Abstract
Cain/cabin1 is an endogenous inhibitor of calcineurin (Cn), a calcium-dependent serine/threonine phosphatase involved in various cellular functions including apoptosis. We show here that during apoptosis cain/cabin1 is cleaved by calpain at the carboxyl terminus to generate a cleavage product with a molecular mass of 32 kDa as a necessary step leading to Cn-mediated cell death. Mouse cain/cabin1 was identified from a thymus cDNA library by an in vitro substrate-screening assay with calpain. Exposure of Jurkat cells to the calcium ionophore, A23187, induced cain/cabin1 cleavage and cell death, accompanied by activation of calpain and Cn. The calpain inhibitors, calpeptin and zLLY, suppressed both A23187-induced cain/cabin1 cleavage and Cn activation, indicating that Cn activation and cain/cabin1 cleavage are calpain-dependent. Expression of cain/cabin1 or a catalytically inactive Cn mutant [CnAβ2(1–401/H160N)] and treatment with FK506 reduced A23187-induced cell death. In vitro calpain cleavage and immunoprecipitation assays with deletion mutants of cain/cabin1 showed that cleavage occurred in the Cn-binding domain of cain/cabin1, indicating that the cleavage at its C terminus by calpain prevented cain/cabin1 from binding to Cn. In addition, in vitro binding assays showed that cain/cabin1 bound to the Cn B-binding domain of Cn A. Taken together, these results indicate that calpain cleaves the calcineurin-binding domain of cain/cabin1 to activate Cn and elicit calcium-triggered cell death.
Calpains are cytosolic calcium-activated neutral cysteine proteases and ubiquitously distributed in all animal cells (1–3). The calpain family has at least six members, which can be divided into two groups on the basis of their tissue distribution: ubiquitous and tissue-specific. The best-characterized calpains are two ubiquitously expressed isozymes, μ- and m-calpain, which can be distinguished by their in vitro requirement for different levels of calcium for activation. μ-Calpain and m-calpain require micro- and millimolar concentrations of calcium, respectively. Calpain-mediated proteolysis proceeds in a limited manner but, unlike caspase, it does not require a specific amino acid residue.
Calpain has been proposed to influence normal signal transduction processes by cleaving cytoskeletal proteins, membrane proteins, and enzymes (2). Calpain also plays important role in forms of cell death that participate in the pathogenesis of several diseases (3). Calpain activation may contribute to neuronal cell death in ischemic brain injury (2), thymocyte cell death (4), Alzheimer's disease (5, 6), and myelin protein degradation in multiple sclerosis (7). Thus, calpain inhibitors exert protective effects in models of focal or global cerebral ischemia (8, 9) and delay neuronal death after transient ischemia (10) and myocardial infarction (11). A key to understanding the molecular basis of calpain-mediated cell death likely lies in the identification and characterization of critical calpain substrates. For example, calpain cleaves and activates caspase-12 and cdk5 to mediate calcium-triggered cell death (12–14).
Recently, cain/cabin1 was identified as a protein that binds to and inhibits calcineurin (Cn) with its C-terminal region (15–17). Cn seems to mediate upstream events in calcium-activated cell death. High levels of Cn activity predispose thymocytes, T cells, Jurkat cells, and neuronal cells to apoptosis induced by T cell receptor activation, calcium ionophores, growth-factor deprivation, L-glutamate, and status epilepticus (18–21). In our screen to identify new calpain substrates by using an in vitro small pool assay, we have identified mouse cain/cabin1 as a putative calpain substrate. In this study, we show that the C terminus of cain/cabin1 is cleaved in Jurkat cells undergoing calcium ionophore-induced apoptosis, generating a cleavage product of 32 kDA. Cain/cabin1 cleavage and Cn activation are suppressed by calpain inhibitors. Expression of the Cn-binding domain of cain/cabin1 suppresses A23187-induced cell death. Thus, we propose that A23187-induced cell death involves cleavage of cain/cabin1 by calpain, which leads to Cn activation.
Materials and Methods
Reagents.
Calpeptin, zLLY, A23187, and FK506 were purchased from Calbiochem. Antibodies against caspase-3 and α-tubulin were purchased from Santa Cruz Biotechnology. Rhodamine-110 was from Molecular Probes. Cn activity assay and TNT Systems were purchased from Promega. A23187, m-calpain, and other molecular biology grade materials were purchased from Sigma or New England Biolabs.
