillin-clavulanate (20 μg/10 μg) (augmentin) and a 30-μg disc containing each third-generation cephalosporin test antibiotic were compared for synergy. The discs were positioned 20 mm apart from one another on an MHA that had been swabbed with the test isolate. A clear enlargement of the margin of the cephalosporin inhibition area near the augmentin disc was regarded as a positive ESBL test result. While using the combination disk test, ceftazidime (30 µg) disks were used alone and in conjunction with clavulanic acid (30 µg/10 µg) to confirm the existence of ESBLs. A zone diameter ≥5 mm increase with ceftazidime/clavulanate disks than in ceftazidime disks was found to produce ESBL [15].
Carbapenemase detection and confirmation
Suspected carbapenemase-producing gram-negative bacteria were confirmed phenotypically using the modified carbapenem inactivation method. Briefly, the method consists of suspending a 1 μl loop of test organisms in 2 mL of trypticase soy broth. A 10 μg meropenem disc was added to the prepared suspension and incubated for 4 hours at 35 °C. A suspension of E. coli ATCC 25922 calibrated to 0.5 McFarland was prepared and inoculated on a Mueller-Hinton agar plate. After 4 hours, the meropenem disc was removed and inserted on an MHA plate previously inoculated with the indicator strain E. coli ATCC 25922. The plates were then incubated for 18–24 h at 35 °C, and the plates were analyzed based on the Clinical and Laboratory Standards Institute’s recommendations [15].
Methicillin-resistant Staphylococcus aureus (MRSA) screening and confirmation
Methicillin-resistant Staphylococcus aureus (MRSA) was identified using the cefoxitin disk diffusion test. Isolate was initially grown on a Mueller-Hinton agar plate using a 30-µg cefoxitin disk and a 0.5 McFarland standard suspension. Every plate was incubated aerobically for the entire night at 37°C. The test was performed according to CLSI guidelines [15].
Data quality assurance
The reliability of the research results was ensured through rigorous quality assurance measures throughout data collection and laboratory procedures. Strict adherence was maintained while collecting, labeling, handling, and transporting biological samples as well as preparing media according to manufacturer guidelines and laboratory Standard Operating Procedures (SOPs). The media’s quality was verified against CLSI standards, ensuring compliance with expiration dates. To minimize contamination, aseptic techniques were employed during sample collection and inoculation onto culture media. Additionally, all pre-analytical procedures adhered to standard operating guidelines. The sterility and performance of the culture media were assessed by incubating them overnight at 37°C. The performance of MacConkey agar and blood agar plates was evaluated using control strains, including Escherichia coli (ATCC 25922), Proteus mirabilis (ATCC 35659), Staphylococcus aureus (ATCC 25923), and Streptococcus pneumoniae (patient strain). Antibiotic efficacy testing was performed using international control bacterial strains such as E. coli (ATCC 25922), S. aureus (ATCC 25923), and Pseudomonas aeruginosa (ATCC 27853). Additionally, biochemical test media were inoculated with known positive and negative bacterial controls to verify accuracy. All isolated bacteria were stored in accordance with the laboratory’s standard operating procedures.
Data analysis and interpretation
Data was analyzed using SPSS version 20.0 to show the incidence of bacterial pathogens on inanimate surfaces as well as their antibiotic resistance trend. The findings were evaluated via descriptive statistics, and the findings were described using tables, graphs, and text.
Ethics approval
The study was approved by the department of research and ethics review committee of the medical Laboratory Sciences, College of Health Sciences; Addis Ababa University (Ref. No. MLS/165/24) and City Government of Addis Ababa health bureau (Ref. No. A/A/H/10432/227). Letter of permission was obtained from Zewditu Memorial Hospital to access wards and collect data and Tikur Anbessa Specialized Teaching Hospital to use the necessary materials and to conduct laboratory analysis.
Results
Prevalence of bacterial isolates from inanimate surfaces and medical equipment at ICUs and ORs
In this study, a total of 204 swab samples were collected from various inanimate objects in the operating rooms (ORs) (n=113) and intensive care units (ICUs) (n=91) of Zewditu Memorial Hospital. Among all cultured samples, the overall bacterial prevalence was 77.5% (n=158/204). A total of 171 bacterial isolates were identified from the collected swabs. Of the 171 bacterial isolates, 71.3% (n=122/171) were Gram-positive bacteria, while Gram-negative bacteria accounted for 28.7% (n=49/171) (Fig 1).
Fig 1. Gram negative and gram positive bacteria isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
N-number of bacteria; CoNs-coagulase negative Staphylococcus aureus.
Gram-positive bacteria identified in this study included Staphylococcus aureus 4.9% (n=6/122), Coagulase-negative Staphylococci (CoNS) 64.7% (n=79/122), and Bacillus spp. 30.3% (n=37/122). Among the Gram-negative bacteria, the most common isolates were Pseudomonas aeruginosa 36.7% (n=18/49), Acinetobacter spp. 16.3% (n=8/49), and Escherichia coli 14.2% (n=7/49). Overall, CoNS was the most frequently isolated bacterium, accounting for 46.1% (n=79/171), followed by Bacillus spp. at 21.6% (n=37/171) and Pseudomonas aeruginosa at 10.5% (n=18/171).
Distribution of bacterial isolates between intensive care units and operation rooms
Of all 171 bacteria, 50.8% (n=87/171) were identified in intensive care units (ICUs). Both Gram-positive and Gram-negative bacteria showed variations across different wards, with operation rooms (ORs) reporting 71.4% (n=60/84) Gram-positive and 28.5% (n=24/84) Gram-negative bacteria, while ICUs had 71.2% (n=62/87) Gram-positive and 28.7% (n=25/87) Gram-negative bacteria.
In ICUs, Gram-positive bacteria were predominantly responsible for contamination, comprising 71.2% (n=62/87). Coagulase-negative Staphylococci (CoNS) were the most common bacteria identified at ICU, accounting for 52.8% (n=46/87), followed by Bacillus spp. at 12.6% (n=11/87). The majority of bacterial isolates in ICUs were from the neonatal intensive care unit (NICU), which accounted for 51.7% (n=45/87) of the cases. Within the NICU, the leading pathogens were CoNS from Gram-positive bacteria and Pseudomonas aeruginosa from Gram-negative bacteria, each with isolation rates of 51.1% (n=23/45) and 13.3% (n=6/45), respectively. In the operation rooms (ORs), contamination was primarily caused by Gram-positive bacteria, which made up 71.4% (n=60/84). The most common pathogens in the ORs were CoNS at 39.2% (n=33/84) followed by Bacillus spp. at 30.9% (n=26/84) (Table 1).
