Abstract
Compartmentation of metabolic reactions and thus transport within and between cells can be understood only if we know subcellular distribution based on nondestructive dynamic monitoring. Currently, methods are not available for in vivo metabolite imaging at cellular or subcellular levels. Limited information derives from methods requiring fixation or fractionation of tissue (1, 2). We thus developed a flexible strategy for designing protein-based nanosensors for a wide spectrum of solutes, allowing analysis of changes in solute concentration in living cells. We made use of bacterial periplasmic binding proteins (PBPs), where we show that, on binding of the substrate, PBPs transform their hinge–bend movement into increased fluorescence resonance energy transfer (FRET) between two coupled green fluorescent proteins. By using the maltose-binding protein as a prototype, nanosensors were constructed allowing in vitro determination of FRET changes in a concentration-dependent fashion. For physiological applications, mutants with different binding affinities were generated, allowing dynamic in vivo imaging of the increase in cytosolic maltose concentration in single yeast cells. Control sensors allow the exclusion of the effect from other cellular or environmental parameters on ratio imaging. Thus the myriad of PBPs recognizing a wide spectrum of different substrates is suitable for FRET-based in vivo detection, providing numerous scientific, medical, and environmental applications.
Keywords: maltose-binding protein (MBP)‖malE‖FRET‖periplasmic binding protein
Compartmentation of metabolic reactions and thus transport within and between cells can be understood only if their subcellular distribution is established by nondestructive dynamic monitoring techniques. Currently, methods are not available for in vivo metabolite imaging at cellular or subcellular levels. Limited information is derived from methods requiring fixation or fractionation of tissue (1, 2). Static analysis of metabolite composition in organs, tissues, and cellular compartments involves cell disruption. Most techniques neither measure metabolite changes in real-time nor account for likely variations in local metabolite concentration at the cellular level. Current methods have low resolution and are prone to artifacts, e.g., contamination by other cell types or subcellular compartments. Thus, little is known about the dynamic changes in concentration of metabolites such as sugars and amino acids/neurotransmitters at the site of transport, i.e., in the synaptic cleft relative to the cytosol of adjacent neurons and glia or at the loading site of the phloem in plants, but also in the distribution of different sugars within cellular compartments. To better understand metabolism and compartmentation, a noninvasive technique would be of significant advantage.
To generate a set of multifunctional nanosensors with specificity to a large number of different compounds, suitable binding proteins fulfilling a number of criteria are required. First, the binding proteins must undergo a conformational change on substrate binding. Ideally, they should belong to a family covering a wide spectrum of substrates. Furthermore, high-affinity binding would be advantageous, because it would provide a comparatively fast way to generate mutants with lower affinities suited for an optimal physiological detection range. Finally, for measurements in eukaryotes, heterologous proteins have a decreased probability of being recognized by endogenous factors. Periplasmic binding proteins (PBPs) match these criteria in that: (i) They constitute a diverse family of proteins being part of transport complexes in Escherichia coli and other Gram-positive bacteria. (ii) PBPs bind many different substrates with ultrahigh affinity. They deliver the solute to a transporter in the cytoplasmic membrane where the interaction of PBP and the transporter leads to release of the substrate from the PBP and activation of transport (3). Moreover, some PBPs are crucially involved as sensors in chemotactic responses of bacteria. (iii) Many PBPs have been well characterized at the structural, biochemical, and molecular levels (4–7). (iv) Also, on binding of their substrate, the two-lobed ellipsoid PBPs close by undergoing a substrate-induced conformational change, which has been compared to a “Venus Flytrap” movement. We have combined these attributes with the visual advantages of green fluorescent protein (GFP) variants to design a nanosensor for the determination of maltose concentration in vitro and in vivo. The maltose nanosensor was expressed in yeast cells allowing direct monitoring of the change in cytosolic maltose concentrations in response to external supply.
Experimental Procedures
Fluorescent Indicator Protein (FLIP) Constructs and Plasmids.
A cassette was constructed by using an enhanced cyan fluorescent protein (ECFP) PCR product followed by a linker and an enhanced yellow fluorescent protein (EYFP) PCR product (CLONTECH). A truncated malE PCR product encoding mature maltose-binding protein (MBP) without N-terminal signal peptide (position 79–1188 relative to the ATG) was fused between the two GFP genes. Subsequently, the chimeric fragment was inserted into pRSET (Invitrogen) and yeast expression vector pDR195 (8) and transferred into E. coli BL21(DE3)Gold (Stratagene), Saccharomyces cerevisiae SuSy7/ura3 expressing StSUT1 (9) and EBY4000 (10). The malE mutations W62A, W230A, and W340A were generated by using the QuikChange kit (Stratagene).
