Abstract
Strains of Toxoplasma gondii can be grouped into three predominant clonal lineages with members of the type I group being uniformly lethal in mice. To elucidate the basis of this extreme virulence, a genetic cross was performed between a highly virulent type I strain (GT-1) and a less-virulent type III strain (CTG), and the phenotypes of resulting progeny were analyzed by genetic linkage mapping. Analysis of independent recombinant progeny identified several quantitative trait loci that contributed to acute virulence. A major quantitative trait locus located on chromosome VII accounted for ≈50% of the virulence phenotype, whereas a minor locus on chromosome IV, linked to the ROP1 gene, accounted for ≈10%. These loci are conserved in other type I strains, indicating that acute virulence is controlled by discrete genes common to the type I lineage.
Keywords: QTL mapping, genetics, parasite, linkage analysis
Toxoplasma gondii is a widespread protozoan parasite that infects most types of warm-blooded mammals and causes opportunistic disease in humans (1). The vast majority of T. gondii strains that have been studied fall into one of three highly clonal, yet closely related, lineages (2–5). These clonal types do not show strong geographic or host boundaries, and each is distributed worldwide and found in a variety of different hosts. However, there are strong phenotypic differences between the lineages, the most notable of which is virulence in the mouse model. Archetypal type I strains share the trait of being uniformly lethal in outbred mice (LD100 = 1); by contrast, types II and III are considerably less virulent (LD100 ≥ 103) (5). By nature, virulent type I strains do not readily give rise to chronic infections in mice, whereas such long-term infections are characteristic of types II and III strains (5). Several studies have observed an increased frequency of type I strains in severe congenital toxoplasmosis in humans, suggesting the type I lineage may be more pathogenic for humans as well (4, 6).
T. gondii is readily propagated in a variety of cultured cell types and is equipped with efficient techniques for molecular genetic analyses that facilitate the introduction of foreign genes and the disruption of endogenous genes (7). Classical genetic crosses can be performed in cats, the definitive host in the life cycle, where a complex developmental program leads to the production of an oocyst that undergoes meiosis after being shed into the environment (8, 9). The genome is haploid and relatively stable, and a molecular karyotype has been developed by hybridization of markers to specific chromosomes separated by pulse-field gel electrophoresis (10). The segregation of restriction fragment length polymorphism (RFLP) markers has previously been used to build a rudimentary genetic linkage map (10, 11). Equipped with both forward and reverse genetics, T. gondii provides an ideal model for genetic analysis of complex biological traits such as virulence (12).
The three lineages of T. gondii differ by only 1–2% at the DNA sequence level (4), and their unusual dimorphic allelic composition indicates they have arisen from a recent common ancestry (13). Consequently, the marked phenotypic differences between strain types likely depend on the small number of genetic differences between them. The role of individual genes in pathogenesis can be established by reverse genetics, as shown by knockouts in the genes encoding the surface antigen 3 (sag3) (14) and a dense granule secretory protein (gra2) (15), both of which are attenuated in the mouse model. Although these results demonstrate a role for these genes in infectivity, they do not directly address the question of what genes are responsible for the naturally occurring differences in virulence between strain types. To identify genes important for pathogenesis, we have crossed a highly virulent type I strain with a less-virulent type III strain of T. gondii. Our results demonstrate that virulence has an underlying genetic basis and reveal that several discrete loci contribute to the acute virulence typical of type I strains.
Materials and Methods
Development of Genetic Markers.
Polymorphic DNA markers were adapted for PCR analysis by sequencing genomic probes that were previously used for molecular karyotype analysis and genetic linkage mapping of T. gondii (10, 11). DNA inserts from the type I RH strain were end-sequenced by using T7 or T3 primers, and the resulting sequences were used as templates to design primers to amplify 300- to 800-bp DNA fragments from the type II strain (PTG) (16) and the type III strain (CTG) (17). Sequences of the same genomic region from all three reference strains were analyzed for single-nucleotide polymorphisms that altered one or more restriction endonuclease recognition sites. PCR amplification of genomic DNAs followed by restriction endonuclease digestion was used to identify RFLP markers. A few markers were obtained from previously described genetic markers (4, 13, 18, 19). In total, 52 RFLP and 10 microsatellite markers were adapted for rapid PCR-RFLP analysis of T. gondii. Details of the markers and analyses are available on request.
Genetic Cross and Isolation of Recombinant Progeny.