Table 1. Distribution of bacteria among wards isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
| Bacterial isolates | Adult ICUs n(%) | Neonatal ICUs n(%) | Major OR n(%) | CS OR n(%) | Gyn & Pedi OR n(%) | Total n(%) |
|---|---|---|---|---|---|---|
| CoNS | 23(29.11) | 23(29.11) | 18(22.7) | 12(15.18) | 3(3.79) | 79(46.1) |
| Bacillus spp. | 5(13.5) | 6(16.2) | 15(40.5) | 9(24.3) | 2(5.4) | 37(21.6) |
| S. aureus | 3(50) | 2(33.33) | 0(0) | 1(16.66) | 0(0) | 6(3.5) |
| P. aeruginosa | 3(16.66) | 5(27.77) | 0(0) | 0(0) | 10(55.55) | 18(10.5) |
| Klebsiella spp. | 2(11.11) | 4(22.2) | 1(12.5) | 0(0) | 1(12.5) | 8(4.67) |
| E. coli | 2(28.5) | 1(14.28) | 1(14.28) | 1(14.28) | 3(42.8) | 7(4.09) |
| Acinetobacter spp. | 2(25) | 0(0) | 2(25) | 4(50) | 0(0) | 8(4.67) |
| Citrobacter spp. | 0(0) | 1(50) | 1(50) | 0(0) | 0(0) | 2(1.16) |
| E. cloacae | 1(25) | 2(50) | 0(0) | 0(0) | 1(25) | 4(2.33) |
| Shigella spp. | 1(50) | 1(50) | 0(0) | 0(0) | 0(0) | 2(1.16) |
| Total | 42(24.5) | 45(26.3) | 38(22.2) | 27(15.7) | 19(11.1) | 171(100) |
n- number of tested strains; %- percentage; ICU-intensive care unit; CoNS- Coagulase-negative Staphylococci; Gyn & Pedi OR- (gyneacology & pediatrics neurology operation room); CSOR- cesarean section operation room.
Distribution of bacterial pathogens over different surfaces
The highest number of bacterial-contaminated samples were collected from environmental surfaces (14%), followed by tables (12.8%) and patient monitors (9.94%). Environmental surfaces were predominantly contaminated with Bacillus spp. 37.5% (n = 9/24), followed by Coagulase-negative Staphylococci (CoNS) (29.1%, n = 7/24). Tables used by healthcare workers in operating rooms and intensive care units were primarily contaminated with CoNS 45.4% (n = 10/22). Patient monitors were mainly colonized by CoNS 47% (n = 8/17), Bacillus spp. 23.5% (n = 4/17), and Acinetobacter spp. 11.7% (n = 2/17) (Table 2).
Table 2. Distribution of bacterial pathogens over different surfaces isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
| Inanimate objects | CoNS n(%) | Bacillus spp. n(%) | S. aureus n(%) | P. aeruginosa n(%) | Klebsiella spp. n(%) | E. coli n(%) | Acinetobacter spp. n(%) | Citrobacter spp. n(%) | E. cloacae n(%) | Shigella spp. n(%) | Total n(%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anastasia machine (7) | 3(3.79) | 1(2.7) | 0(0) | 1(5.5) | 0(0) | 1(14.28) | 1(1.25) | 0(0) | 0(0) | 0(0) | 7(4.09) |
| Bed (11) | 8(10.1) | 1(2.7) | 0(0) | 0(0) | 2(25) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 11(6.4) |
| Linens (15) | 6(7.5) | 4(10.8) | 2(33.3) | 1(5.5) | 2(25) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 15(8.77) |
| Monitor (17) | 8(10.1) | 4(10.8) | 0(0) | 1(5.5) | 0(0) | 1(14.2) | 2(25) | 1(50) | 0(0) | 0(0) | 17(9.94) |
| Suction Machine (8) | 3(3.7) | 2(5.4) | 0(0) | 0(0) | 0(0) | 1(14.2) | 0(0) | 0(0) | 1(25) | 1(50) | 8(4.67) |
| OR Table (6) | 3(3.7) | 3(8.1) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 6(3.5) |
| Tables (22) | 10(12.6) | 4(10.8) | 0(0) | 5(27.77) | 0(0) | 3(42.8) | 0(0) | 0(0) | 0(0) | 0(0) | 22(12.8) |
| Work Station (6) | 3(3.79) | 0(0) | 0(0) | 2(11.11) | 1(12.5) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 6(3.5) |
| Oxygen Cylinder (12) | 7(8.8) | 3(8.1) | 0(0) | 1(5.5) | 0(0) | 1(14.28) | 0(0) | 0(0) | 0(0) | 0(0) | 12(7.01) |
| Bed Trails (10) | 6(7.59) | 1(2.7) | 1(16.66) | 0(0) | 1(5.55) | 0(0) | 1(12.5) | 0(0) | 0(0) | 0(0) | 10(5.8) |
| OR light (11) | 5(6.3) | 3(8.1) | 0(0) | 2(11.1) | 0(0) | 0(0) | 1(12.5) | 0(0) | 0(0) | 0(0) | 11(6.4) |
| IV Stand (12) | 6(7.5) | 1(2.7) | 1(16.66) | 3(16.66) | 0(0) | 0(0) | 1(12.5) | 0(0) | 0(0) | 0(0) | 12(7.01) |
| Environmental surfaces (24) | 7(8.8) | 9(24.3) | 2(33.33) | 2(11.11) | 1(5.55) | 0(0) | 0(0) | 1(50) | 1(25) | 1(50) | 24(14.03) |
| Others (10) | 4(5.06) | 1(2.7) | 0(0) | 0(0) | 1(12.5) | 0(0) | 2(25) | 0(0) | 2(50) | 0(0) | 10(5.8) |
| Total | 79(46.19) | 37(21.6) | 6(3.5) | 18(10.5) | 8(4.67) | 7(4.09) | 8(4.67) | 2(1.16) | 4(2.3) | 2(1.16) | 171(100) |
Notes: Others- (ventilators, phototherapy machines, Electro-surgical unit generators, pulse oximeter); Environmental surfaces- (floors, walls, door knobs); n- number of tested strains; %-percentage; CoNS- Coagulase-negative Staphylococci.
Antimicrobial resistance patterns of gram-negative bacteria
The majority of gram-negative bacteria exhibited significantly high resistance to most of the tested antibiotics. For instance, resistance rates were 67.3% for ampicillin, 61.2% for amoxicillin and clavulanic acid, 63.2% for ciprofloxacin, 63.2% for sulfamethoxazole-trimethoprim, 57.1% for cefepime, and 55.1% for piperacillin-tazobactam. Similarly, considerable resistance was observed for amikacin (53.06%), chloramphenicol (53.06%), gentamicin (46.9%), ceftriaxone (34.6%), and ertapenem (32.6%). Lower resistance levels were noted for ceftriaxone (30.6%), cefotaxime (16.3%), and meropenem (16.3%). The highest resistance levels in Acinetobacter spp. were seen against penicillin, cephalosporins, carbapenems, and monobactams, with resistance rates of 100% for ampicillin, ceftazidime, and amoxicillin and clavulanic acid, 97.4% for ceftriaxone, and 92.3% for cefotaxime. Acinetobacter spp. showed a relatively low resistance to amikacin (35.9%) (Table 3).