Expression and Purification of FLIPs.
BL21(DE3)Gold were grown for 2 days at 21°C in the dark. Cells were harvested by centrifugation, resuspended in 20 mM Tris⋅Cl, pH 7.9, and disrupted by ultrasonication. FLIPs were purified by His-Bind affinity chromatography (Novagen). Binding to the resin was performed in batch at 4°C for 4 h, washed in a column with 20 mM Tris⋅HCl and 20 mM Tris⋅HCl containing 20 mM imidazole at pH 7.9, and eluted with 200 mM imidazole in Tris⋅HCl, pH 7.9.
In Vitro Characterization of FLIPs.
Emission spectra of purified proteins were recorded spectrofluorometrically (SFM25, Kontron, Zürich) after exciting ECFP at 433 nm at horizontal analyzer and polarizer positions. Substrate titration curves were obtained by using a microtiter fluorimeter (FL600, Bio-Tek, Highland Park, VT), a 440/20-nm filter for ECFP excitation, and 530/20- and 485/30-nm filters for EYFP and ECFP emission. All in vitro analyses were performed in 20 mM sodium phosphate buffer at pH 7. Fluorescence resonance energy transfer (FRET) was determined as EYFP–ECFP emission intensity ratio. By using the change in ratio on ligand binding, binding constants (Kd) were determined by fitting the substrate titration curves to the equation for the binding of a ligand to a protein: S = (r − Rmin)/(Rmax − Rmin) = [S]bound/[P]total = n[S]/(Kd + [S]), where [S], substrate concentration; [S]bound, concentration of bound substrate; n, number of equal binding sites; [P]total, total concentration of binding protein; r, ratio; Rmin, minimum ratio in the absence of ligand; and Rmax, maximum ratio at saturation with ligand. Ratiometric measurements were performed on three independent protein extracts. Hill coefficients were determined by using the Hill equation S = (n[S]n)/(Kd + [S]n).
Beer Analysis.
Titrations were performed as for in vitro characterization described above by using different dilutions of beer (Tannenzäpfle, Badische Staatsbräuerei Rothaus, Rothaus, Germany) in 20 mM sodium phosphate buffer (pH 7). The beer dilution at 50% saturation, which equals the maltose concentration at 50% saturation (Kd), was determined by nonlinear regression. Equation: [M] = Kd/f (where [M], maltose concentration; f, dilution at half saturation) was used to calculate the maltose concentration in beer. HPLC analysis was performed by using Aminex 87-H column (Bio-Rad) and Rezek 10-μm column (Phenomenex, Torrance, CA) and a differential refractometer (LKB).
Imaging.
SuSy7/ura3 expressing StSUT1 and EBY4000 transformed with FLIPmal-25μ and FLIPmal-control in pDR195 were grown for 3–5 days in SD medium with 2% ethanol as the sole carbon source. For EBY4000 20 mg/liter of histidine, 20 mg/liter of tryptophan, and 30 mg/liter of leucine were added to the medium. Confocal images were taken on a Leica DMRE microscope equipped with a confocal head TCS SP (Leica, Wetzlar, Germany). For imaging, yeast cells were transferred to a polylysine-coated cover slide and immobilized by using 2% alginate and Ca2+ in a total volume of 100 μl. Sugar solutions were added as volumes of 5 μl on top of the alginate-embedded yeast. Imaging was performed on a fluorescence microscope (DMIRB, Leica) with a cooled charge-coupled device camera (Sensys Photometrics, Tucson, AZ). Dual emission intensity ratio was recorded by using metafluor 4.5 software (Universal Imaging, Media, PA) with 436/20 excitation and two emission filters (480/40 for ECFP and 535/30 for EYFP) and a neutral density filter (1% transmission) on the excitation port.
Results
Construction of a Maltose Sensor.