The type I strain GT-1 (20) was mutagenized with ethyl nitrosourea as described previously (21). Mutants resistant to 5-fluoro-2-deoxyuridine (FUDRR) were selected by growth on human foreskin fibroblast monolayer cells in 1 × 10−5 M FUDR. An FUDRR clone of GT-1 (F3) was crossed with a clone of the type III strain CTG that was doubly resistant to adenine arabinoside (AraAR) and sinefungin (SNFR) (17). Genetic crosses were performed by the method described previously (11). In brief, two parallel crosses were performed by separately, coinfecting two specified pathogen-free cats with a mixture of parasite tissue cysts that had been grown in mice (22). Oocysts were collected after shedding in cat feces as described previously (22). Doubly resistant progeny clones (AraAR/FUDRR or SNFR/FUDRR) were selected by expansion in the presence of either 3 × 10−4 M AraA and 1 × 10−5 M FUDR or 3 × 10−7 M SNF and 1 × 10−5 M FUDR and then cloned by limiting dilution. Alternatively, random progeny clones were obtained by limiting dilution in the absence of any drug selection. To identify unique clones, recombinants were genotyped by using 17 genetic markers that are distributed on 10 of the 11 known T. gondii chromosomes.
Determination of Acute Virulence in Mice.
Eight-week-old female CD-1 outbred mice (Charles River Breeding Laboratories) were used for experimental infections. For each parasite clone, mice were infected by i.p. injection of 10, 100, or 1,000 tachyzoites (five animals/dose) (23). The accumulative percent mortality was recorded for 30 days postinfection. At the end of the 30-day observation period, blood samples were collected from surviving mice, and the sera were tested by Western blotting for antibodies against T. gondii RH strain lysate (23). Mouse sera were diluted 1:1,000 in PBS containing 1% nonfat dry milk and used as the primary antibody. Horseradish peroxidase-conjugated goat anti-mouse IgG (Amersham Pharmacia) was diluted 1:10,000 and used as the secondary antibody. Antibody binding was detected by using enhanced chemiluminescent substrate (Pierce).
Genetic Mapping and Linkage Analysis.
Segregation analysis of RFLP markers among the 26 genetically unique progeny analyzed here was carried out by using the MAPMAKER/EXP 3.0 package (24). The segregation of RFLP markers in the present cross was pooled with data from a previous cross that was also analyzed by segregation of RFLP markers (11). Chromosome maps were created by using specific markers that were mapped previously by karyotype (10) or linkage analysis (11) to anchor the map (underlined in Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). The composite maps generated in MAPMAKER/EXP have a minimum logarithm of odds (LOD) score of 3.0. Linkage analysis was performed by using MAPMANAGER QTX following the instructions of the authors (25). The linkage between virulence, based on accumulative mortality or lethality in mice, and genetic markers was analyzed by single-locus association and interval mapping by using MAPMANAGER QTX. Quantitative trait loci (QTL) were measured by the likelihood ratio statistic, and the significance level (P value) was determined based on the X2 statistic. Genome-wide significance was determined by performing 1,000 permutations by using mapmanager qtx.
Results
Isolation of Recombinant Progeny and Genotypic Analysis.
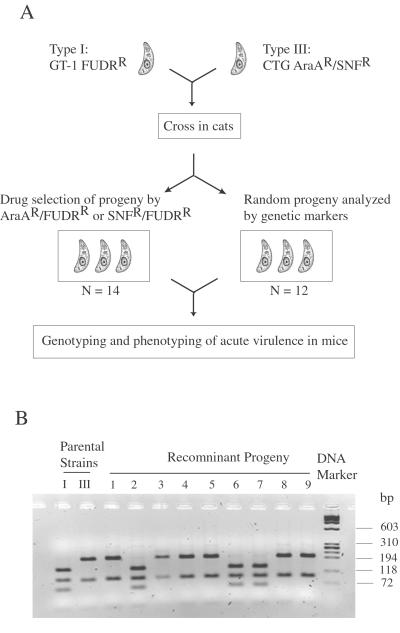
The virulent strain GT-1 (FUDRR) was crossed with the less-virulent strain CTG (AraAR/SNFR), and oocysts were recovered and allowed to sporulate and undergo meiosis in vitro (diagramed in Fig. 1A). The resulting progeny are haploid and can be directly analyzed for segregation of polymorphic DNA markers (11). Previous studies have indicated that T. gondii is capable of self-mating or of crossing, and that these events occur with roughly equal frequency (9). Thus, to be assured of obtaining recombinants, 28 clones were initially selected for resistance to a combination of FUDR with either SNF or AraA. Separately, 69 clones were cloned randomly and then subjected to genotyping by using a set of 17 independent DNA markers. Surprisingly, all but one of these randomly selected clones was recombinant, suggesting there was a strong bias for cross-fertilization in this cross. Recombinants were genotyped by using 53 PCR-based markers, an example of which is shown in Fig. 1B. In total, 26 clones proved to be genetically unique, including 14 clones obtained by drug selection and 12 clones obtained by random selection.