Table 3. AMR pattern for gram negative bacteria isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
| Isolated bacteria | AST pattern | AMI n(%) | AMC n(%) | GEN n(%) | AMP n(%) | CTX n(%) | CTZ n(%) | CTR n(%) | CMP n(%) | CIP n(%) | CPM n(%) | ERT n(%) | MEM n(%) | SXT n(%) | PPT n(%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa (18) | R | 17 (94.4) | 11(61.1) | 9(50) | 10(55.5) | 2(11.1) | 5(27.7) | 3(16.6) | 6(33.3) | 6(33.3) | 9(50) | 7(38.8) | 0(0) | 12(66.6) | 9(50) |
| S | 1(5.5) | 7(38.88) | 9(50) | 8(44.4) | 16(88.8) | 13(72.2) | 15(83.3) | 12(66.6) | 12(66.6) | 9(50) | 11(61.1) | 18(100) | 6(33.3) | 9(50) | |
| E. coli (7) | R | 3(42.8) | 6(85.7) | 3(42.8) | 6(85.7) | 2(28.5) | 3(42.8) | 2(28.5) | 4(57.1) | 6(85.7) | 3(42.8) | 2(28.5) | 0(0) | 4(57.1) | 3(42.8) |
| S | 4(57.1) | 1(14.28) | 4(57.1) | 1(14.28) | 5(71.4) | 4(57.1) | 5(71.4) | 3(42.8) | 1(14.28) | 4(57.1) | 5(71.4) | 7(100) | 3(42.8) | 4(57.1) | |
| E. cloacae (4) | R | 2(50) | 4(100) | 4(100) | 4(100) | 0(0) | 2(50) | 2(50) | 3(75) | 4(100) | 4(100) | 1(25) | 0(0) | 3(75) | 2(50) |
| S | 2(50) | 0(0) | 0(0) | 0(0) | 4(100) | 2(50) | 2(50) | 1(25) | 0(0) | 0(0) | 3(75) | 4(100) | 1(25) | 2(50) | |
| Acinetobacter spp. (8) | R | 1(12.5) | 3(37.5) | 2(25) | 6(75) | 3(37.5) | 4(50) | 4(50) | 7(87.5) | 3(37.5) | 4(50) | 2(25) | 2(25) | 4(50) | 5(62.5) |
| S | 7(87.5) | 5(62.5) | 6(75) | 2(25) | 5(62.5) | 4(50) | 4(50) | 1(12.5) | 5(62.5) | 4(50) | 6(75) | 6(75) | 4(50) | 3(37.5) | |
| Citrobacter spp. (2) | R | 1(50) | 1(50) | 1(50) | 0(0) | 0(0) | 1(50) | 1(50) | 1(50) | 2(100) | 1(50) | 0(0) | 0(0) | 1(50) | 1(50) |
| S | 1(50) | 1(50) | 1(50) | 2(100) | 2(100) | 1(50) | 1(50) | 1(50) | 0(0) | 1(50) | 2(100) | 2(100) | 1(50) | 1(50) | |
| K. pneumoniae (3) | R | 0(0) | 1(33.33) | 0(0) | 1(33.33) | 0(0) | 0(0) | 0(0) | 1(33.330) | 3(100) | 0(0) | 0(0) | 2(66.66) | 0(0) | 1(33.33) |
| S | 3(100) | 2(66.66) | 3(100) | 2(66.66) | 3(100) | 3(100) | 3(100) | 2(66.66) | 0(0) | 3(100) | 3(100) | 1(33.3) | 3(100) | 2(66.66) | |
| K. oxytoca (3) | R | 1(33.33) | 1(33.33) | 1(33.33) | 3(100) | 1(33.33) | 1(33.33) | 2(66.66) | 2(66.66) | 3(100) | 3(100) | 1(33.33) | 1(33.33) | 3(100) | 2(66.66) |
| S | 2(66.66) | 2(66.66) | 2(66.66) | 0(0) | 2(66.66) | 2(66.66) | 1(33.33) | 1(33.33) | 0(0) | 0(0) | 2(66.66) | 2(66.66) | 0(0) | 1(33.33) | |
| K. ozaenae (2) | R | 1(50) | 2(100) | 2(100) | 2(100) | 0(0) | 1(50) | 1(50) | 1(50) | 2(100) | 2(100) | 1(50) | 1(50) | 2(100) | 2(100) |
| S | 1(50) | 0(0) | 0(0) | 0(0) | 2(100) | 1(50) | 1(50) | 1(50) | 0(0) | 0(0) | 1(50) | 1(50) | 0(0) | 0(0) | |
| Shigella spp (2) | R | 0(0) | 1(50) | 1(50) | 1(50) | 0(0) | 0(0) | 0(0) | 1(50) | 2(100) | 2(100) | 2(100) | 0(0) | 2(100) | 2(100) |
| S | 2(100) | 1(50) | 1(50) | 1(50) | 2(100) | 2(100) | 2(100) | 1(50) | 0(0) | 0(0) | 0(0) | 2(100) | 0(0) | 0(0) |
n- number; R-resistant; S-sensitive; %- percentage; AST- antimicrobial susceptibility test; AMI- amikacin; AMC- amoxicillin-clavulanic acid (AMC); GEN- gentamicin; AMP- ampicillin; CTX- cefotaxime; CTZ- ceftazidime; CTR- ceftriaxone; CMP- chloramphenicol; CIP- ciprofloxacin; CPM- cefepime; ERT- ertapenem; MEM- meropenem; SXT- trimethoprim-sulfamethoxazole (SXT); PPT- piperacillin-tazobactam.
The antibiotic resistance rate among gram-positive bacteria was notably high, with 100% resistance to penicillin, azithromycin, clindamycin, and erythromycin. A low resistance level was observed for gentamicin (33.3%) and sulfamethoxazole-trimethoprim (33.3%). Using cefoxitin as a surrogate marker, 50% (n=3/6) of S. aureus isolates were identified as MRSA (Table 4).