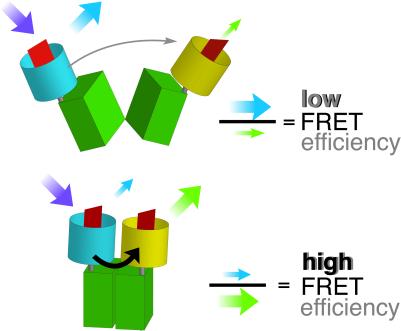
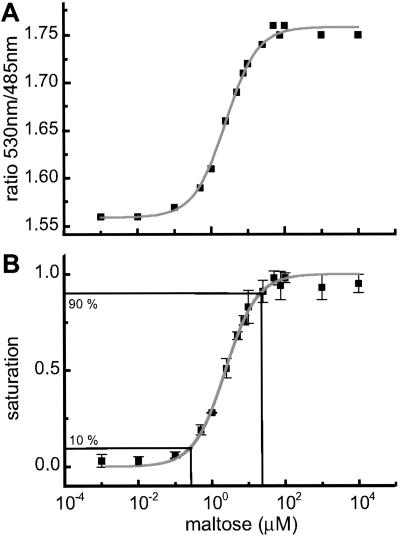
GFP variants with different spectral properties have been developed that can be used as fluorescent partners in ratiometric FRET measurements to quantify second messengers and to monitor changes in vivo (11, 12). A major part of the MBP was flanked with GFP variants for use as a FRET-based nanosensor (FLIPmal). The donor chromophore, an ECFP, was fused to the N terminus, and an EYFP was attached to the C terminus of MBP as an acceptor chromophore. Binding of maltose should bring the N and C termini, which are located at the distal ends of the two lobes, respectively, closer together, thereby increasing FRET. According to the Förster equation (13), the distance R0 at which energy transfer is 50% efficient and where a given change in distance produces the highest possible change in FRET is ≈5 nm for ECFP and EYFP as partners (11). Because MBP is relatively small, with dimensions of 3 × 4 × 6.5 nm (14), the distance between the chromophores is calculated as being smaller than R0. Furthermore, comparison of the bound and unbound states of crystal structures shows that the change in distance between the N and C termini translates into a relative movement of the chromophores of only 1 nm (15). However, in addition to the change in distance, one of the MBP lobes rotates by 35° and twists laterally by 8° relative to the other lobe (5), translating into a movement that could affect the relative orientation of the transition dipoles of the attached chromophores and lead to a change in FRET (Fig. 1). When the MBP was fused, purified, and analyzed for FRET, no change in ratio was observed, probably due to the irreversibility of maltose binding (16, 17) (Table 1). In contrast, a deletion of the first five amino acids of the mature MPB (FLIPmal-2μ) enabled determination of a maximum FRET change of 0.2 when maltose was added. The binding constant (Kd) for maltose was determined optically as 2.3 μM (Table 1; Fig. 2). Reverse titration showed that the binding of maltose was reversible, resulting in the measurement of an identical Kd. Spectra at three different maltose concentrations (zero, Kd, saturation) showed an isosbestic point at 511 nm (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
Figure 1.
Substrate-induced conformational changes. The ECFP donor chromophore was fused to the N terminus of MBP; the EYFP acceptor chromophore was attached to the C terminus. Binding of maltose brings the N and C termini, which are located at the distal ends of the two lobes, respectively, closer together, thereby increasing FRET.
Table 1.
Properties of the nanosensors
| Nanosensor | Kd, μM, of nanosensor* | Kd, μM, of binding protein† | Range for quantification,‡ μM | ΔRmax§ |
|---|---|---|---|---|
| FLIPmal¶ | – | 3 | – | – |
| FLIPmal-2μ | 2.3 ± 0.14 | – | 0.26–21.12 | 0.2 |
| FLIPmal-25μ | 25 ± 1.3 | 37 | 2.78–225.13 | 0.2 |
| FLIPmal-225μ | 226 ± 24 | 200 | 25.17–2038.98 | 0.1 |
| FLIPmal-control‖ | >100,000 | >1,000 | – | – |
Kd of the nanosensors determined by FRET.
Kd of the binding protein alone (18, 32).
Range of concentration for which a nanosensor can be used. The range for quantification was defined as the range between 10 and 90% saturation (as in Fig. 2B).
ΔRmax, the maximum change in ratio between absence and saturation of the binding protein by its substrate (Rmax − Rmin).
Because FLIPmal could not be purified without bound maltose, the Kd could not be determined.
FLIPmal-control does not show any increase in emission intensity ratio up to 100 mM substrate and thus can be used as negative control.
Figure 2.
