Abstract
Mycobacterium tuberculosis and Mycobacterium bovis bacillus Calmette–Guérin produce a heparin-binding hemagglutinin adhesin (HBHA) required for extrapulmonary dissemination and a laminin-binding protein (LBP) involved in cytoadherence through laminin recognition. These adhesins bear posttranslational modifications that are not present when the proteins are produced in a recombinant (r) form in Escherichia coli. Mass spectrometry analysis of HBHA revealed that the posttranslational modifications are borne by the C-terminal moiety, which comprises the heparin-binding domain made of repeated lysine-rich motifs. Amino acid sequencing showed that these modifications consist of mono- and dimethyllysines within these motifs. The methyllysine-containing repeats were recognized by mAb 4057D2 and were also detected in LBP, which is equally recognized by mAb 4057D2. This Ab does not recognize the recombinant forms of these proteins. However, when rHBHA and rLBP were subjected to NaBH4 and formalin treatment to induce lysine methylation, reactivity with mAb 4057D2 was recovered. Methylated rHBHA displayed enhanced resistance to proteolysis compared with rHBHA, as previously observed for native HBHA. S-adenosylmethionine-dependent HBHA methyltransferase activity was detected in the cell-wall fractions of M. bovis bacillus Calmette–Guérin and of Mycobacterium smegmatis, a species that produces LBP but naturally lacks hbhA, suggesting that the same enzyme(s) methylate(s) both LBP and HBHA. This hypothesis was confirmed by the fact that HBHA produced by recombinant M. smegmatis was also methylated. These results show that mycobacteria use enzymatic methylation of lysines to ensure greater stability of their adhesins.
Although tuberculosis remains a major cause of morbidity and mortality worldwide with 3 million deaths and 10 million new cases per year (1), relatively little is known about the virulence factors expressed by its etiologic agent, Mycobacterium tuberculosis. M. tuberculosis infection begins by inhalation of droplet nuclei smaller than 0.5 μm and containing one to three bacilli that reach the lung alveoli (2, 3). Dissemination of viable bacilli from the alveolus lumen into the lymph or circulatory systems is an important step in the pathogenesis of tuberculosis (4, 5). Because M. tuberculosis exhibits a tropism for macrophages and has evolved mechanisms to survive and multiply within the macrophage phagosome, it is generally accepted that these professional phagocytes are the main target cells of the tubercle bacillus (6–8).
However, besides its interaction with macrophages, M. tuberculosis is also able to invade and replicate within other cell types, including epithelial cells (9–11). Adherence of the tubercle bacillus to epithelial cells is predominantly mediated by the heparin-binding hemagglutinin adhesin (HBHA), a 199-residue surface-exposed protein that is also involved in mycobacterial agglutination (12). The adherence mediated by HBHA relies on the interaction of its C-terminal domain composed of lysine-rich motifs with heparan sulfate-containing proteoglycans present on the surface of epithelial cells (13–15). M. tuberculosis mutants lacking HBHA are impaired in epithelial cell adherence but display wild-type levels of adherence to macrophages (16). These mutants are severely affected in their ability to disseminate after intranasal infection, indicating that HBHA is an extrapulmonary dissemination factor (16). Extrapulmonary dissemination of wild-type M. tuberculosis can be inhibited by preincubation with mAbs that recognize the HBHA C-terminal heparin-binding domain, suggesting that this domain plays a key role in extrapulmonary dissemination (16). Previous studies have demonstrated that HBHA undergoes posttranslational modifications that are not performed when the adhesin is produced in a recombinant form (rHBHA) by Escherichia coli (13). One of the mAbs that block extrapulmonary dissemination of M. tuberculosis specifically recognizes the modified form of HBHA. In this study, we demonstrate that this modification consists of a complex methylation pattern of the lysine residues within the HBHA heparin-binding domain. Moreover, we show that this modification is performed by enzyme(s) associated with the mycobacterial cell wall and confers increased protease resistance to HBHA. Finally, we show that a similar modification is also borne by the laminin-binding protein (LBP), another mycobacterial adhesin that mediates adherence through cell-surface laminin recognition (17, 18).
Materials and Methods
Bacterial Strains and Growth Conditions.
M. tuberculosis (strain 103), Mycobacterium bovis bacillus Calmette–Guérin (strain 1173P2; World Health Organization), and Mycobacterium smegmatis (strain mc2155) were grown in Sauton medium as described (12). E. coli BL21(DE3)(pET-HBHA) (15) and E. coli BL21(DE3) (pET-MSLBP) (17) were grown in LB broth supplemented with 30 μg/ml of kanamycin (19).
Expression of the M. tuberculosis hbhA in M. smegmatis.
The 0.9-kb SalI fragment containing the full-length M. tuberculosis hbhA gene with its promoter was excised from pPK1 (16), treated with T4 DNA polymerase (Roche, Mannheim, Germany) to generate blunt ends, and subsequently inserted into the ScaI site of pRR3, giving rise to pPK5. M. smegmatis was electroporated (2.5 kV, 800 Ω, 25 μF) with 1 μg of pPK5, followed by plating onto 7H10 Middlebrook supplemented with 20 μg/ml of kanamycin. Kanamycin-resistant clones were grown in Sauton medium containing 20 μg/ml of kanamycin and tested for HBHA production by immunoblotting.