Table 4. AMR pattern for gram-positive bacteria isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
| Isolated bacteria | AST Pattern | GEN n(%) | CIP n(%) | OXA n(%) | SXT n(%) | PEN n(%) | AZI n(%) | CLI n(%) | ERY n(%) | TET n(%) |
|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | R | 2(33.33) | 3(50) | 3(50) | 2(33.33) | 6(100) | 6(100) | 6(100) | 6(100) | 1(16.66) |
| S | 4(66.66) | 3(50) | 3(50) | 4(66.66) | 0(0) | 0(0) | 0(0) | 0(0) | 5(83.33) |
n- Number of tested strains; R-resistant; S-sensitive; %- percentage; GEN- gentamicin; CIP-ciprofloxacin; OXA-oxacillin; SXT- sulfamethoxazole + trimethoprim; PEN- penicillin; AZI- azithromycin; CLI- clindamcin; ERY- erythromycin; TET- tetracycline.
MDR level of gram-negative and gram-positive bacteria
Among the 55 bacterial isolates selected for antimicrobial susceptibility testing, multidrug resistance (defined as resistance to at least one antibiotic from three or more different classes) was observed in 61.8% (n=34/55) of the isolates (Table 5).
Table 5. Multi-drug resistance pattern of bacteria isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
| Gram negative bacteria | Isolated bacteria | R0 n(%) |
R1 n(%) |
R2 n(%) |
R3 n(%) |
R4 n(%) |
R5 n(%) |
MDR | |
|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa (18) | 5(27.77) | 1(5.55) | 1(5.55) | 4(22.22) | 0(0) | 6(50) | 10(55.55) | ||
| E. coli (7) | 1(14.2) | 0(0) | 2(28.5) | 1(14.2) | 0(0) | 3(42.8) | 4(57.14) | ||
| E. cloacae (4) | 0(0) | 0(0) | 0(0) | 1(25) | 0(0) | 3(75) | 4(100) | ||
| Acinetobacter spp. (8) | 1(12.5) | 1(12.5) | 2(25) | 1(12.5) | 1(12.5) | 2(25) | 4(50) | ||
| Citrobacter spp. (2) | 0(0) | 1(33.33) | 0(0) | 0(0) | 0(0) | 1(33.33) | 1(33.33) | ||
| K. pneumoniae (3) | 0 (0) | 1(33.33) | 1 (33.33) | 1(33.33) | 0(0) | 0(0) | 1(33.33) | ||
| K. oxytoca (3) | 0 (0) | 0 (0) | 0 (0) | 1(33.33) | 1(33.33) | 1(33.33) | 3(100) | ||
| K. ozanae (2) | 0(0) | 0(0) | 0(0) | 0(0) | 1(33.33) | 1(33.33) | 2(66.66) | ||
| Shigella spp. (2) | 0(0) | 0(0) | 0(0) | 1(33.33) | 0(0) | 1(33.33) | 2(66.66) | ||
| Gram positive bacteria | S. aureus (6) | 0(0) | 0(0) | 3(50) | 1(16.66) | 0(0) | 1(16.66) | 1(16.66) | 3(50) |
n- Number of tested strains; %.-Percentage; R0- sensitive to all classes of antibiotics; R1-resistant to one class of antibiotics; R2- resistant to two classes of antibiotics; R3- resistant to three classes of antibiotics; R4- resistant to four classes of antibiotics; R5- resistant to five classes of antibiotics; R6- resistant to six classes of antibiotics; MDR- multidrug resistant.
Extended-spectrum β-lactamase (ESBL) producing bacteria
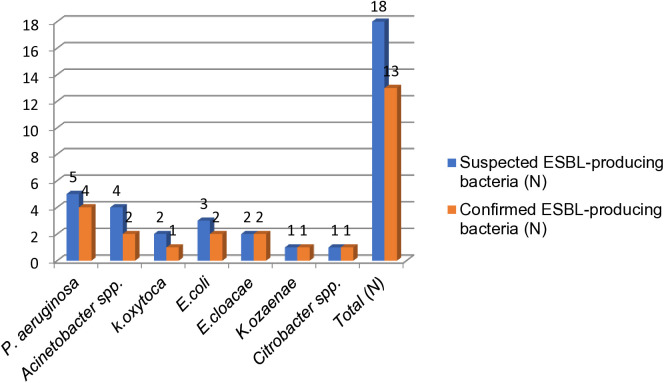
A total of 18 isolates were suspected to be ESBL producers from 49 Gram-negative bacteria identified. These included P. aeruginosa 27.7% (n = 5/18), Acinetobacter spp. 22.2% (n = 4/18), E. coli 16.6% (n = 3/18), E. cloacae 11.1% (n = 2/18), K. oxytoca 11.1% (n = 2/18), K. ozaenae 5.5% (n = 1/18) and Citrobacter spp. 5.5% (n = 1/18). Using the combination disc test, the overall prevalence of ESBL-producing bacteria was 26.5% (n = 13/49). The breakdown of ESBL positivity by species was as follows: K. ozaenae 100% (n = 1/1), E. coli 66.6% (n = 2/3), K. oxytoca 50% (n = 1/2), Acinetobacter spp. 50% (n = 2/4), E. cloacae 50% (n = 1/2), Citrobacter spp. 100% (n = 1/1), and P. aeruginosa 80% (n = 4/5) (Fig 2).
Fig 2. ESBL-producing bacteria confirmed with combination disk method isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
7N-number; ESBL-extended-spectrum beta-lactamases.
The double disk synergy method, another phenotypic confirmatory technique, was used to test all 18 isolates for ESBL production. According to this method, 24.4% (n=12/49) of ESBL cases were confirmed. Among the 13 isolates that tested positive using the combination disc test (CDT), 7.69% (n=1/13) were negative, while 92.3% (n=12/13) were positive. The following bacteria were confirmed as ESBL-positive using the double disk method: E. coli 33.33% (n=1/3), K. ozaenae 100% (n=1/1), K. oxytoca 100% (n=1/1), Acinetobacter spp. 50% (n=2/4), P. aeruginosa 80% (n=4/5), E. cloacae 100% (n=2/2), and Citrobacter spp. 100% (n=1/1). Among the bacteria initially suspected of producing ESBL, only 33.3% (n=6/18) were confirmed as ESBL-negative using this approach (Fig 3).
Fig 3. ESBL-producing bacteria confirmed with Double disk synergy method isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
8N-number; ESBL-extended-spectrum beta-lactamases.