In vitro substrate titrations of purified nanosensors. (A) FLIPmal-2μ: the emission intensity ratio (530/485-nm ratio) increases by 0.2 with increasing maltose concentration. (B) By transforming the maltose-dependent ratio change into saturation of the sensor with maltose, the Kd was determined as 2.3 μM by using nonlinear regression. The saturation curve represents the titration of three independent protein extracts. The range for quantification was defined as the range between 10 and 90% saturation.
Affinity Mutants of the Sensor.
To expand the range of the nanosensor for physiological measurements, the MBP was mutagenized to decrease substrate affinity. MBPs bind maltose via hydrogen bonding and by stacking interactions with aromatic residues in the binding pocket (4, 14). Mutations of two tryptophan residues involved in maltose binding to alanine reduced the affinity of wild-type MBP for maltose (18). Mutagenized MBP-based maltose nanosensors have Kds of 25 μM (FLIPmal-25μ; W230A) and 226 μM (FLIPmal-225μ; W62A; Table 1). For all maltose sensors, the fluorescence response fits to a Hill coefficient equal to 1, consistent with the formation of a 1:1 FLIPmal/maltose complex. As compared with wild-type MBP, the MBP fusion proteins show affinities similar to values reported in the literature (18), indicating that the GFP moieties did not affect the overall MBP structure. The similarity of data obtained with different methods proves the suitability of FRET measurements for determination of binding constants. Analyte titration curves indicate that the three FLIPmal nanosensors allow maltose quantification in a broad concentration range between 0.3–21 μM (FLIPmal-2μ; Fig. 2) and 3–225 μM (FLIPmal-25μ) (Table 1). In the case of FLIPmal-225μ, the maximum change in ratio decreased by 0.1, indicating that the mutation affects not only binding affinity but also protein structure and hinge–twist motion. Nevertheless, FLIPmal-225μ allowed in vitro maltose quantification from 25 to 2,000 μM (Table 1). The spectra of FLIPmal-25μ at three different maltose concentrations (zero, Kd, saturation) showed the same isosbestic point as those of FLIP-2μ, whereas the crossover point for FLIPmal-225μ was at 504 nm (Fig. 6).
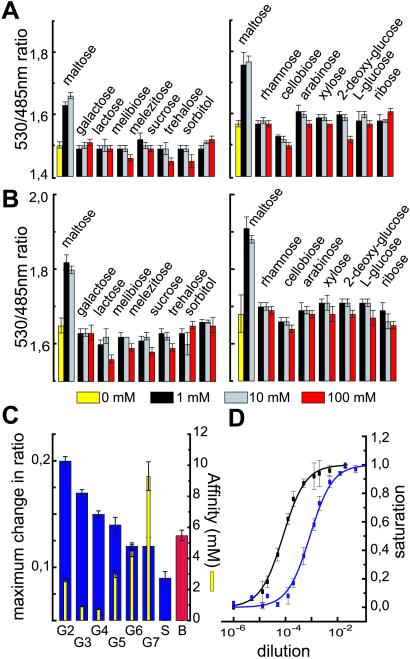
In principle, the PBPs should be highly specific for their substrate, because bacteria also use them as sensors for chemotaxis. However, it is known that MBP can bind maltotriose, maltotetraose, and other maltooligosaccharides (MOS) in addition to maltose with Kds as low as 0.2 μM in the case of maltotriose (19). Nonetheless, mutants may posses an altered specificity. The FRET-based detection of conformational changes allows rapid determination of the substrate spectrum. Therefore, 14 sugars were analyzed by using a microtiter plate assay (Fig. 3 A and B). As compared with published data, FLIPmal-2μ and FLIPmal-25μ were unaltered regarding their specificity. Both specifically recognize maltose but none of the tested pentoses, hexoses, sugar alcohols, disaccharides, or trisaccharides, which lack the α-1,4-glucosidic link present in maltose (Fig. 3 A and B). Both mutants also did not bind d-glucose (data not shown). As expected, the FLIPmal nanosensors recognize maltotriose and longer-chain MOS (Fig. 3C). In agreement with the reduced closing movement in the presence of longer α-1,4-oligomaltoside chains observed in spectroscopic analyses (15, 20), the maximum change in ratio (at saturation) decreased with the length of the maltose chain, soluble starch giving the lowest ratio, whereas the affinity remained at a similar range (Fig. 3C). The effects on maximum change in ratio indicate that also the FLIPmals in solution are present in two conformations, defined as R and B modes, and that increasing the maltoside chain length shifts the equilibrium toward the B mode (20, 21).
Figure 3.