Purification of Native and Recombinant Forms of HBHA and M. smegmatis LBP (MS-LBP).
Native HBHA (nHBHA) and rHBHA were purified by chromatography on heparin–Sepharose CL-6B (Pharmacia) as described (12, 13) followed by reverse-phase HPLC by using a Nucleosil C18 column, as described elsewhere (20). HBHA produced by recombinant M. smegmatis (MS-HBHA) was purified as for nHBHA. Native M. smegmatis LBP (nMS-LBP) and MS-LBP produced by E. coli (rMS-LBP) were purified as described (17).
Chemical Methylation of rHBHA and rMS-LBP.
Chemical methylation of the lysine residues borne by rHBHA and rMS-LBP was carried out as described (21). Briefly, purified rHBHA or rMS-LBP was dialyzed for 1 hr at 4°C against 250 volumes of 100 mM borate buffer (pH 9.0). After dialysis, 3 ml of protein at 1 mg/ml was transferred into a capped glass tube containing 70 μl of a freshly prepared NaBH4 solution at 40 mg/ml and 6 μl of formalin (Sigma). The tubes were kept on ice, and 200-μl samples were taken every 10 min to check, by immunoblotting and mass spectrometry analyses, the completion of the methylation reaction.
Enzymatic Methylation of rHBHA.
M. smegmatis or M. bovis bacillus Calmette–Guérin grown in 100 ml of Sauton medium at an OD600 of 0.8 was harvested by centrifugation at 10,000 × g for 15 min and resuspended in 10 ml of 50 mM Hepes buffer (pH 7.4) containing 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (Pefabloc Sc, Roche, Mannheim, Germany) and 15% (vol/vol) glycerol (Buffer A). The cells were sonicated for 10 min at 4°C by using a Branson sonifier at an output of 5 delivered to the microtip, and the whole-cell lysates were centrifuged at 20,000 × g for 15 min at 4°C. For the methylation assay, 300 μl of clarified mycobacterial whole-cell lysate at 1 mg of protein/ml was mixed with 40 μl (1 μCi) S-adenosyl-l-[methyl-14C]methionine (AdoMet; 60 mCi/mmol, Amersham Pharmacia Biosciences)/100 μl of purified rHBHA at 0.5 mg/ml/5 μl of 1 M MgCl2/55 μl of Buffer A. The methylation assay was carried out at 37°C, and 100-μl samples were taken at different time points, mixed with 1/3 (vol/vol) of Laemmli buffer (22), heated at 95°C, and subjected to SDS/PAGE followed by autoradiography.
Enzymatic methylation was also performed by using mycobacterial cell wall or cytoplasmic fractions prepared as described (23). Briefly, the mycobacterial cells were suspended in ice-cold PBS and disrupted by sonication. The lysates were centrifuged for 30 min at 1,000 × g. The supernatants were centrifuged for 20 min at 20,000 × g. The resulting pellets consist of the cell-wall fractions, whereas the supernatants contain mixtures of cytoplasmic components and cell membranes. The cytosolic fractions contain isocitrate dehydrogenase as a marker, whereas the cell-wall fractions contain arabinomannan and arabinogalactan (23). Methyltransferase reactions using these fractions were carried out for 3 hr as described above. Then, 200 μl of 1% BSA and 300 μl of 40% trichloroacetic acid (TCA) were added, and the mixtures were chilled on ice for 30 min. After centrifugation at 25,000 × g for 30 min at 4°C, the pellets were washed thrice with 800 μl of ice-cold TCA. The radioactivity associated with the precipitated proteins was measured by using a liquid scintillation counter (Beckman Instruments, Carlsbad, CA), and the results are expressed as cpm/mg of protein.
Mass Spectrometry Analysis.
Protein and peptide samples (0.1–10 pmol) were prepared by the dry droplet method. Peptide or protein solution (0.5 μl) was mixed with freshly dissolved α-cyano-4-hydroxycinnaminic acid or sinapinic acid, respectively, each at 10 mg/ml in 50% CH3CN and 0.1% trifluoroacetic acid. After spotting and drying, mass spectrometry analyses were performed by using a matrix-assisted laser desorption ionization/time-of-flight Voyager-DE-STR (Applied Biosystems). Peptides below 3,000 Da were analyzed by using the following setting parameters: positive and reflector modes, acceleration voltage of 20 kV, grid voltage of 61%, 90 ns of delayed extraction time, and low mass gate of 500 Da. For peptides between 3,000 and 10,000 Da, the setting parameters were: positive and reflector modes, acceleration voltage of 25 kV, grid voltage of 65%, 250 ns of delayed extraction, and low mass gate of 1,000 Da. Proteins were analyzed by using the following setting parameters: positive and linear modes, acceleration voltage of 25 kV, grid voltage of 92%, 750 ns of delayed extraction time, and low mass gate of 1,000 Da. The spectra were calibrated externally by using the [M+H+] monoisotopic ions of different peptides or average masses of E. coli thioredoxin and horse apomyoglobin (Applied Biosystems).