Carbapenemase-producing bacteria
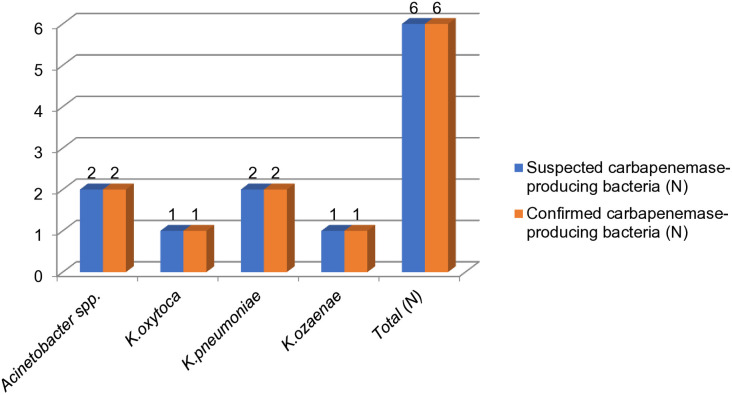
Out of the 49 bacterial isolates, 18.2% (n=6/49) exhibited intermediate or resistant patterns to imipenem and/or meropenem, suggesting possible carbapenemase production. These isolates were further tested for carbapenemase production and confirmed phenotypically using the Modified Carbapenemase Inactivation Method (MCIM). The suspected carbapenem-resistant isolates included K. pneumoniae 33.3% (n=2/6), Acinetobacter spp. 33.3% (n=2/6), K. ozaenae 16.6% (n=1/6) and K. oxytoca 16.6% (n=1/6). Of the 6 isolates that showed resistance or intermediate zones for meropenem, 100% (n=6/6) were positive for carbapenemase production using the MCIM, resulting in an overall prevalence of 12.22% (n=6/49) for carbapenemase-producing bacteria among the total gram-negative isolates. The predominant carbapenemase-producing organisms were Klebsiella species, accounting for 8.16% (n=4/49), with K. pneumoniae 66.6% (n=2/3), K. ozaenae 50% (n=1/2), and K. oxytoca 33.3% (n=1/3) making up the majority. The second most common carbapenemase producer was Acinetobacter spp., which accounted for 25% (n=2/8) (Fig 4).
Fig 4. Carbapenemase-producing bacteria confirmed with MCIM isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
N-number.
Methicillin-resistant Staphylococcus aureus (MRSA)
Out of the 6 S. aureus isolates, 50% (n=3/6) were identified as MRSA using cefoxitin as a surrogate marker. These confirmed MRSA cases were distributed as follows: 33.33% (n=1/3) in the adult intensive care unit (AICU), 33.33% (n=1/3) in the neonatal intensive care unit (NICU), and 33.33% (n=1/3) in the cesarean section operating room (CS OR) (Fig 5).
Fig 5. Methicillin Resistance S. aureus (MRSA) isolated from environmental surfaces and medical equipment at Zewditu Memorial Hospital.
N-number.
Discussion
A previous study has employed molecular typing to examine the clonal relationships between clinical strains and inanimate surfaces, revealing that hospital environments, healthcare workers’ (HCWs’) hands, and clinical specimens from admitted patients are sources of nosocomial infections [16]. Furthermore, if terminal cleaning is ineffective, patients admitted to rooms previously occupied by individuals infected or colonized with multidrug-resistant (MDR) strains may be up to three times more likely to contract healthcare-associated infections (HCAIs) from contaminated environmental surfaces or equipment [17].
In this study, 158 out of 204 environmental specimens (77.4%) tested positive for bacterial growth and overall, 171 bacterial isolates were found. This finding was consistent with results from similar studies, including a study from Tikur Anbessa Hospital (86%) [18], a nearby hospital to the study site, and another from Mekelle in Northern Ethiopia (88.5%) [19]. A similar result was reported in a study conducted in Nigeria (70.3%) [20]. Other studies from Cameroon (50.4%) [21], Nigeria (46.3.4%) [22], and India (43.3%) [23] reported much lower bacterial contamination rates. These differences could be attributed to variations in ventilation systems, hand hygiene practices, and sterilization and disinfection methods.
The higher bacterial contamination levels observed in this study may be due to ineffective disinfectants, poor adherence to basic precautions like hand washing and contact precautions, and the movement of microorganisms through airflow. Hospitals with inadequate waste management, insufficient awareness of contamination levels, ineffective disinfectants, and a reluctance to invest in contamination control measures (such as proper ventilation systems) are all strongly linked to this situation [24].
These findings indicate substantial contamination of inanimate objects by both Gram-positive (71.3%) and Gram-negative (28.7%) bacteria. Similar results were reported in other studies, such as one conducted at Tikur Anbessa Hospital in Addis Ababa, which found a distribution of 56.3% Gram-positive and 43.7% Gram-negative bacteria [18], as well as studies from Korea (73.2% vs 26.8%) [25], India (56.99% vs 43.01%) [26], and Nigeria (52.2% vs 47.8%) [27]. The prevalence of Gram-positive bacteria could be due to their ability to survive in abiotic hospital environments for several days to months, as they lack the lipid-dominant, desiccation-prone outer membrane found in Gram-negative bacteria [18]. However, the growth and resistance patterns of Gram-positive bacteria are diverse [28]. In contrast, studies conducted in India [23], Indonesia [29], and Morocco [30] found Gram-negative bacteria to be the predominant environmental isolates. These discrepancies may result from differences in sampling times, methods, and culture techniques, as well as variations in the items sampled.
In this study, Coagulase-negative Staphylococci (CoNS) were the most frequently isolated bacteria (46.1%), followed by Bacillus spp. (21.6%) and P. aeruginosa (10.5%) across the wards, which is consistent with findings from various studies worldwide [25, 26]. Although CoNS are part of the normal human skin flora, they can cause clinically significant infections, such as bloodstream and tissue infections [31]. Risk factors for CoNS infections include immune impairment and the use of prosthetic materials, such as intravascular catheters [32].
Among the many surfaces and inanimate objects analyzed in this study, the highest levels of bacterial contamination were found on tables, environmental surfaces, monitors, and bed linens, which aligns with prior research conducted in Ethiopia and other countries [18,33,34,35]. Environmental surfaces and tables were primarily contaminated by CoNS (29.1% and 45.45%), Bacillus spp. (37.5% and 18.18%), and P. aeruginosa (8.33% and 22.7%), respectively. Similar findings were observed for linens and bed samples in studies from Mizan Tepi, Ethiopia [33]. Cross-contamination from patient flora, healthcare workers’ hands, or contaminated patient and healthcare worker footwear may be the source of this contamination.
Klebsiella spp. were predominantly found in both adult and neonatal intensive care units (ICUs), where they are associated with ventilator-associated pneumonia and bloodstream infections. A similar result was documented in a study conducted in Iran [36]. The majority of Gram-negative bacteria (GNB) in this study showed high resistance to most tested antibiotics, including ampicillin (97.5%), ceftriaxone (91.3%), ceftazidime (91.3%), amoxicillin-clavulanic acid (85%), cefotaxime (83.8%), and cefoxitin (76.3%). This high resistance is likely due to the distinctive structure of Gram-negative bacteria. Those results align with similar resistance levels observed in studies from Ethiopia and other African nations, including Tikur Anbessa [18] and Sudan [37]. The high resistance to β-lactam antibiotics is likely caused by selective pressure exerted by these antibiotics [38]. Since these antibiotics are commonly prescribed, their overuse presents significant challenges [18]. On the other hand, resistance to non-β-lactam antibiotics, such as ciprofloxacin (30.6%), meropenem (16.3%), and cefotaxime (16.3%), was lower. This is consistent with findings from Sudan, where ciprofloxacin resistance was 42.7% and amikacin resistance was 23.5% [37].