Fig 1.
(A) Isolation of recombinant progeny for genetic mapping and virulence linkage analysis. Clonal isolates of the type I strain GT-1 resistant to 5-fluoro-2-deoxyuridine (FUDRR) and type III strain CTG resistant to adenine arabinoside and sinefungin (AraAR/SNFR) were crossed, and recombinant progeny clones were selected with drug selections (AraAR/FUDRR or SNFR/FUDRR combination). Alternatively, progeny clones were chosen at random and genotyped to obtain recombinant ones. (B) An example of genotyping using PCR/RFLP analysis for the SAG3 gene amplified from the parental strains (indicated as I, III) and the progeny (indicated as 1–9). PCR products were digested with restriction enzyme NciI and resolved in 3% agarose gel in the presence of ethidium bromide. DNA marker = φ174 DNA digested with HaeIII.
Determination of Virulence in Mice.
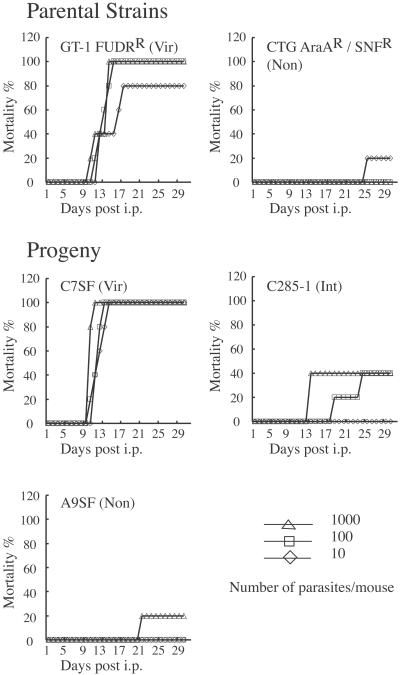
The acute virulence of recombinant clones was determined by cumulative mortality and serological status of surviving mice after i.p. inoculation in outbred mice. For this analysis, all 28 of the drug-selected clones and the 12 genetically unique clones that were obtained randomly were included. The parental clone GT-1 resulted in 100% mortality of all infected mice (no seropositive survivors), whereas the parental clone CTG was significantly less virulent with an approximate LD50 of >103 (Fig. 2). Among the recombinant progeny, three distinct phenotypes were observed; a representative of each is shown in Fig. 2. Highly virulent clones such as C7SF resulted in uniformly high mortality (no seropositive survivors), whereas less-virulent clones such as A9SF had high LD50s, and survivors were typically seropositive (Fig. 2 and Table 1). A third class of clones with an intermediate phenotype, characterized by high mortality yet with seropositive survivors, was also observed, as typified by clone C285–1 (Fig. 2 and Table 1).
Fig 2.
Mortality of mice inoculated with parental strains or one of three representative phenotypes observed in recombinant progeny. Recombinant progeny C7SF has a virulent phenotype similar to GT-1 FUDRR, whereas A9SF has a nonvirulent phenotype similar to CTG AraAR/SNFR. The cloneC285–1 has an intermediate phenotype (Int, 103 ≈LD50 >1, surviving mice sera positive). Mice were infected by i.p. of 10, 100, or 1,000 of tachyzoites (five animals/dose), and mortality was recorded for 30 days postinfection. Days post-i.p. = days after i.p. injection.
Table 1.
Summary of linkage analysis for virulence on chromosome VII
| Clone | Serum test | Vir | Mortality % | SAG4 (VII) | M95 (VII) | cS10-A6 (VII) | M102 (VII) |
|---|---|---|---|---|---|---|---|
| A9SF | + | Non | 6.7 | III | III | III | III |
| E5SF | + | Non | 26.7 | III | III | III | III |
| G2AF | + | Non | 13.3 | III | III | III | III |
| C295-9 | + | Non | 6.7 | III | III | III | III |
| E4SF | + | Int | 36.4 | III | III | III | III |
| G7SF | + | Int | 43.5 | III | III | III | III |
| C285-1 | + | Int | 40 | III | III | III | III |
| C285-4 | + | Int | 66.7 | III | III | III | III |
| C285-11 | + | Int | 41.7 | III | III | I | I |
| A9AF | + | Int | 71.4 | III | III | III | III |
| A6AF | + | Int | 92.6 | III | III | I | I |
| G2SF | + | Int | 92.9 | I | I | I | I |
| C11AF | − | Vir | 100 | III | III | III | III |
| D9AF | − | Vir | 100 | III | III | III | III |
| E7SF | − | Vir | 100 | I | I | I | I |
| B4SF | − | Vir | 100 | I | I | I | I |
| H7AF | − | Vir | 100 | I | I | I | I |
| E8SF | − | Vir | 100 | III | I | I | I |
| B10AF | − | Vir | 100 | I | I | I | I |
| C285-10 | − | Vir | 100 | I | I | I | III |
| C295-31 | − | Vir | 100 | I | I | I | I |
| C295-3 | − | Vir | 100 | I | I | III | III |
| C7SF | − | Vir | 100 | I | I | I | I |
| C295-5 | − | Vir | 100 | I | I | I | III |
| C295-27 | − | Vir | 100 | I | I | I | I |
| C295-29 | − | Vir | 100 | I | I | I | I |
| B5AF | − | Vir | 100 | III | III | I | I |
| E6AF | − | Vir | 100 | III | I | I | III |
| C285-13 | − | Vir | 100 | I | I | I | I |
| C295-2 | − | Vir | 100 | I | I | I | I |
| LOD | 3.9 | Linkage | 2 × 10−4 | 1 × 10−5 | 2 × 10−5 | 2 × 10−3 |
I, Genotype of type I parental strain; III, Genotype of type III parental strain.