Protease Digestions and Peptide Separation.
One nanomol of purified lyophilized nHBHA, rHBHA, or nMS-LBP was digested overnight with 5% endoproteinase Glu-C (Roche) or 1% endoproteinase Arg-C (Roche) in 100 mM phosphate buffer (pH 8.0). The resulting peptides were separated by reverse-phase HPLC by using a Beckman Ultrasphere ODS column (2 × 200 mm) and a linear 0–60% acetonitrile elution gradient prepared in 0.1% trifluoroacetic acid and directly analyzed by mass spectrometry as described above.
Amino Acid Analysis and Sequencing.
Purified nHBHA or nMS-LBP was hydrolyzed by boiling overnight at 110°C in 6 N HCl, and their amino acid composition was determined by using a Beckman Gold System amino acid analyzer. N-terminal peptide sequencing was performed by automated Edman degradation by using a pulsed liquid sequenator (Procise 492, Applied Biosystems) equipped with a 120A amino acid analyzer. Samples of 10–20 μl corresponding to 250–500 pmol of peptide were used per sequencing run.
Proteolytic Resistance Assay.
Purified rHBHA or chemically methylated rHBHA, each at 1 mg/ml, was incubated at 37°C in a total volume of 1.5 ml of PBS in the presence of 0.1 μg/ml of porcine pancreas trypsin (Sigma) or 400 μl of mouse bronchoalveolar lavage fluid. At different time points, 200-μl aliquots were mixed with 100 μl of Laemmli buffer (22) and analyzed by SDS/PAGE. To collect bronchoalveolar lavage fluids, mice under anesthesia were killed and trachea cannulated. Bronchoalveolar fluid was recovered by three consecutive lavages with 0.5 ml of PBS and centrifuged at 5,000 × g to remove cellular debris.
Other Techniques.
SDS/PAGE was performed as described by Laemmli (22), and proteins were stained with Coomassie blue R-250 (ICN) or transferred onto nitrocellulose membranes (Schleicher & Schuell) as described (24). Immobilized proteins were probed with anti-HBHA mAb 4057D2 (13, 25). Immune complexes were developed with alkaline phosphatase-linked goat anti-mouse IgG (Promega). Protein concentrations were determined by using the bicinchoninic acid method (BCA Protein Assay; Pierce) and BSA (Sigma) as a standard.
Results
Posttranslational Modification of the C-Terminal Domain of nHBHA.
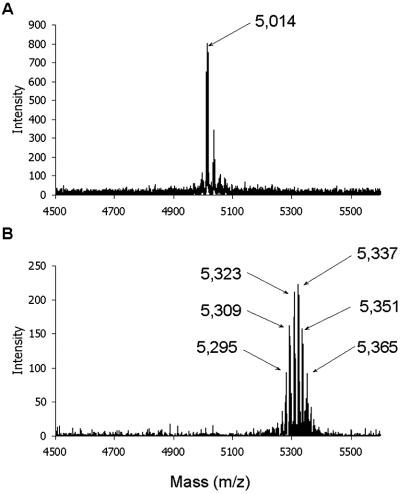
The expression of hbhA in E. coli leads to a recombinant protein with an apparent Mr somewhat lower than that of nHBHA (13), most likely due to posttranslational modifications of the native protein that are not performed by E. coli. To measure the mass difference between the native and recombinant proteins, nHBHA and rHBHA were subjected to mass spectrometry analyses. For rHBHA, a molecular mass of 21,475 Da was measured, which corresponds to the mass calculated on the basis of the amino acid sequence deduced from the hbhA sequence (13). The molecular mass of nHBHA was 21,796 Da, demonstrating that nHBHA bears a 321-Da posttranslational modification. To map the modification site, nHBHA and rHBHA were digested with endoproteinase Glu-C. After separation by reverse-phase HPLC, the masses of the resulting peptides were determined by mass spectrometry analysis. The mass fingerprintings obtained with the native and recombinant proteins were identical for all peptides, except for the peptide exhibiting a mass of 5,014 Da in rHBHA and a median mass of 5,337 Da in nHBHA (Fig. 1). Because this mass difference was similar to that observed with the full-length proteins, the HBHA posttranslational modification is entirely borne by this peptide. N-terminal sequencing revealed that it corresponds to the C-terminal lysine-rich domain (residues 150–199) of HBHA (14, 15). Whereas the mass spectrometry analysis of this domain in rHBHA gave a single peak, six major peaks separated by a mass of 14 Da each were detected for the C-terminal domain of nHBHA, indicating the presence of a heterogeneous population of modified peptides (Fig. 1).
Fig 1.
Mass spectrometry analysis of the C-terminal domain (residues 150–199) of rHBHA (A) and nHBHA (B). The masses of the major peaks are indicated by arrows.
Methyllysines in nHBHA.