Gram-positive bacteria in this study exhibited high resistance to azithromycin (100%), penicillin (100%), clindamycin (100%), and erythromycin (100%). Notably, 50% of Staphylococcus aureus isolates were methicillin-resistant. This high resistance is consistent with a meta-analysis study in Ethiopia, which reported pooled resistance rates of 97.2% for erythromycin and 99.1% for penicillin [39]. A similar level of penicillin resistance (92.8%) was found at Tikur Anbessa Hospital [18].
Extended-spectrum beta-lactamase (ESBL) production by nosocomial pathogens, such as E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp., is concerning due to its association with increased hospital costs, prolonged hospital stays, and higher mortality rates [40]. In this study, ESBL-producing bacteria were phenotypically confirmed using the combination disk and double-disc synergy methods. The overall prevalence of ESBL bacteria was 26.5% (n=13/49), which was higher than other studies, such as one by Muzslay et al. (3.1%) [41], Tikur Anbessa Hospital (15.3%), and 9% [42,43], and 20.1% in Tanzania [44]. These differences could be attributed to variations in patient numbers, sample sizes, methodologies, and geographic differences.
This study found that P. aeruginosa, Klebsiella spp., E. coli, and Acinetobacter spp. exhibited ESBL production. This was consistent with findings from Tikur Anbessa Hospital, where Klebsiella spp. was the most common ESBL producer, followed by Acinetobacter spp. and E. coli [42]. In Nepal, Acinetobacter spp. and E. coli were reported as dominant ESBL producers [45], with differences possibly due to variations in the prevalence of Gram-negative bacteria and hygiene practices.
The highest contamination of ESBL-producing bacteria was found on environmental surfaces, followed by oxygen cylinders. In contrast, another study found that the highest contamination occurred on chairs, pooled samples, and sinks [42], with differences likely due to cleaning practices at the hospitals.
Carbapenemase-producing bacterial infections are a significant cause of morbidity and mortality. Carbapenem resistance has been reported in various locations worldwide, including Korea [46]. In this study, carbapenemase-producing bacteria, including K. ozaenae, K. pneumoniae, K. oxytoca, and Acinetobacter spp., were analyzed. The current study found that these bacteria were 100% resistant to meropenem, indicating a serious concern regarding carbapenem drugs. Carbapenemase-producing isolates often develop resistance to other antibiotic classes due to gene transfer or mutations in genetic loci [47, 48]. The prevalence of carbapenemase-producing bacteria in this study was 12.2%, which is higher than a study in Korea (0.4%) [46]. The difference may be due to geographical variation, sample sizes, and differences in hygiene practices.
S. aureus is a well-known nosocomial pathogen linked to numerous clinical issues in ICUs and operating rooms. In this study, ICUs were most frequently contaminated with S. aureus, and 50% of the isolates were MRSA. The overall prevalence of MRSA was 2.45%, which was significantly lower than studies conducted in Ethiopia (85.7%) [18], Nepal (33.3% and 54.4%) [24, 49], and Tanzania (19.5%) [50]. This could be due to differences in sample sizes, hygiene practices, and geographical factors. Compared to other locations, the highest number of S. aureus isolates was found on bed linens and bedrails. These surfaces are frequently touched by patients, visitors, and healthcare professionals. Patients in ICUs and operating rooms are particularly vulnerable to nosocomial infections. MRSA and S. aureus contamination on these surfaces increases the risk of transmission and can lead to conditions such as pneumonia and septicemia. MRSA-associated infections are difficult to treat due to limited treatment options, highlighting the need for further research in this area.
Hospital design and hygiene practices should primarily focus on managing nosocomial pathogens and resistant strains, which can contaminate surfaces, hands, equipment, and air. A deeper understanding of bacterial contamination mechanisms is essential for developing evidence-based prevention strategies [19]. This study’s findings underscore the importance of raising awareness among infection control teams and healthcare workers regarding bacterial contamination in ICUs and operating rooms and its potential link to nosocomial infections. However, the results may not be generalizable due to the study being conducted at a single center, and the relationship between nosocomial infections and contaminated objects or devices was not explored. Therefore, further research in this field is needed.
Conclusion
Acinetobacter spp., P. aeruginosa and S. aureus were the most commonly identified pathogenic bacteria from inanimate objects at Zewditu Memorial Hospital, all of which are potential causes of healthcare-associated infections. The antimicrobial resistance profile of these isolates was found to increase in clean, inanimate hospital environments. This study highlights a concerning level of multidrug-resistant bacteria, including ESBL and carbapenemase producers, contamination on hospital surfaces. These findings underscore the urgent need for robust infection prevention measures and continuous surveillance of the hospital environment to prevent the spread of resistant bacteria. This is especially important for vulnerable patient groups, such as neonates and those in intensive care units (ICUs) and operating rooms (ORs). Furthermore, advanced research is needed to explore the clonal relationship between clinical strains and inanimate surfaces.
Supporting information
(XLSX)
Acknowledgments
We would like to express our sincere gratitude to Zewditu Memorial Hospital and Tikur Anbessa Specialized Teaching Hospital for allowing us to conduct this study.