+, seropositive survivors; −, no serum positive survivors.
Virulence phenotypes; Non, nonvirulent; Int, intermediate and Vir, virulent.
Genetic locus. Letters in parentheses denote chromosomes.
A9SF, E4SF, and G7SF are apparently genetically identical clones.
A9AF and C11AF are apparently genetically identical clones.
A6AF and B5AF are apparently genetically identical clones.
Comparison of serum positivity to M95.
The P value of for single-locus association.
Among the 28 drug selected clones, 14 appeared to be genetically redundant based on the typing of 53 independent markers. Ten of these clones, corresponding to six distinct genotypes, had virulence phenotypes that matched closely to that of genetically similar progeny. However, four clones that appeared genetically identical to at least one other clone differed in mouse virulence by more than 5% cumulative mortality. These clones may have additional genetic differences not detected here, and because of their dissimilar phenotypes they were included in linkage analysis shown in Table 1.
Genetic Mapping and Linkage Analysis.
The segregation of 53 independent genetic markers was analyzed by using the 26 genetically unique clones isolated here, and ordered linkage groupings were assembled by using MAPMAKER/EXP. Data from the present cross were pooled with those obtained from a previous cross (11) to create a composite genetic linkage map with the threshold LOD score of 3.0 (shown in Fig. 4). The map is comprised of 112 markers including 62 PCR-based markers and 50 genomic probes mapped by Southern blot that collectively identify 57 unique loci. This map is in close agreement with the previous identification of 11 distinct linkage groups in T. gondii (11) and extends the total genome map to include ≈400 centimorgans (cM). The average map unit is estimated to be approximately 215 kb/cM, based on positioning the outermost markers at the ends of chromosomes and estimating their physical sizes by gel separation (10). The relative density of markers was not always proportional to physical size. Notably, chromosome IV has a combined genetic linkage map of 73.2 cM and is ≈3 Mb in size, whereas chromosome VII has a combined map unit size of only 26.5 cM despite being 4.5 Mb in size (10). These differences may arise from a nonrandom distribution of markers or from different rates of recombination between different chromosomes.
There are presently two linkage groups that have not been assigned to specific chromosomes based on the LOD ≥3.0 criteria (Fig. 4). Unknown linkage group 1 consists of several markers (L353, M163, and L31-T7) that were most closely associated with chromosome V (LOD 2.1), whereas a second group of markers (M144 and SAG3) were most closely associated with chromosome IX (LOD = 2.5). Resistance to SNF was closely linked to the B1 marker on chromosome IX; resistance to FUDR showed perfect agreement with the known target of this inhibitor, the uracil phosphoribosyl transferase (UPRT) gene on chromosome IV; and resistance to AraA showed perfect agreement with its known target, the adenosine kinase (AK) gene located on unknown group 1.