Comparison of the substrate specificity of two FLIPmal mutants. The ratio change of purified mutants FLIPmal-2μ (A) and FLIPmal-25μ (B) was tested in the presence of various pentoses, hexoses, sugar alcohols, and di- and trisaccharides at three different concentrations. A significant increase in ratio was observed only in the presence of maltose. (C) The maximum change in ratio (Left) and the affinity constant (Right) of FLIPmal-2μ was analyzed in the presence of different MOS, soluble starch, and beer. The maximum change in ratio decreased with increasing chain length, whereas the Kd remained similar. (D) Purified FLIPmal-2μ (black) and FLIPmal-25μ (blue) were titrated with different dilutions of beer. The dilution at half saturation equals the Kd and can be used to determine the maltose concentration.
Analysis of Maltose in Complex Mixtures.
To test whether the system can be used for rapid analysis of complex solutes, we measured a local beer by using a microtiter plate assay. The beer dilution at 50% saturation equals the maltose concentration at 50% saturation (Kd). Titrations of two different FLIP mutants with different dilutions of beer allowed determination of analyte concentrations of 29.6 ± 1.8 mM (Fig. 3D). To confirm the data, HPLC analysis was performed on the same sample; however, maltose concentration was measured as only 3.5 mM (data not shown). As shown above, the maltose sensors also bind MOS, whereas quantification by HPLC specifically detected maltose. Because after fermentation beer contains significant amounts of MOS and soluble starch (22), the discrepancy must be due to MOS detected by the sensors. The presence of MOS in beer is supported by the finding that, as in case of pure MOS and soluble starch, the maximum change in ratio determined in beer was reduced (ΔRmax = 0.13). Addition of 5 mM maltose restored a ΔRmax of 0.2 (Fig. 7, which is published as supporting information on the PNAS web site). The measured values are consistent with enzymatic measurements of MOS and with determinations using MBP-based bioelectronic nanosensors (23).
In Vivo Imaging of Maltose Uptake.
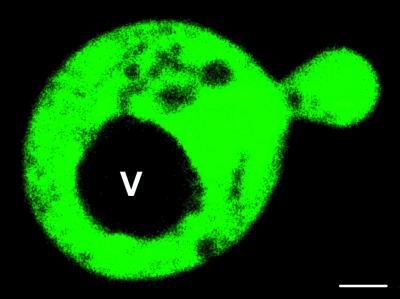
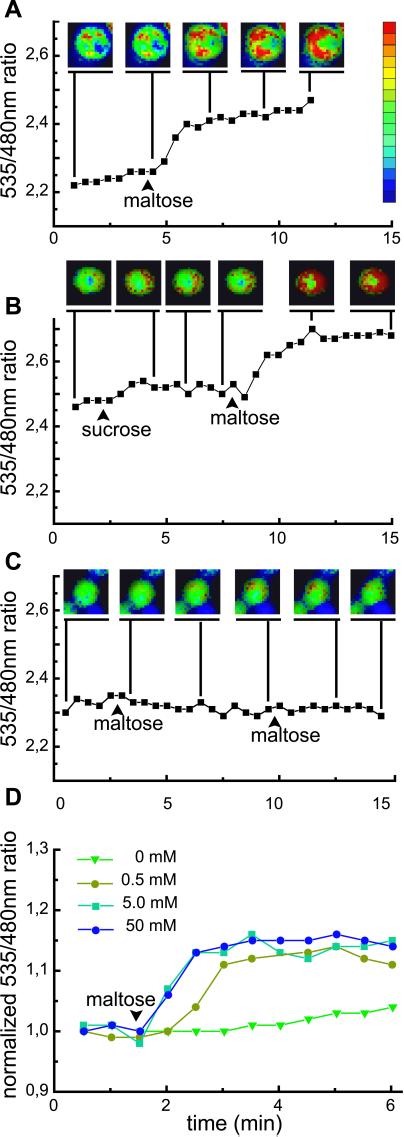
To test the FLIP nanosensors in vivo, dynamic changes of cytosolic maltose concentration were visualized in single yeast cells. Yeast was chosen as a model system, because maltose uptake and hydrolysis in this system can be controlled, and because maltose is not directly phosphorylated after import. A yeast strain deficient in maltose uptake and cleavage, SuSy7/ura3, expressing StSUT1, a proton sucrose cotransporter able to mediate also proton-coupled maltose transport (9), was transformed with FLIPmal-25μ. Confocal imaging of FLIPmal-25μ expressing yeast cells showed that the fluorescent fusion protein is expressed in the cytosol, whereas no signal was detected in the vacuole (Fig. 4). Thus, FLIPmal-25μ should allow direct monitoring of maltose uptake into the cytosol with a subcellular resolution. To keep initial cytosolic maltose concentration low and to monitor the highest possible change in FRET, yeast was grown on ethanol as the sole carbon source. On addition of 50 mM extracellular maltose, the 535/480-nm emission intensity ratio increased by 0.15–0.2, indicating that maltose is transported into the yeast cytosol, where it is recognized by FLIPmal-25μ (n = 69; Fig. 5A). The response was specific, because addition of sucrose had no effect (n = 38; Fig. 