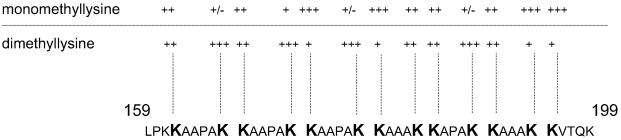
To identify the amino acid(s) that bear(s) the modification(s), the peptide spanning residues 150–199 was sequenced by automated Edman degradation, thereby revealing that the modification is borne by some of the lysine residues. The lysines at positions 153, 161, and 199 displayed a reverse-phase HPLC retention time identical to that of the PTH-Lys standard, indicating that they were not modified (not shown). In contrast, the other 13 lysines displayed retention times close to those of the phenylthiohydantoin (PTH)-Arg or the PTH-Leu standard, indicating the presence of at least two types of lysine modification. The molecular mass difference of 14 Da observed between the various peaks resulting from the mass spectrometry analysis of the HBHA C-terminal peptide suggests that the modifications could be attributed to methylations, potentially generating mono-, di-, and/or trimethyllysines (26). Using mono-, di-, and trimethyllysines as references, all of the modified lysines of nHBHA were found to behave like mono- or dimethyl-PTH-Lys during the reverse-phase HPLC. The distribution of the modified lysines within the C terminus of nHBHA as well as their methylation pattern are shown in Fig. 2. Most lysines are either preferentially mono- or dimethylated, whereas for some lysines, equivalent numbers of mono- and dimethyl groups were found. These proportions varied slightly from one preparation to another. Fig. 2 shows a general consensus obtained after three independent analyses. This methylation pattern leads to the addition of up to 26 methyl groups on nHBHA, which corresponds to a molecular mass increase of up to 364 Da, compatible with the heterogeneity of the C-terminal domain of nHBHA revealed by the mass spectrometry analyses.
Fig 2.
Distribution pattern of the mono- and dimethyllysines within the C-terminal domain of nHBHA (residues 159–199). The modified lysines appear in bold, and the symbols (±) indicate the prevalence of the methylation types associated with each modified lysine.
In Vitro Methylation of rHBHA and Antigenicity.
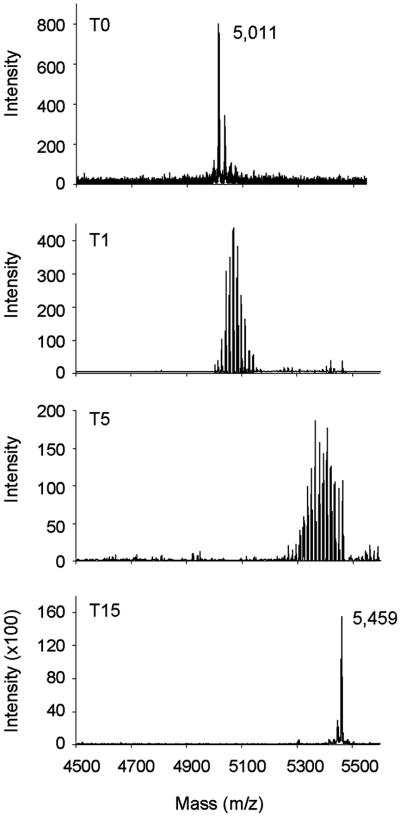
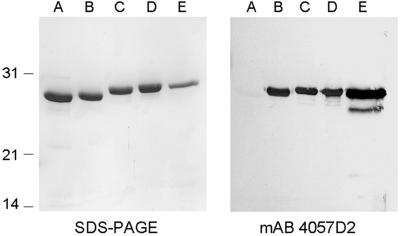
The anti-HBHA mAb 4057D2 has been shown to not recognize rHBHA (13), suggesting that the posttranslational modification constitutes part of its epitope. To confirm that methyllysines are required for mAb 4057D2 binding, rHBHA was methylated by using NaBH4 and formalin and then analyzed by immunoblotting by using mAb 4057D2. Mass spectrometry analysis of the peptide 150–199 derived from chemically treated rHBHA was used to follow the methylation. After only 1 min of treatment, mass heterogeneity was observed for the peptide, as demonstrated by the numerous peaks with increments of 14 Da that reflect the progression of the methylation reaction (Fig. 3). After 15 min, the peptide exhibited a single mass peak of 5,459 Da, indicating that the reaction was complete. The difference between the masses of fully methylated and nonmethylated rHBHA was 448 Da, which could correspond to the presence of 32 methyl residues. Amino acid sequencing of the chemically treated peptide revealed that all 16 lysines present in the peptide were dimethylated. SDS/PAGE analysis revealed that the apparent Mr of the methylated rHBHA forms increased gradually over time to eventually reach that of nHBHA (Fig. 4 Left). Probing these proteins by immunoblotting with mAb 4057D2 showed that the 4057D2 epitope appears already after 1 min of in vitro methylation (Fig. 4 Right). The mAb reacted as strongly to partially methylated rHBHA as it did to fully methylated rHBHA, whereas it did not recognize the nonmethylated protein, indicating that the methyl groups are crucial determinants of this epitope.
Fig 3.
Mass spectrometry analysis of the C-terminal domain (residues 150–199) of rHBHA (T0), and rHBHA subjected to chemical lysine methylation for 1 min (T1), 5 min (T5), or 15 min (T15). The single mass peaks at T0 and T15 are indicated.
Fig 4.