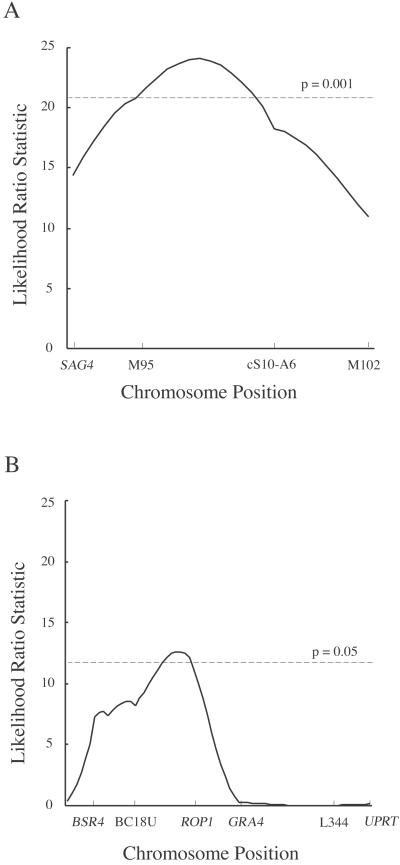
Acute virulence as defined by cumulative mortality was tightly linked to loci on chromosome VII as shown by single-locus association analyses of the segregation of chromosomal markers (Table 1). Although there are several exceptions to a strict association of virulence with loci on chromosome VII, the majority of virulent clones have type I alleles at markers M95 and cS10-A6 (Table 1). Interval mapping confirmed that a QTL responsible for cumulative mortality lies between markers M95 and cS10-A6 on chromosome VII in a region that comprises approximately 14 map units. This QTL has a significance level of P ≤ 0.001 for a genome-wide analysis (adjusted for the size of the genome) and is estimated to account for ≈50% of the variance seen in virulence between the progeny (Fig. 3A). The region of chromosome VII that exceeded the highly significant level (P ≤ 0.001) is ≈8 cM in length, which is estimated to be between 1.3 and 1.7 Mb in physical distance (estimated on the basis of the rate of recombination for chromosome VII versus the whole genome). Furthermore, when considered as a strict Mendelian trait (i.e., ± scoring), the phenotype of uniform lethality (i.e., no serum-positive survivors) had a LOD score of 3.9 with marker M95 on chromosome VII, a value that is considered highly statistically significant (Table 1). In summary, these analyses indicate that acute virulence seen in type I strains maps to a specific locus on chromosome VII that lies between markers M95 and cS10-A6.
Fig 3.
Interval mapping of a virulence-associated QTLs. (A) A major QTL is located between the markers M95 and cS10-A6 on chromosome VII. The dotted line represents the highly significant linkage threshold (P = 0.001) for a genome-wide analysis. (B) A second QTL observed on chromosome IV lies near ROP1. The dotted line represents the significance level (P = 0.05) for a genome-wide analysis. Markers are indicated at the base of map and are separated by distance proportional to map units.
No other QTLs were identified by whole-genome analysis; however, when the variance associated with the QTL on chromosome VII was fixed, two other minor loci were identified as being associated with cumulative mortality. A locus on chromosome Ib showed an inverse correlation between the type I genotype and virulence. A second, independent QTL on chromosome IV, where the type I allele was positively correlated with virulence, was most closely associated with the ROP1 gene. Each of these QTLs was estimated to account for ≈10% of the phenotype. Both of these secondary QTLs are considered only suggestive on the basis of a genome-wide analysis (0.1 ≥ P ≥ 0.05); however, when both the chromosome VII and Ib QTLs were fixed, the ROP1-associated locus was considered significant at the P ≤ 0.05 level on the basis of a genome-wide analysis. Interval mapping of the ROP1-associated QTL revealed a tightly spaced interval that was significantly associated with virulence, as shown in Fig. 3B.
To determine whether the DNA polymorphisms associated with the QTLs identified in the progeny examined here were also found in other virulent strains, we analyzed 45 naturally occurring strains isolated from humans and animals, as described previously (4, 5) (Table 2, which is published as supporting information on the PNAS web site). Included in this group were 19 type I strains, all of which are acutely virulent in mice as defined by the LD100 = 1 viable organism criteria described above. A combination of two closely linked polymorphisms detected by the probe cS10-A6 was used to analyze the QTL on chromosome VII. These two polymorphisms are each biallelic, yet together they provide distinct patterns for each of the three major strain types. Although all 19 virulent strains shared the same type I genotype, 17 of 17 type II strains and 6 of 6 type III strains contained the type II and type III genotypes, respectively (Table 2). A similar set of strains was analyzed by using the ROP1 marker on chromosome IV. This marker distinguishes only two alleles, one found in type I strains and a second shared by type II and III strains. Again, all 19 type I strains had the type I allele, whereas all 17 type II and 6 of 6 type III strains possessed the second allele (Table 2). These data indicate that the genes associated with QTLs for virulence on chromosomes VII and IV are likely conserved among all type I strains.
Discussion
We have taken advantage of the ability to perform genetic linkage analysis in the parasite T. gondii to investigate the complex trait of naturally occurring virulence. Our findings reveal several parasite QTLs that are associated with acute virulence in the mouse model of toxoplasmosis. The major contribution to acute virulence was associated with a locus on chromosome VII, with minor contributions from loci on chromosomes Ib and IV. These studies demonstrate that forward genetics offers a powerful means for analyzing the molecular basis of acute virulence in toxoplasmosis. Similar approaches are likely to be useful in related parasitic infections and, combined with molecular genetic approaches, should aid in the identification of genes important for pathogenesis.