5B). Furthermore, yeast extracts did not seem to contain substances binding to FLIPmals, whereas maltose-spiked extracts gave results similar to pure maltose (Fig. 8, which is published as supporting information on the PNAS web site). Imaging showed that the increase was rapid in the first 2 min after addition of maltose, followed by a sustained accumulation in the cytosol increasing over the next several minutes, and in most cases leaving a central section unstained, probably representing the central vacuole. Because SuSy7/ura3 lacks maltase activity, the emission intensity ratio remains constant after reaching its maximum. When FLIPmal-25μ was expressed in EBY4000, a yeast strain with constitutive maltase activity, the full change in ratio was detected, indicating that the enzyme, which has a KM of 16.6 mM (24) for maltose, is unable to reduce cytosolic maltose levels below saturation of FLIPmal-25μ (Fig. 5D). A reduction of the external maltose concentration to 0.5 mM led to a delayed accumulation but not to a reduction of the maximum ratio, indicating that also in this case, cytosolic maltose concentrations increased to levels at which all nanosensors were in the bound state (Fig. 5D).
Figure 4.
Confocal imaging of FLIPmal-25μ expressed in yeast. FLIPmal-25μ is detected in the cytosol, whereas no signal was found in the vacuole (V). (Bar = 1 μm.)
Figure 5.
Visualizing dynamic maltose concentration change in the cytosol of yeast. (A, B) SuSy7/ura3 expressing StSUT1 for maltose uptake into the cytosol and FLIPmal-25μ (A, n = 32; B, n = 43). Each graph indicates the emission intensity ratio (535/480-nm ratio) for a single yeast cell. Addition of maltose increased the ratio by 0.15 to 0.2, whereas addition of sucrose had no effect on the emission intensity ratio. (C) SuSy7/ura3 expressing StSUT1 and FLIPmal-control. Addition of extracellular maltose or sucrose did not increase the ratio (n = 19). Yeast images are pseudocolored to demonstrate the ratio change. Extracellular sugar solutions were added at the indicated time points at a final concentration of 50 mM (arrowhead). (D) EBY4000 strain: each graph indicates the average emission intensity ratio of four to seven cells. Addition of low levels of maltose (0.5 mM, olive) led to a retarded change in ratio as compared with higher levels (5 mM, aqua; 50 mM, blue); no change was observed with addition of water (green).
Development of a Control Sensor.
FRET efficiency and analyte quantification critically depend on analyte-induced conformational changes. However, in addition to the maltose-induced hinge–twist motion, the overall protein structure of the nanosensor may be affected by other factors, such as ionic conditions. Furthermore, EYFP is highly sensitive to halides and acidic pH, because both parameters favor protonation of the chromophore anion (25). Although introduction of a Q69M mutation in EYFP reduces sensitivity (26), nonspecific conformational changes still cannot be overcome. To exclude artifacts, a mutant with a Kd for maltose higher than 100 mM was generated (FLIPmal-control). FLIPmal-control displayed no increase in emission intensity ratio up to 100 mM maltose, thus it could be used as a control for the imaging of maltose uptake into yeast. On addition of maltose, the ratio remained constant, indicating that a change neither in chloride concentration nor in pH or any other maltose-independent effect affected FLIPmal conformation under the conditions tested (Fig. 5C).