Plasmid Construction.
Rat cain/cabin1 and cain/cabin1-CΔb (2073–2173) preparations were described (15). Cain/cabin1-CΔa (1842–2077) was amplified by PCR and subcloned into pcDNA3-hemagglutinin (HA) (pHA-cain/cabin1-CΔa). Human pCain/cabin1, pCnAβ2 (1–401/H160N), pHA-CnB, and pNF-AT-luc (NF-AT, nuclear factor of activated T cells) were described (16). To construct expression plasmids, a cDNA spanning residues 1842–2182 of rat cain/cabin1 was amplified by PCR and subcloned into pGEX4T-3 (pGexCain/cabin1-C) (Amersham Pharmacia) and pcDNA3-HA (pHA-cain/cabin1). The same fragment was subcloned into pBabe to construct the mammalian expression plasmid (pBabe-cain/cabin1). CnA, CnAΔCaMAI (1–398), and CnAΔeD (1–329) were amplified by PCR and subcloned into pcDNA3-HA (pHA-CnA, pHA-CnAΔCaMAI, and pHA-CnΔAeD).
In Vitro Screening for New Calpain Substrate Genes: Calpain Cleavage Assay.
Proteins from small pools (50–150 cDNAs) of a mouse thymus cDNA library were prepared by in vitro translation by using a TNT System (Promega) in the presence of [35S]methionine (Amersham Pharmacia). In vitro calpain cleavage reactions were performed in PBS containing 1 mM CaCl2 for 60 min at 30°C. The reaction mixtures were then separated by SDS/PAGE, after which the dried gel was exposed to x-ray film. Secondary and tertiary screenings were performed until a single putative, positive clone was obtained. The isolated clones were then subjected to DNA-sequencing analysis.
Antibody Generation and Western Blot Analysis.
pGEXCain/cabin1-C (1842–2182) was transformed into BL21(DE3) and its expression was induced with 0.2 mM isopropyl-1-thio-α-D-galactopyranoside. Glutathione S-transferase (GST)-cain/cabin1-C was purified by using glutathione Sepharose 4B beads (Amersham Pharmacia) and injected four times into a rabbit. Anti-cain/cabin1 antibody was purified from serum by using antigen-affinity chromatography. Western blot analysis was performed as described by Jung et al. (22).
Cell Culture and DNA Transfection.
Jurkat (human T lymphoma) cells were grown in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% FBS. B103 (rat neuroblastoma) and HEK 293T (human embryonic kidney) cells were grown in DMEM containing 10% FBS. DNA transfection was performed by using LipofectAMINE PLUS reagent according to the methods recommended by the manufacturer (GIBCO/BRL).
Cell Viability and DNA Fragmentation Assay.
Cell viability was assessed by exclusion of 0.04% trypan blue, and DNA fragmentation was assayed as described by Eastman (23).
Calpain and Cn Activity Assay.
Cells were incubated in Hepes-buffered saline [5 mM Hepes/0.15 M NaCl (pH 7.35)] containing 2 mM EDTA and incubated at 37°C for 20 min in the presence of 10 μM Rhodamine-110-based substrate [(t-BOC-Leu-Met)2-R110]. After washing, fluorescence was measured at 498 nm for excitation and at 521 nm for emission by using a fluorescence microplate reader (FL-600) (Bio-Tek, Burlington, VT). Cn assays were performed according to the manufacturer's instruction (Promega, Ser/Thr PPTase assay system) (34).
NF-AT Reporter Gene Assays.
Cells transiently transfected with pNF-AT-Luc were assayed for the luciferase activity according to the manufacturer's instructions (Promega) (16).
In Vitro Binding Assay and Immunoprecipitation.
In vitro binding assays were performed as described by Lai et al. (15). For intracellular binding assay, cells were lysed in RIPA buffer [50 mM Tris⋅HCl (pH 7.4)/1% Nonidet P-40/0.25% Na-deoxycholate/50 mM NaCl/1 mM EDTA/1 mM PMSF/1 μg/ml each of aprotinin, leupeptin, and pepstatin/1 mM Na3VO4/1 mM NaF]. Cain/cabin1 was immunoprecipitated from cell lysates after incubation with anti-cain/cabin1 antibody and protein-A-coupled Sepharose CL-4B at 4°C for 2 h.