Apicomplexan parasites have complex life cycles involving meiosis in a definitive host and asexual propagation in a variety of intermediate hosts. Genetic crosses have been performed in only a few apicomplexan species including Plasmodium (27), Toxoplasma (9, 11) and Eimeria (28). Genetic linkage analyses of simple Mendelian traits have been used to examine the basis of chloroquine resistance in Plasmodium (29) and drug resistance in Eimeria (28) and Toxoplasma (11). Classical genetic analyses are also well suited for identifying genes that contribute to natural variation in complex biological traits, including: virulence, infectivity, growth, differentiation, and drug resistance.
Previous studies have indicated that strains of T. gondii are equally capable of self fertilization or of crossing when fed simultaneously to a cat (9). In the present cross, the frequency of cross-fertilization was almost 100%, indicating one or both of the parental strains may suffer from deficiencies in self-mating or that there is a strong bias for the progeny of cross-fertilization. Previous studies have shown that GT-1 is capable of self-mating (26), and the recovery of at least one progeny with a parental genotype of the CTG strain indicates this strain is also capable of self-fertilization. Although the bias toward recombinants is unlikely to have influenced the mapping of virulence traits here, it may be highly significant to the role recombination plays in natural populations. The population structure of T. gondii is extremely clonal and, whereas crosses do not happen frequently in the wild (2–5), when they do occur they may contribute substantially to the transfer of virulence traits within the population (13).
Analysis of complex biological traits has been greatly enhanced by methods for mapping loci for quantitative traits that vary in expression along a continuum rather than being simple presence or absence (30). Host susceptibility to infection is ideally suited for such analysis, because this trait is typically multigenic and quantitative in nature. QTL mapping has been used to identify genes responsible for susceptibility and resistance to parasitic infections in insect and mammalian hosts (31–34). Resistance to chronic toxoplasmosis in the mouse model is primarily controlled by the Ld gene in the H-2 region of the major histocompatability locus (35–37). By contrast, the NRAMP1 locus plays a less direct role in resistance, and its effect is highly dependent on the genetic background of the host strain (38). It is important to note that, whereas these studies define the basis of resistance to relatively nonvirulent type II strains of T. gondii, the acute virulence of type I strains in the mouse is independent of host genotype (5). Recent studies have shown that the extreme virulence of type I strains of T. gondii in the mouse is due to overstimulation of a Th-1 cytokine response, leading to a lethal cascade of proinflammatory cytokines (39, 40).
By analyzing the progeny from a cross between virulent type I and less-virulent type III strains, we have identified several QTLs in the parasite that collectively account for a majority of acute virulence due to toxoplasmosis. The primary locus controlling acute virulence lies on chromosome VII between the microsatellite marker M95 and cS10-A6, a region that spans ≈1.5 Mb. Isolating a gene(s) responsible for virulence may eventually be possible using a combination of finer linkage analysis combined with physical mapping techniques that will be facilitated by a newly initiated genome project for T. gondii. Additionally, the excellent tools for reverse genetics that are available in T. gondii (7) will allow direct validation of the importance of particular genes in mediating enhanced virulence by either transgenic expression in a less-virulent background or by gene disruption in a virulent background.
Two other QTLs implicated in acute virulence were identified here, including one on chromosome Ib that was negatively associated with the type I genotype. This result may indicate that alleles from the normally less-virulent type III strain may also contribute to virulence. However, the relatively weak statistical support for this locus combined with the small sample size suggests that this association may be simply due to chance. Stronger support was observed for the association of virulence with a type I allele linked to the ROP1 gene on chromosome IV. ROP1 encodes a protein secreted from the rhoptries during host cell invasion, a process that may contribute to intracellular survival (41, 42). However, at present, it is not possible to determine whether the locus identified by QTL mapping is due to ROP1 or another nearby gene. Because of the small sample size used for analysis here, the actual contributions of the various loci identified here may be less than the estimates provided by mapmanager. Furthermore, it is likely that additional loci contributing to virulence may have been overlooked in this analysis. Although not a trivial undertaking, these limitations could be resolved by analysis of further progeny from this cross or additional crosses in future studies.