SDS/PAGE and immunoblot analyses of rHBHA, chemically methylated rHBHA and nHBHA. Purified rHBHA (A), purified rHBHA subjected to chemical lysine methylation for 1, 5, and 15 min (B, C, and D, respectively), and purified nHBHA (E) were analyzed by SDS/PAGE followed by Coomassie blue staining (Left) or by immunoblotting using mAb 4057D2 (Right). The sizes of the Mr markers given in kDa are shown in the left margin.
Methyllysines in LBP.
Although M. smegmatis does not produce HBHA, a mAb 4057D2-reactive antigen has been detected on its surface and identified as a laminin-binding protein (MS-LBP) (17). Like HBHA, this protein exhibits a lysine-rich C-terminal domain that is also posttranslationally modified. Moreover, its recombinant form produced in E. coli (rMS-LBP) is not recognized by mAb 4057D2. To determine whether nMS-LBP also contains methyllysines that could explain its reactivity with mAb 4057D2, the purified protein was subjected to endoproteinase Arg-C digestion, and the masses of the resulting peptides were determined by mass spectrometry analyses. As observed for nHBHA, most of the peptides derived from the C-terminal domain of nMS-LBP (residues 110–208) displayed a mass heterogeneity, with mass peaks exhibiting increments of 14 Da (not shown). The presence of mono- and dimethyllysines within MS-LBP was confirmed by total amino acid analysis after acid hydrolysis of the protein. To determine whether nMS-LBP recognition by mAb 4057D2 also depends on the presence of methylated lysines, rMS-LBP was subjected to chemical lysine methylation as described for rHBHA. As shown in Fig. 5, mAb 4057D2 reactivity was recovered after such treatment, indicating that as for nHBHA, the reactivity of nMS-LBP with this mAb depends on the presence of methyllysines.
Fig 5.
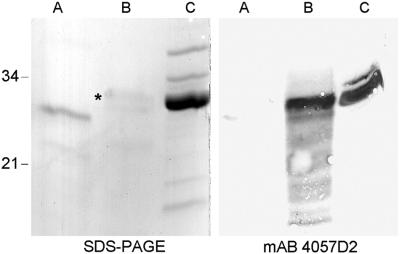
SDS/PAGE and immunoblot analyses of rMS-LBP, chemically methylated rMS-LBP, and nMS-LBP. rMS-LBP (A), rMS-LBP subjected for 15 min to chemical lysine methylation (B) and nMS-LBP (C) were analyzed by SDS/PAGE followed by Coomassie blue staining (Left) or by immunoblotting by using mAb 4057D2 (Right). The sizes of the Mr markers given in kDa are shown in the left margin, and the band corresponding to chemically methylated rMS-LBP is indicated by *.
Resistance of Methylated rHBHA to Proteolysis.
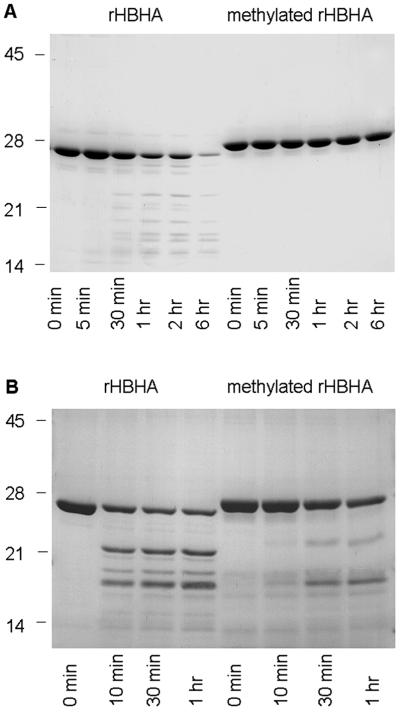
It has been shown that compared with that of nHBHA, the C-terminal domain of rHBHA is highly susceptible to proteolysis (13). To determine whether the methylation of lysines increases the resistance of rHBHA to proteolysis, rHBHA and chemically methylated rHBHA were incubated at 37°C in the presence of trypsin. As shown in Fig. 6A, rHBHA was almost completely degraded after 6 hr of incubation, whereas methylated rHBHA showed no significant degradation, indicating that the methylation of lysines is responsible for HBHA resistance to proteolysis. Partially methylated rHBHA exhibits reduced resistance to trypsin compared with fully methylated rHBHA (not shown).
Fig 6.
SDS/PAGE analysis of the proteolytic degradation of rHBHA and methylated rHBHA. rHBHA and fully methylated rHBHA were incubated with trypsin (A) or with mouse bronchoalveolar fluid (B). At the indicated time points, the proteins were analyzed by SDS/PAGE and Coomassie blue staining. The sizes of the Mr markers given in kDa are shown in the left margin.
Because trypsin-like proteases are normal constituents of the airways (27), rHBHA and methylated rHBHA were incubated in the presence of mouse bronchoalveolar lavage fluid to mimic the environment encountered by M. tuberculosis during infection. As shown in Fig. 6B, methylated rHBHA exhibited also higher resistance to degradation on incubation with bronchoalveolar lavage fluid. Incubation for more than 1 hr resulted in substantial degradation even of methylated rHBHA (not shown). Interestingly, the degradation products observed on treatment of rHBHA were different from those found after treatment of methylated rHBHA, indicating that the methylation of lysines may affect the proteolytic pattern of HBHA.