Results
Identification of Cain/Cabin1 as m-Calpain Substrate by in Vitro Screening.
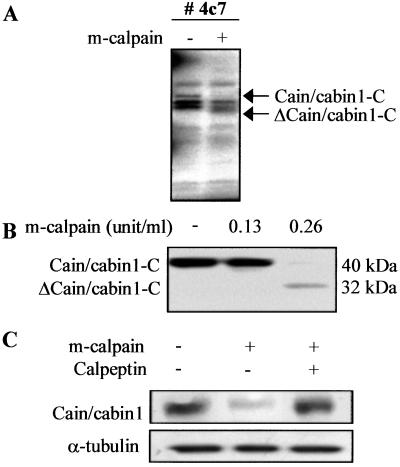
To identify new calpain substrates, we have developed an assay system, which is a modification of a method reported by Chung et al. (24), and screened a mouse thymus cDNA library by using purified m-calpain. Proteins were translated from small cDNA pools with TNT-coupled rabbit reticulocyte lysate in the presence of [35S]methionine, and then incubated with m-calpain (Fig. 1A). One such pool (no. 4c7) contained a putative calpain substrate, which after secondary screening and DNA-sequencing analysis, turned out to be the C-terminal region of mouse cain/cabin1 spanning residues 1852–2195 (cain/cabin1-C). Cleavage of this truncated cain/cabin1-C was confirmed to generate a 32-kDa protein by in vitro cleavage with the indicated amounts of m-calpain (Fig. 1B).
Figure 1.
In vitro cleavage of cain/cabin1 by purified m-calpain. (A) Small pools of cDNA (approximately 100) were translated in vitro in the presence of [35S]methionine, and then incubated with m-calpain (0.165 unit/ml). The reaction products and untreated controls were separated by SDS/PAGE and then exposed to x-ray film. Arrows indicate Cain/cabin1 and its cleavage product. (B) The C terminus of mouse cain/cabin1 (cain/cabin1-C) was labeled with [35S]methionine and incubated with the indicated amounts of m-calpain at 30°C for 1 h. ΔCain/cabin1-C indicates a cleavage product of cain/cabin1-C. (C) Cain/cabin1 was transiently expressed in HEK293T cells. One day later, cell extracts were prepared and incubated with m-calpain (0.75 unit/ml) in the presence or absence of calpeptin (20 μg/ml). The reaction products were analyzed by Western blotting with anti-cain/cabin1 antibody.
Full-length cain/cabin1 labeled with [35S]methionine was not synthesized by using the in vitro transcription/translation system, probably because of its large size (230 kDa). Instead, cain/cabin1 cleavage was indirectly assessed. After transient expression of cain/cabin1 in HEK293T cells, cell extracts were prepared and incubated with purified m-calpain. Subsequent Western blot analysis of the reaction products showed that the level of cain/cabin1 was reduced by treatment with m-calpain. This effect was blocked by calpeptin, a calpain inhibitor (Fig. 1C), indicating that cain/cabin1 cleavage is calpain-dependent.
Cain/Cabin1 Is Cleaved by Calpain During Calcium Ionophore-Induced Cell Death.
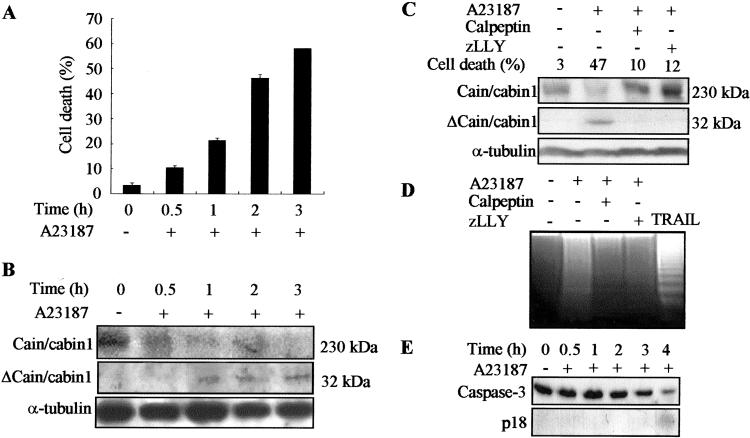
We then examined cain/cabin1 cleavage in the cells. Calcium ionophore increases intracellular calcium levels, leading to calpain activation and cell death. When Jurkat cells were exposed to the calcium ionophore A23187, cell death gradually increased (from 11% at 30 min to 58% at 3 h) (Fig. 2A). Examination of the cell extracts by Western blot analysis showed that by 30 min after treatment with A23187, full-length cain/cabin1 levels were reduced substantially. After 1 h of treatment, a 32-kDa protein was detected with anti-cain/cabin1 antibody and continued to accumulate further (Fig. 2B). Because anti-cain/cabin1 antibody was generated against purified cain/cabin1-C and recognized the C terminus of cain/cabin1, the 32-kDa protein detected by the antibody is the cleavage product containing the C-terminal region of cain/cabin1-spanning residues 1842–2182.