Abbreviations
- CDC
Centers for Disease Control and Prevention
- CLSI
Clinical Laboratory Standard Institute
- CONS
Coagulase-Negative Staphylococcus
- ESBL
Extended Spectrum β-Lactamase
- GNB
Gram-Negative bacteria
- GPB
Gram-Positive bacteria
- HCWs
Healthcare workers
- HCAI
Health care-associated infection
- HAI
Hospital Acquired Infection
- ISO
International Organization for Standardization
- MRSA
Methicillin Resistant Staphylococcus aureus
- MDR
Multi-Drug Resistance
- MHA
Mueller Hinton agar
- ICUs
Intensive Care Units
- ORs
Operation Rooms
- SPSS
Statistical Package for Social Science
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Hammuel C, Jatau ED, Whong CM. Prevalence and antibiogram pattern of some nosocomial pathogens isolated from hospital environment in Zaria, Nigeria. Aceh International J Sci Technol. 2014;3(3). [Google Scholar]
- 2.Alphons KS, Fortune TV, Haindongo E, Guillaume AY. Bacterial contamination and antimicrobial susceptibility from the hands of health care workers (HCWs) and inanimate surfaces in the neonatal intensive care unit (NICU) at the Windhoek Central Hospital (WCH). Microbiology and Nature. 2020;1(3):83–95. [Google Scholar]
- 3.Ayalew W, Mulu W, Biadglegne F. Bacterial contamination and antibiogram of isolates from health care workers’ fomites at Felege Hiwot Referral Hospital, northwest Ethiopia. Ethiopian J Health Development. 2019;33(2). [Google Scholar]
- 4.Porter L, Sultan O, Mitchell BG, Jenney A, Kiernan M, Brewster DJ, et al. How long do nosocomial pathogens persist on inanimate surfaces? A scoping review. Journal of Hospital Infection. 2024;147:25–31. [DOI] [PubMed] [Google Scholar]
- 5.Debela GA, Tesfaye BT, Yizengaw MA. Risk Factors for Inappropriate Antimicrobial Therapy Among Patients with Hospital-Acquired Infection at Jimma Medical Center: A Prospective Observational Study. Infect Drug Resist. 2022;15:837–50. doi: 10.2147/IDR.S349358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segujja F, Mwambi B, Drago C, Lubowa N, Mugambwa J, Wabuyi P. Characterization and Antimicrobial Susceptibility Patterns of Isolates from Ward Fomites. BBJ. 2016;14(4):1–16. doi: 10.9734/bbj/2016/27861 [DOI] [Google Scholar]
- 7.Vasoo S, Barreto JN, Tosh PK. Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin Proc. 2015;90(3):395–403. doi: 10.1016/j.mayocp.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 8.CDC A. Antibiotic resistance threats in the United States. Washington, DC, USA: US Department of Health and Human Services. 2019. [Google Scholar]
- 9.Bush K, Bradford PA. β-lactams and β-lactamase inhibitors: an overview. Cold Spring Harbor Perspectives in Medicine. 2016;6(8):a025247. doi: 10.1101/cshperspect.a025247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alebel M, Mekonnen F, Mulu W. Extended-Spectrum β-Lactamase and Carbapenemase Producing Gram-Negative Bacilli Infections Among Patients in Intensive Care Units of Felegehiwot Referral Hospital: A Prospective Cross-Sectional Study. Infect Drug Resist. 2021;14:391–405. doi: 10.2147/IDR.S292246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farooq S, Rishi S, Dewani S, Bashir L, Mahnoor M. Drug resistant bacterial contamination of inanimate surfaces, equipment and health care workers in ICU of a tertiary care hospital in North India. International Journal of Health Sciences. 2022;6(S4):4214–20. [Google Scholar]
- 12.Barma MM, Nasir IA, Babayo A. Bacterial pathogens and their antibiotic susceptibility pattern in Intensive Care Units of the University of Maiduguri Teaching Hospital, Nigeria. J Med Tropics. 2017;19(1):16–20. [Google Scholar]
- 13.Ngolet LO, Bolenga Liboko AF, Ossibi Ibara BR, Elira Dokekias A. Hospital acquired infection in a department of hematology-oncology care in the Congo. Am J Blood Res. 2021;11(2):191–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Cleanrooms and associated controlled environments — Bio contamination control. 2003. https://doi.org/ISO14698-1:2003
- 15.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 2024.
- 16.Suleyman G, Alangaden G, Bardossy AC. The Role of Environmental Contamination in the Transmission of Nosocomial Pathogens and Healthcare-Associated Infections. Curr Infect Dis Rep. 2018;20(6):12. doi: 10.1007/s11908-018-0620-2 [DOI] [PubMed] [Google Scholar]
- 17.Rawlinson S, Ciric L, Cloutman-Green E. How to carry out microbiological sampling of healthcare environment surfaces? A review of current evidence. J Hosp Infect. 2019;103(4):363–74. doi: 10.1016/j.jhin.2019.07.015 [DOI] [PubMed] [Google Scholar]
- 18.Sebre S, Abegaz WE, Seman A, Awoke T, Desalegn Z, Mihret W, et al. Bacterial Profiles and Antimicrobial Susceptibility Pattern of Isolates from Inanimate Hospital Environments at Tikur Anbessa Specialized Teaching Hospital, Addis Ababa, Ethiopia. Infect Drug Resist. 2020;13:4439–48. doi: 10.2147/IDR.S286293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darge A, Kahsay AG, Hailekiros H, Niguse S, Abdulkader M. Bacterial contamination and antimicrobial susceptibility patterns of intensive care units medical equipment and inanimate surfaces at Ayder Comprehensive Specialized Hospital, Mekelle, Northern Ethiopia. BMC Res Notes. 2019;12(1):621. doi: 10.1186/s13104-019-4658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ndu IK, Asinobi IN, Ekwochi U, Nduagubam OC, Amadi OF, Okeke IB, et al. The bacterial profile and sensitivity patterns of isolates from medical equipment and surfaces in the children’s emergency room of a Nigerian hospital: The bacterial profile and sensitivity patterns of isolates from medical equipment and surfaces. Medical Sci Discovery. 2019;6(9):192–7. [Google Scholar]
- 21.Merlin TW, Clement AN, Jérôme A, George EO, Francois-Xavier MK, Marcelin NN. Pattern of antibiotics resistance and phenotypic characterization of multidrug resistant bacteria isolates in four hospitals of Littoral region, Cameroon. J Drug Delivery Therapeutics. 2021;11. [Google Scholar]
- 22.Omololu-Aso J, Ukwuoma PC. Surface colonization of health care facilities in hematology by methicillin resistance Staphylococcus aureus (MRSA) at Obafemi Awolowo University Teaching Hospital Complex, Nigeria. Arch Med. 2017;9(4). [Google Scholar]
- 23.Saikeerthana D, Prabha P, Vijayashree V, Krishna GN. Overview of pathogens causing nosocomial infections in various wards of a tertiary health care hospital and their antimicrobial susceptibility pattern-a prospective surveillance study. J Pure & Applied Microbiology. 2021;15(4). [Google Scholar]