Enzymatic Methylation of rHBHA.
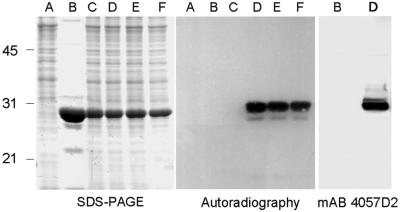
To investigate whether mycobacteria contain HBHA methyltransferases, purified rHBHA was incubated with M. bovis bacillus Calmette–Guérin lysates in the presence of radiolabeled AdoMet, the universal donor of methyl groups in enzymatic methylations (28). As shown in Fig. 7, the M. bovis bacillus Calmette–Guérin lysate induced covalent radiolabeling of rHBHA after 10 min of incubation, indicating the transfer of 14C-methyl groups from AdoMet to rHBHA. In vitro methylation by the bacillus Calmette–Guérin extract was confirmed by the recovery of the mAb 4057D2 reactivity after 10 min of incubation (Fig. 7). No labeling was observed when rHBHA was incubated in the absence of lysate or in the presence of lysate previously boiled for 5 min, consistent with the methylation being catalyzed by heat-sensitive enzyme(s). Extended incubation times up to 3 hr did not further increase the level of labeling, suggesting that the methylation reaction was complete after 10 min. When nHBHA was used as a substrate instead of rHBHA, no radiolabel was incorporated, indicating that the native protein cannot be overmethylated in the presence of excess bacillus Calmette–Guérin enzymes.
Fig 7.
SDS/PAGE, autoradiography, and immunoblot analyses of rHBHA incubated with bacillus Calmette–Guérin lysate in the presence of radiolabeled AdoMet. Clarified whole-cell lysate was incubated for different times with radiolabeled AdoMet in the absence (A) or in the presence of rHBHA (C–F). The incubation times tested were 3 hr (A, C, and F), 1 hr (E), and 10 min (D). In C, the WCL was boiled for 5 min before incubation with rHBHA. B shows rHBHA incubated with radiolabeled AdoMet in the absence of WCL. Left shows the polyacrylamide gel stained with Coomassie blue, Center shows the autoradiography, and Right shows the immunoblot by using mAb 4057D2. The sizes of the Mr markers given in kDa are shown in the left margin.
In vitro methylation of rHBHA was also observed by using lysates of M. tuberculosis and M. smegmatis (not shown), suggesting that the HBHA methyltransferase(s) are also present in these species, although the latter lacks hbhA.
When the HBHA methyltransferase activity was fractionated into cytosol and cell-wall fractions derived from bacillus Calmette–Guérin or M. smegmatis, most of the activity was found in the cell-wall fraction in both species (Table 1), suggesting that HBHA may be modified during its export process.
Table 1.
[methyl-14C]AdoMet-dependent methyltransferase activities for rHBHA in the cytosol and cell wall fractions of M. bovis bacillus Calmette–Guérin (BCG) and M. smegmatis
| Cytosol | Cell wall | |
|---|---|---|
| M. bovis BCG | 945 ± 105 | 10,477 ± 768 |
| M. smegmatis | 28,267 ± 1,468 | 83,791 ± 2,569 |
The activities expressed in cpm/mg of protein represent averages of triplicate experiments with standard deviations.
Expression of the M. tuberculosis hbhA in M. smegmatis.
Because the M. smegmatis lysate was able to methylate rHBHA in vitro, hbhA from M. tuberculosis was expressed under the control of its own promoter in M. smegmatis to know whether the recombinant protein (MS-HBHA) was also methylated in vivo. SDS/PAGE and immunoblot analyses of M. smegmatis (pPK5) harboring M. tuberculosis hbhA revealed the production of a recombinant protein comigrating with nHBHA and being recognized by mAb 4057D2, indicating that MS-HBHA is methylated in vivo by M. smegmatis (not shown). Mass spectrometry analysis of purified MS-HBHA revealed a mass of 21,790 Da, which is very close to that of HBHA. Endoproteinase Glu-C treatment of MS-HBHA followed by mass spectrometry analysis of the resulting peptides indicated that the posttranslational modification was limited to the C-terminal domain (residues 150–199). Amino acid sequencing of this domain demonstrated that the methylation pattern of MS-HBHA was similar to that of nHBHA, confirming that although M. smegmatis lacks hbhA, it produces the HBHA methyltransferase(s).