Discussion
One of the most significant advances in the analysis of signal transduction was the development of ion-selective dyes that allowed monitoring, e.g., calcium changes in living cells in response to a stimulus. These devices were further refined by the development of proteinaceous calcium sensors, including FRET-based imaging systems (27, 28). Despite the importance for the understanding of metabolism, transport, sensing and homeostasis, no such dyes were available for organic molecules such as sugars. It thus seems crucial to be able to determine whether different cell types differ with respect to sugar metabolism and to directly visualize changes with a subcellular localization. As a prototype, a proteinaceous nanosensor was developed for detecting maltose. Maltose as a degradation product of starch hydrolysis serves as a carbon source for bacteria, fungi, and animal nutrition. In plants, hydrolytic degradation of transient starch in leaf chloroplasts takes place at night but also plays a crucial role during reserve mobilization in germinating seedlings. Maltose can be exported from chloroplasts (29), a process which is currently not fully understood. For the analysis of these physiological processes, a maltose nanosensor was developed on the basis of the bacterial MBP. Despite the comparatively small movement of the two lobes toward each other, fusion of two different GFPs to the extremities of a shortened MBP allowed maltose-dependent FRET measurements. Mutants were generated with differing binding constants, providing a set of nanosensors with a broad concentration range between 0.3 and 2,000 μM (Table 1). These nanosensors, together with a mutant that was unable to bind maltose, were used to follow the uptake of externally supplied maltose into the cytosol of yeast. For exact in vivo determination of cytosolic maltose concentrations at the equilibrium between uptake and degradation by maltase, a maltose nanosensor with a Kd closer to the KM of maltase is required. Unfortunately, the maximum ratio change for FLIPmal-225μ, which might sense at this range, was too low for obtaining significant data. To generate more sensitive fluorescent indicators, e.g., with higher maximum changes in ratio, modification of the linkers between the fluorescent proteins and MBP in combination with mutagenesis of the binding protein will be used. Taken together, the set of three maltose nanosensors is suited for analyzing dynamic changes in maltose concentration, e.g., in plastids of roots or leaves to follow starch turnover after targeting the nanosensor to the chloroplast or to determine the efficiency and regulation of maltose metabolism in brewers yeast.
It is obvious that FLIPmals similar to the native MBPs bind not only maltose but also MOS. Thus, in vivo imaging will allow determination of the presence of maltose and MOS. The ability to detect both maltose and MOS will be interesting, e.g., with respect to determining the presence of soluble starch and maltose in mesophyll cytosol or to follow starch turnover in chloroplasts (29). Mutagenesis and fluorescence screening may be means for identifying FLIPmals selective for maltose.
Because the relative movement of the two MBP lobes is comparatively small, we have to assume that the hinge–twist movement contributes to the measured FRET change. The microtiter plate-based assay will therefore also permit a detailed analysis of the MBP movement, as well as rapid screening of potential mutants and their substrate specificity if modified in relation to other mutants. The system is therefore also suitable for a detailed analysis of dynamics of MBP.
Bacteria possess a large number of PBPs with specificity for different substrates such as sugars, amino acids including neurotransmitters, peptides, organic acids and vitamins, and also for inorganic ions (e.g., phosphate or nitrate) and metals (e.g., iron or zinc) (30) (http://www.biology.ucsd.edu/%7eipaulsen/ transport). For many of the well characterized PBPs, crystal structures are available that may help in designing nanosensors for imaging diverse analytes in vivo, ex vivo, and in vitro, provided the GFP moieties can be attached in suitable positions and that similar movements translate into significant FRET changes.
Taken together, a prototype for a novel set of nanosensors was established, potentially enabling us to monitor dynamic changes in a wide spectrum of inorganic and organic analytes with subcellular resolution in vivo and ex vivo. These tools will help us to better understand transport processes within and between cells. For instance, many transporters have been characterized at the molecular level; however, their actual function can be determined only if there is sufficient information about the concentrations in the compartments, especially in knockout mutants. Furthermore, the system can be used to develop rapid devices monitoring solute composition of liquids, as exemplified for MOS analysis in beer, similar to the bioelectric nanodevices that were also developed on the basis of PBPs (23). Obviously, the advent of a set of novel nanosensors will provide us with the tools for numerous scientific, medical, and environmental applications.
Supplementary Material
Acknowledgments
We thank Robert Gauss and Bettina Stadelhofer for excellent assistance, Harald Stransky for sugar analysis, and Rama Panford and Felicity deCourcy for critical reading of the manuscript. This project was supported by grants to W.B.F. from the Gottfried-Wilhelm Leibniz Award and the Deutsche Forschungsgemeinschaft (SFB 446).
Abbreviations
- PBP
periplasmic binding protein
- GFP
green fluorescent protein
- MBP
maltose-binding protein
- ECFP
enhanced cyan fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- MOS
maltooligosaccharides
- FRET
fluorescence resonance energy transfer
- FLIP
fluorescent indicator protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 9614.