- 24.Bhatta DR, Koirala S, Baral A, Amatya NM, Parajuli S, Shrestha R, et al. Methicillin-Resistant Staphylococcus aureus Contamination of Frequently Touched Objects in Intensive Care Units: Potential Threat of Nosocomial Infections. Can J Infect Dis Med Microbiol. 2022;2022:1023241. doi: 10.1155/2022/1023241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi HJ, Kim JH, Kim NY, Lee JB, Eom JS. Environmental Culture of Bacteria at the Intensive Care Unit of a Tertiary Hospital in Korea: A Consideration for Improving Medical Environmental Safety and Healthcare-associated Infection. Korean J healthc assoc Infect Control Prev. 2020;25(2):105–14. doi: 10.14192/kjicp.2020.25.2.105 [DOI] [Google Scholar]
- 26.Purav P, Komal P, Payal R. Bacterial Contaminants and their Antimicrobial Profile from Hospital Surfaces and Equipments of Various Areas in a Tertiary Care Hospital of Gujarat, India. JCDR. 2022. doi: 10.7860/jcdr/2022/55783.16399 [DOI] [Google Scholar]
- 27.Aminu M, Usman-Sani H, Usman MA. Characterization and determination of antibiotic susceptibility pattern of bacteria isolated from some fomites in a teaching hospital in northern Nigeria. Afr J Microbiol Res. 2014;8(8):814–8. doi: 10.5897/ajmr2013.6512 [DOI] [Google Scholar]
- 28.Sizar O, Leslie S, Unakal C. Gram-Positive Bacteria. StatPearls. 2024. [PubMed] [Google Scholar]
- 29.Radji M, Fauziah S, Aribinuko N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pac J Trop Biomed. 2011;1(1):39–42. doi: 10.1016/S2221-1691(11)60065-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lalami AEO, Touijer H, Ettayebi M, Benchemsi N. Microbiological monitoring of environment surfaces in a hospital in Fez city, Morocco. J Materials and Environmental Science. 2016;7(1):123–30. [Google Scholar]
- 31.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. doi: 10.1128/CMR.00109-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dadi NCT, Radochová B, Vargová J, Bujdáková H. Impact of Healthcare-Associated Infections Connected to Medical Devices-An Update. Microorganisms. 2021;9(11):2332. doi: 10.3390/microorganisms9112332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worku T, Derseh D, Kumalo A. Bacterial Profile and Antimicrobial Susceptibility Pattern of the Isolates from Stethoscope, Thermometer, and Inanimate Surfaces of Mizan-Tepi University Teaching Hospital, Southwest Ethiopia. Int J Microbiol. 2018;2018:9824251. doi: 10.1155/2018/9824251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekrami A, Kayedani A, Jahangir M, Kalantar E, Jalali M. Isolation of common aerobic bacterial pathogens from the environment of seven hospitals, Ahvaz, Iran. Jundishapur Journal of Microbiology. 2011;4(2). [Google Scholar]
- 35.Bakkali M, Hmid K, Kari K, Zouhdi M, Mzibri M, Laglaoui A. Characterization of bacterial strains and their resistance status in hospital environment. J Trop Dis. 2015;4(180):2. [Google Scholar]
- 36.Tajeddin E, Rashidan M, Razaghi M, Javadi SSS, Sherafat SJ, Alebouyeh M, et al. The role of the intensive care unit environment and health-care workers in the transmission of bacteria associated with hospital acquired infections. J Infect Public Health. 2016;9(1):13–23. doi: 10.1016/j.jiph.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 37.Nurain AM, Bilal NE, Ibrahim ME. The frequency and antimicrobial resistance patterns of nosocomial pathogens recovered from cancer patients and hospital environments. Asian Pacific Journal of Tropical Biomedicine. 2015;5(12):1055–9. doi: 10.1016/j.apjtb.2015.09.015 [DOI] [Google Scholar]
- 38.Uddin TM, Chakraborty AJ, Khusro A, Zidan BRM, Mitra S, Emran TB, et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J Infect Public Health. 2021;14(12):1750–66. doi: 10.1016/j.jiph.2021.10.020 [DOI] [PubMed] [Google Scholar]
- 39.Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. BMC Infect Dis. 2016;16(1):689. doi: 10.1186/s12879-016-2014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muzslay M, Moore G, Alhussaini N, Wilson APR. ESBL-producing Gram-negative organisms in the healthcare environment as a source of genetic material for resistance in human infections. J Hosp Infect. 2017;95(1):59–64. doi: 10.1016/j.jhin.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 42.Jemberu AA, Tullu KD, Mulate YW. Magnitude and drug resistance profile of Extended Spectrum β-Lactamase (ESBL) producing gram-negative bacteria from different inanimate objects at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Ethiopian J Health Development. 2023;37(1). [Google Scholar]
- 43.Araya S, Desta K, Woldeamanuel Y. Extended-spectrum beta-lactamase-producing gram-negative bacteria on healthcare workers’ mobile phones: evidence from Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Risk management and healthcare policy. 2021;:283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moremi N, Claus H, Silago V, Kabage P, Abednego R, Matee M, et al. Hospital surface contamination with antimicrobial-resistant Gram-negative organisms in Tanzanian regional and tertiary hospitals: the need to improve environmental cleaning. J Hosp Infect. 2019;102(1):98–100. doi: 10.1016/j.jhin.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 45.Bhatta DR, Hamal D, Shrestha R, Hosuru Subramanya S, Baral N, Singh RK, et al. Bacterial contamination of frequently touched objects in a tertiary care hospital of Pokhara, Nepal: how safe are our hands?. Antimicrob Resist Infect Control. 2018;7:97. doi: 10.1186/s13756-018-0385-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SH, Kim GR, Kim E-Y, Jeong J, Kim S, Shin JH. Carbapenemase-producing Eenterobacterales from hospital environment and their relation to those from patient specimens. J Infect Public Health. 2022;15(2):241–4. doi: 10.1016/j.jiph.2022.01.002 [DOI] [PubMed] [Google Scholar]
- 47.Hasan SA, Raoof WM, Ahmed KK. Antibacterial activity of deer musk and Ziziphus spina-christi against carbapebem resis-tant gram negative bacteria isolated from patients with burns and wounds. Regul Mech Biosyst. 2024;15(2):267–78. doi: 10.15421/022439 [DOI] [Google Scholar]
- 48.Hasan SA, Raoof WM, Ahmed KK. First report of co-harboring bleomycin resistance gene (Blembl) and carbapenemase resistance gene (Blandm-1) Klebsiella pneumoniae in Iraq with comparison study among the sensitivity test, the BD Phoenix CPO Detect test, and the Rapidec® Carba NP test. Siberian Journal of Life Sciences and Agriculture. 2024;16(4):208–37. [Google Scholar]
- 49.Bhatta DR, Hosuru Subramanya S, Hamal D, Shrestha R, Gauchan E, Basnet S. Bacterial contamination of neonatal intensive care units. Antimicrobial Resistance & Infection Control. 2021;10:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nkuwi EJ, Kabanangi F, Joachim A, Rugarabamu S, Majigo M. Methicillin-resistant Staphylococcus aureus contamination and distribution in patient’s care environment at Muhimbili National Hospital, Dar es Salaam-Tanzania. BMC Res Notes. 2018;11(1):484. doi: 10.1186/s13104-018-3602-4 [DOI] [PMC free article] [PubMed] [Google Scholar]