Discussion
nHBHA of M. tuberculosis and M. bovis bacillus Calmette–Guérin has been demonstrated to be posttranslationally modified (13). This modification results in an increase in apparent Mr and is not performed when the protein is produced by E. coli (13). Here, we show that this modification consists of mono- and dimethylations of lysines present in the C-terminal heparin-binding domain of nHBHA. This was evidenced by comparative mass spectrometry analysis of the native and recombinant forms of HBHA and by amino acid sequencing of the C-terminal domain of nHBHA. Previous studies have suggested that nHBHA (13), as well as LBP (17), may be glucosylated, which may be crucial for the recognition of both proteins by mAb 4057D2. The results reported here provide no evidence for glucose covalently bound to nHBHA or nMS-LBP. Instead, the increase in Mr and the reactivity with mAb 4057D2 can be accounted for entirely by a complex methylation pattern of either protein. Immunoelectron microscopy studies (12, 17) have shown that both proteins are present in the outermost layer of the mycobacterial cell wall, which contains large amounts of D-glucan (23). It is therefore likely that the glucose detected in the previous studies was a contaminant that strongly but noncovalently bound to HBHA and MS-LBP.
Methyllysines have been described in eukaryotic proteins, including histones, cytochromes c, ribosomal proteins, and muscle proteins (29). In some proteins, such as the calf thymus histone III, mono-, di-, and trimethyllysines have been detected, whereas in other proteins only one or two of these derivatives is present (29, 30). The methylation of histones seems to be involved in epigenic control of gene expression (31). However, for most proteins, the role of their methylation is unknown. Even less is known about lysine methylation of bacterial proteins. The Salmonella typhimurium flagellin is one of the few proteins that have been shown to be methylated at lysine residues (32). However, the role of the methylation in the S. typhimurium flagellin is unclear, because even mutants that lack the methyltransferase have flagella that function normally, and the mutants swim as actively as the wild type (33). Nevertheless, lysine methylation of the Salmonella flagellin seems to stabilize the protein against tryptic cleavage (34). A similar function could be assigned to the methylation of HBHA, because nHBHA or in vitro-methylated rHBHA were more resistant to tryptic digestion than nonmethylated rHBHA. In addition, methylation appeared to protect HBHA from proteolytic degradation by proteases present in bronchoalveolar lavage fluids, which may be of particular relevance with respect to the biological function of HBHA, and especially of its C-terminal domain. HBHA has been shown to be responsible for binding of M. tuberculosis to host cells other than macrophages (12, 16), such as epithelial cells, and for extrapulmonary dissemination after intranasal infection (16), qualifying it as an important virulence factor of the tubercle bacillus. The epithelial cell-binding activity of HBHA has been mapped to its C-terminal lysine-rich domain, which recognizes heparan–sulfate receptors (15). Although methylation should increase the hydrophobicity of this domain, it does not appear to modify its affinity for sulfated glycosaminoglycans (not shown). Because lysine-rich domains are particularly prone to degradation by trypsine-like proteases, and because trypsine-like proteases are abundant in the airways (27), it follows that methylation-mediated protection of HBHA against proteolytic degradation may be very useful for the efficient completion of the M. tuberculosis infectious cycle.
In addition, the lysine-rich repeats of LBP also appear to be important for its function (17) and would therefore also be useful to protect it against proteolysis. Mass spectrometry analysis, protein sequencing data, and differential recognition by mAb 4057D2 provide strong evidence that nMS-LBP is also methylated in its lysine-rich domain, with a methylation pattern very similar to that of nHBHA. It is likely that nMS-LBP and nHBHA are methylated by the same methyltransferase(s), because extracts from bacillus Calmette–Guérin, a species that produces HBHA, and from M. smegmatis, a species that naturally lacks HBHA but produces LBP, are both able to catalyze AdoMet-dependent methylation of rHBHA. In addition, when the bacillus Calmette–Guérin hbhA gene was introduced into M. smegmatis, the recombinant organism was able to produce HBHA with a methylation pattern very similar to that of nHBHA, as evidenced by mass spectrometry analysis and amino acid sequencing of MS-HBHA.
Both HBHA and LBP are surface-associated proteins and must therefore cross the mycobacterial plasma membrane during biosynthesis. However, neither of these two proteins is synthesized with an N-terminal signal sequence typical for secreted proteins, suggesting that they are secreted via a novel mechanism. Whether the methylation of these two proteins plays a role in their secretion remains to be investigated. That most of the HBHA methyltransferase activities of bacillus Calmette–Guérin and M. smegmatis are associated with the cell-wall fractions suggests that HBHA and LBP are methylated during secretion and that therefore methylation might play a role in their secretion. Answers to these questions will depend on the identification of the methyltransferase(s) involved and the construction of isogenic strains specifically lacking the enzyme(s) to investigate the role of the methylation in HBHA and LPB secretion and biological functions.
Acknowledgments
This work was supported by Institut Pasteur de Lille, Institut National de la Santé et de la Recherche Médicale, the Ministère de la Recherche, and the Fondation pour la Recherche Médicale (fellowship to K.P.).
Abbreviations
HBHA, heparin-binding hemagglutinin adhesin
r, recombinant
LBP, laminin-binding protein
n, native
M. smegmatis LBP, MS-LBP
AdoMet, S-adenosyl-l-[methyl-14C]methionine
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Raviglione M. C., Snider, D. E. & Kochi, A. (1995) J. Am. Med. Assoc. 273, 220-226. [PubMed] [Google Scholar]
- 2.Riley R. L., Mills, C. C., Nyka, W., Weinstock, N., Storey, P. B., Sultan, L. U., Riley, M. C. & Wells, W. F. (1959) Am. J. Hyg. 70, 185-196. [Google Scholar]
- 3.Riley R. L., Mills, C. C., O'Grady, F., Sultan, L. U., Wittstadt, F. & Shivpuri, D. N. (1962) Am. Rev. Respir. Dis. 85, 511-525. [DOI] [PubMed] [Google Scholar]
- 4.Hopewell P. C. (1994) in Tuberculosis, ed. Bloom, B. R. (Am. Soc. Microbiol., Washington, DC), pp. 25–46.