Importantly, the QTLs identified here do not map to loci previously implicated in virulence based on reverse genetic approaches. Deletion of the genes for sag3 (14) and gra2 (15) have been shown to decrease virulence in the mouse model, yet they were not associated with acute virulence in the present study. Thus, whereas GRA2 and SAG3 may be necessary for productive infection, they are not responsible for the natural variation in acute virulence of T. gondii. Previous studies on naturally occurring recombinant strains of T. gondii suggested an association between acute virulence and markers on chromosome VIII including a locus near the SAG1 gene (23). In separate studies that analyzed the progeny of a cross between relatively nonvirulent type II and type III strains, it was suggested that a combination of alleles on chromosomes III and IV may contribute to virulence (13). Unfortunately, neither of these studies is directly amenable to statistical analysis and therefore the relative confidence level of these associations is uncertain. In the present study, we did not find evidence for either association being involved in the acute virulence of type I strains. Nonetheless, it is likely that other loci distinct from those mapped here could contribute to acute virulence, particularly in different genetic backgrounds. As we demonstrate here, identification of such loci should be readily achievable by backcrossing and genetic linkage analysis.
We have demonstrated that forward genetics provides a powerful means for mapping genes involved in complex traits such as virulence. Acute virulence in type I strains of T. gondii, as defined by enhanced mortality with an absence of seropositive survivors, is due to a small number of discrete loci. The QTLs identified here on chromosomes IV and VII are also conserved in other type I strains, including recent clinical isolates from human infection. These results strongly support the idea that type I strains are acutely virulent because they share a small number of genes that are unique to this lineage. Identification of the specific genes that mediate enhanced virulence of the type I lineage should provide insight into the molecular basis of pathogenesis in toxoplasmosis.
Supplementary Material
Acknowledgments
We are grateful to Tovi Lehman and Michael Grigg for personal communications, to Elmer Pfefferkorn for helpful discussions, and to Jim Cheverud for critical review of the manuscript. This work was supported in part by a grant from the National Institutes of Health (AI 36629) (to L.D.S.) and the Biotechnology and Biological Sciences Research Council (to J.W.A.). L.D.S. is the recipient of a Scholars Award in Molecular Parasitology from the Burroughs Wellcome Fund.
Abbreviations
AraA, adenine arabinoside
FUDR, 5-fluoro-2-deoxy uridine
QTL, quantitative trait locus
RFLP, restriction fragment length polymorphism
SNF, sinefungin
LOD, logarithm of odds
R, resistant
cM, centimorgan
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dubey J. P. (1994) J. Am. Vet. Med. Assoc. 205, 1593-1598. [PubMed] [Google Scholar]
- 2.Dardé M. L., Bouteille, B. & Pestre-Alexandre, M. (1992) J. Parasitol. 78, 786-794. [PubMed] [Google Scholar]
- 3.Howe D. K., Honoré, S., Derouin, F. & Sibley, L. D. (1997) J. Clin. Microbiol. 35, 1411-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe D. K. & Sibley, L. D. (1995) J. Infect. Dis. 172, 1561-1566. [DOI] [PubMed] [Google Scholar]
- 5.Sibley L. D. & Boothroyd, J. C. (1992) Nature (London) 359, 82-85. [DOI] [PubMed] [Google Scholar]
- 6.Fuentes I., Rubio, J. M., Ramírez, C. & Alvar, J. (2001) J. Clin. Microbiol. 39, 1566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos D. S., Donald, R. G. K., Morrissette, N. S. & Moulton, A. L. (1994) Methods Cell Biol. 45, 28-61. [DOI] [PubMed] [Google Scholar]
- 8.Dubey J. P. & Frenkel, J. F. (1972) J. Protozool. 19, 155-177. [DOI] [PubMed] [Google Scholar]
- 9.Pfefferkorn L. C. & Pfefferkorn, E. R. (1980) Exp. Parasitol. 50, 305-316. [DOI] [PubMed] [Google Scholar]
- 10.Sibley L. D. & Boothroyd, J. C. (1992) Mol. Biochem. Parasitol. 51, 291-300. [DOI] [PubMed] [Google Scholar]