References
- 1.Farré E M, Tiessen A, Roessner U, Geigenberger P, Trethewey R N, Willmitzer L. Plant Physiol. 2001;127:685–700. doi: 10.1104/pp.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borisjuk L, Walenta S, Weber H, Müller-Klieser W, Wobus U. Plant J. 1998;15:583–591. doi: 10.1046/j.1365-313x.2002.01222.x. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. FEBS Lett. 1994;346:55–58. doi: 10.1016/0014-5793(94)00315-7. [DOI] [PubMed] [Google Scholar]
- 4.Vyas N K, Vyas M N, Quiocho F A. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- 5.Scharff A J, Rodseth L E, Spurlino J C, Quiocho F A. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 6.Quiocho F A, Pflugrath J W. J Biol Chem. 1980;255:6559–6561. [PubMed] [Google Scholar]
- 7.Fukami-Kobayashi K, Tateno Y, Nishikawa K. J Mol Biol. 1999;286:279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 8.Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer W B. FEBS Lett. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- 9.Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward J M, Frommer W B. Plant Cell. 2000;12:1153–1164. doi: 10.1105/tpc.12.7.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg C P, Boles E. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 11.Miyawaki A, Tsien R Y. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 12.Heim R. Methods Enzymol. 1999;302:408–423. doi: 10.1016/s0076-6879(99)02036-4. [DOI] [PubMed] [Google Scholar]
- 13.Förster T. Ann Phys. 1948;2:55–75. [Google Scholar]
- 14.Spurlino J C, Lu G-Y, Quiocho F A. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 15.Hall J A, Thorgeirsson T E, Liu J, Shin Y-K, Nikaido H. J Biol Chem. 1997;272:17610–17614. doi: 10.1074/jbc.272.28.17610. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Sharma S, Quiocho F A, Davidson A L. Proc Natl Acad Sci USA. 2001;98:1525–1530. doi: 10.1073/pnas.041542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silhavy T J, Szmelcman S, Boos W, Schwartz M. Proc Natl Acad Sci USA. 1975;72:2120–2124. doi: 10.1073/pnas.72.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martineau P, Szmelcman S, Spurlino J C, Quiocho F A, Hofnung M. J Mol Biol. 1990;214:337–352. doi: 10.1016/0022-2836(90)90165-I. [DOI] [PubMed] [Google Scholar]
- 19.Ferenci T, Muir M, Lee K-S, Maris D. Biochim Biophys Acta. 1986;860:44–55. doi: 10.1016/0005-2736(86)90496-7. [DOI] [PubMed] [Google Scholar]
- 20.Duan X, Hall J A, Nikaido H, Quiocho F A. J Mol Biol. 2001;306:1115–1126. doi: 10.1006/jmbi.2001.4456. [DOI] [PubMed] [Google Scholar]
- 21.Hall J A, Gehring K, Nikaido H. J Biol Chem. 1997;272:17605–17609. doi: 10.1074/jbc.272.28.17605. [DOI] [PubMed] [Google Scholar]
- 22.Thomas B R, Brandley B K, Rodriguez L. J Am Soc Brew Chem. 2000;58:124–127. [Google Scholar]
- 23.Benson D E, Conrad D W, de Lorimier R M, Trammell S A, Hellinga H W. Science. 2001;293:1641–1644. doi: 10.1126/science.1062461. [DOI] [PubMed] [Google Scholar]
- 24.Vanoni M, Sollitti P, Goldenthal M, Marmur J. Prog Nucleic Acid Res Mol Biol. 1989;37:281–322. doi: 10.1016/s0079-6603(08)60701-1. [DOI] [PubMed] [Google Scholar]
- 25.Wachter R M, Yarbrough D, Kallio K, Remington S J. J Mol Biol. 2000;301:157–171. doi: 10.1006/jmbi.2000.3905. [DOI] [PubMed] [Google Scholar]
- 26.Griesbeck O, Baird G S, Campbell R E, Zacharias D A, Tsien R Y. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 27.Miyawaki A, Llois J, Heim R, McCaffery J M, Adams J A, Ikura M, Tsien R Y. Nature (London) 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 28.Miyawaki A, Griesbeck O, Heim R, Tsien R Y. Proc Natl Acad Sci USA. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleucher J, Vanderveer P J, Sharkey T D. Plant Physiol. 1998;118:1439–1445. doi: 10.1104/pp.118.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier M H., Jr Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.