- 5.Weir M. R. & Thornton, G. F. (1985) Am. J. Med. 79, 467-478. [DOI] [PubMed] [Google Scholar]
- 6.Riley L. W. (1995) Trends Microbiol. 3, 27-31. [DOI] [PubMed] [Google Scholar]
- 7.Schlessinger L. S. (1996) Curr. Top. Microbiol. Immunol. 215, 71-96. [DOI] [PubMed] [Google Scholar]
- 8.Pieters J. & Gatfield, J. (2002) Trends Microbiol. 10, 142-146. [DOI] [PubMed] [Google Scholar]
- 9.Shepard C. C. (1957) J. Exp. Med. 105, 39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonough K. A. & Kress, Y. (1995) Infect. Immun. 63, 4802-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bermudez L. E. & Goodman, J. (1996) Infect. Immun. 64, 1400-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menozzi F. D., Rouse, J. H., Alavi, M., Laude-Sharp, M., Muller, J., Bischoff, R., Brennan, M. J. & Locht, C. (1996) J. Exp. Med. 184, 993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menozzi F. D., Bischoff, R., Fort, E., Brennan, M. J. & Locht, C. (1998) Proc. Natl. Acad. Sci. USA 95, 12625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delogu G. & Brennan, M. J. (1999) J. Bacteriol. 181, 7464-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pethe K., Aumercier, M., Fort, E., Gatot, C., Locht, C. & Menozzi, F. D. (2000) J. Biol. Chem. 275, 14273-14280. [DOI] [PubMed] [Google Scholar]
- 16.Pethe K., Alonso, S., Biet, F., Delogu, G., Brennan, M. J., Locht, C. & Menozzi, F. D. (2001) Nature (London) 412, 190-194. [DOI] [PubMed] [Google Scholar]
- 17.Pethe K., Puech, V., Daffé, M., Josenhans, C., Drobecq, H., Locht, C. & Menozzi, F. D. (2001) Mol. Microbiol. 39, 89-99. [DOI] [PubMed] [Google Scholar]
- 18.Shimoji Y., Ng, V., Matsumura, K., Fischetti, V. A. & Rambukkana, A. (1999) Proc. Natl. Acad. Sci. USA 96, 9857-9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Masungi C., Temmerman, S., Van Vooren, J. P., Drowart, A., Pethe, K., Menozzi, F. D., Locht, C. & Mascart, F. (2002) J. Infect. Dis. 185, 513-520. [DOI] [PubMed] [Google Scholar]
- 21.Means G. E. (1977) Methods Enzymol. 47, 469-478. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U. K. (1970) Nature (London) 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 23.Ortalo-Magné A., Dupont, M.-A., Lemassu, A., Andersen, A. B., Gounon, P. & Daffé, M. (1995) Microbiology 141, 1609-1620. [DOI] [PubMed] [Google Scholar]
- 24.Towbin H., Staehlin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouse D. A., Morris, S. L., Karpas, A. B., Mackall, J. C., Probst, P. G. & Chaparas, S. D. (1991) Infect. Immun. 59, 2595-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLange R. J., Glazer, A. N. & Smith, E. L. (1970) J. Biol. Chem. 245, 3325-3327. [PubMed] [Google Scholar]
- 27.Takahashi M., Sano, T., Yamaoka, K., Kamimura, T., Umemoto, N., Nishitani, H. & Yasuoka, S. (2001) Histochem. Cell. Biol. 115, 181-187. [DOI] [PubMed] [Google Scholar]
- 28.Cantoni G. L. (1975) Annu. Rev. Biochem. 44, 435-451. [DOI] [PubMed] [Google Scholar]
- 29.Paik W. K. & Kim, S. (1971) Science 174, 114-119. [DOI] [PubMed] [Google Scholar]
- 30.DeLange R. J. & Smith, E. L. (1971) Annu. Rev. Biochem. 40, 279-314. [DOI] [PubMed] [Google Scholar]
- 31.Peters A. H. F. M., Mermoud, J. E., O'Carroll, D., Pagani, M., Schweizer, D., Brockdorff, N. & Jenuwein, T. (2002) Nat. Genet. 30, 77-80. [DOI] [PubMed] [Google Scholar]
- 32.Tronick S. R. & Martinez, R. J. (1971) J. Bacteriol. 105, 211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stocker B. A. D., McDonough, M. W. & Ambler, R. P. (1961) Nature (London) 189, 556-558. [Google Scholar]
- 34.Joys T. M. & Kim, H. (1979) Biochim. Biophys. Acta 581, 360-362. [DOI] [PubMed] [Google Scholar]