- 11.Sibley L. D., LeBlanc, A. J., Pfefferkorn, E. R. & Boothroyd, J. C. (1992) Genetics 132, 1003-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boothroyd J. C., Black, M., Kim, K., Pfefferkron, E. R., Seeber, F., Sibley, L. D. & Soldati, D. (1995) in Methods in Molecular Genetics, ed. Adolph, K. (Academic, New York), Vol. 6, pp. 3–29. [Google Scholar]
- 13.Grigg M. E., Bonnefoy, S., Hehl, A. B., Suzuki, Y. & Boothroyd, J. C. (2001) Science 294, 161-165. [DOI] [PubMed] [Google Scholar]
- 14.Dzierszinski F., Mortuaire, M., Cesbron-Delauw, M. F. & Tomavo, S. (2000) Mol. Microbiol. 37, 574-582. [DOI] [PubMed] [Google Scholar]
- 15.Mercier C., Howe, D. K., Mordue, D., Lingnau, M. & Sibley, L. D. (1998) Infect. Immun. 66, 4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasper L. H. & Ware, P. L. (1985) J. Clin. Invest. 75, 1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfefferkorn E. R. & Kasper, L. H. (1983) Exp. Parasitol. 55, 207-218. [DOI] [PubMed] [Google Scholar]
- 18.Blackstone C. R., Dubey, J. P., Dotson, E., Su, C., Thulliez, P., Sibley, D. & Lehmann, T. (2001) J. Parasitol. 87, 1472-1475. [DOI] [PubMed] [Google Scholar]
- 19.Grigg M. E., Ganatra, J., Boothroyd, J. C. & Margolis, T. P. (2001) J. Infect. Dis. 184, 633-639. [DOI] [PubMed] [Google Scholar]
- 20.Dubey J. (1980) Am. J. Vet. Res. 41, 427-429. [PubMed] [Google Scholar]
- 21.Pfefferkorn E. R. & Pfefferkorn, L. C. (1977) Exp. Parasitol. 42, 44-55. [DOI] [PubMed] [Google Scholar]
- 22.Dubey J. P. (1995) J. Parasitol. 81, 410-415. [PubMed] [Google Scholar]
- 23.Howe D. K., Summers, B. C. & Sibley, L. D. (1996) Infect. Immun. 64, 5193-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E. & Newburg, L. (1987) Genomics 1, 174-181. [DOI] [PubMed] [Google Scholar]
- 25.Manly K. F., Cudmore, J., R. H. & Meer, J. M. (2001) Mamm. Genome 12, 930-932. [DOI] [PubMed] [Google Scholar]
- 26.Dubey J. P. (1988) Am. J. Vet. Res. 49, 910-913. [PubMed] [Google Scholar]
- 27.Walliker D., Quakyi, I. A., Wellems, T. E., McCutchan, T. F., Szarfman, A., London, W. T., Corcoran, L. M., Burkot, T. R. & Carter, R. (1987) Science 236, 1661-1666. [DOI] [PubMed] [Google Scholar]
- 28.Shirley M. W. & Harvey, D. A. (2000) Genome Res. 10, 1587-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellems T. E., Walker-Jonah, A. & Panton, L. J. (1991) Proc. Natl. Acad. Sci. USA 88, 3382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander E. S. & Botstein, D. (1989) Genetics 121, 185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman M. J., Severson, D. W., Cornel, A. J., Collins, F. H. & Paskewitz, S. M. (1997) Genetics 146, 965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng L., Cornel, A. J., Wang, R., Erfle, H., Voss, H., Ansorge, W., Kafatos, F. C. & Collins, F. H. (1997) Science 276, 425-428. [DOI] [PubMed] [Google Scholar]
- 33.Blackwell J. M., Goswami, T., Evans, C. A. W., Sibthorpe, D., Papo, N., White, J. K., Searle, S., Miller, E. N., Peacock, C. S., Mohammed, H. & Ibrahim, M. (2001) Cell. Microbiol. 3, 773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebastiani G., Olien, L., Gauthier, S., Skamene, E., Morgan, K., Gros, P. & Malo, D. (1998) Genomics 47, 180-186. [DOI] [PubMed] [Google Scholar]
- 35.McLeod R., Skamene, E., Brown, C. R., Eisenhauer, P. B. & Mack, D. G. (1989) J. Immunol. 143, 3031-3034. [PubMed] [Google Scholar]
- 36.Blackwell J. M., Roberts, C. W. & Alexander, J. (1993) Parasite Immunol. (Oxf.) 15, 317-324. [DOI] [PubMed] [Google Scholar]
- 37.Brown C. R., Hunter, C. A., Estes, R. G., Beckmann, E., Forman, J., David, C., Remington, J. S. & McLeod, R. (1995) Immunology 85, 419-428. [PMC free article] [PubMed] [Google Scholar]
- 38.Blackwell J. M., Roberts, C. W., Roach, T. I. A. & Alexander, J. (1994) Clin. Exp. Immunol. 97, 107-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gavrilescu L. C. & Denkers, E. Y. (2001) J. Immunol. 167, 902-909. [DOI] [PubMed] [Google Scholar]
- 40.Mordue D. G., Monroy, F., La Regina, M., Dinarello, C. A. & Sibley, L. D. (2001) J. Immunol. 167, 4574-4584. [DOI] [PubMed] [Google Scholar]
- 41.Saffer L. D., Mercereau-Puijalon, O., Dubermetz, J. F. & Schwartzman, J. D. (1992) J. Protozool. 39, 526-530. [DOI] [PubMed] [Google Scholar]
- 42.Håkansson S., Charron, A. J. & Sibley, L. D. (2001) EMBO J. 20, 3132-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.