Figure 2.
Calpain-dependent cleavage of cain/cabin1 during A23187-induced cell death. (A) Jurkat cells were incubated with A23187 (1 μM) for the indicated times and cell viability (percent of cell death) was determined by trypan blue exclusion. Bars represent means ± SD from at least four independent experiments. (B) Cell extracts were analyzed by Western blotting with anti-cain/cabin1 antibody. Cain/cabin1 and Δcain/cabin1 indicate full-length (230 kDa) and a cleavage product (32 kDa) of cain/cabin1, respectively. (C) Jurkat cells were exposed to A23187 (1 μM) for 3 h in the presence or absence of the calpain inhibitors, calpeptin (40 μg/ml) and zLLY (20 μM), and analyzed by Western blotting. (D) Internucleosomal DNA fragmentation assays. Jurkat cells were exposed to A23187, and tumor necrosis factor-related apoptosis-inducing ligand (100 ng/ml) was used as a positive control of apoptosis. (E) Proteolytic activation of caspase-3 during A23187-induced cell death was assessed by Western blot analysis. P18 indicates the processed subunit (18 kDa) of caspase-3.
When Jurkat cells were preincubated for 30 min with the calpain inhibitors including calpeptin and zLLY, exposure to A23187 did not induce cain/cabin1 cleavage, indicating that calpain cleaves cain/cabin1 in Jurkat cells (Fig. 2C). Calpain inhibitors also suppressed A23187-induced cell death (47%) to the control levels (10% and 12%). Calcium ionophore treatment elicited cell death with characteristics of apoptosis; Jurkat cells exposed to calcium ionophore became rounded, shrank with condensed nuclei, and showed internucleosomal DNA cleavage and caspase-3 activation (Fig. 2 D and E). In addition, calpain inhibitors suppressed DNA fragmentation, indicating that calpain plays a crucial role in A23187-induced cell death.
Calpain activation was then addressed in Jurkat cells exposed to A23187 by using Rhodamine-110, a cell-permeable fluorogenic substrate (Fig. 3A). Quantitative determination of the fluorescence emitted as a result of calpain activation showed that calpain was activated as early as 30 min in Jurkat cells after exposure to A23187 and remained activated during the treatment (Fig. 3A). Calpain activation was further confirmed by Western blot analysis showing proteolytic degradation of the small subunit of calpain (data not shown) (25).
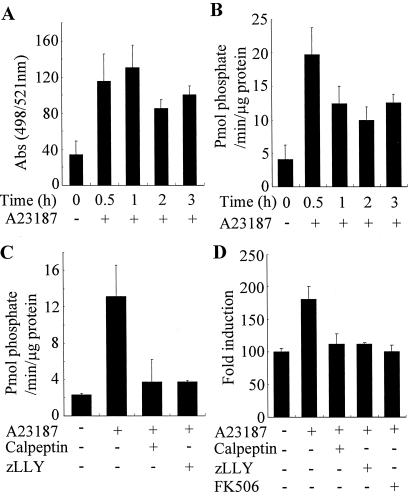
Figure 3.
Suppression of A23187-induced Cn activation by calpain inhibitors in vitro and in vivo. (A) Activity assay of calpain in vivo. Jurkat cells were exposed to A23187 (1 μM) for the indicated times, and calpain activation was then examined by the cell-permeable fluorogenic calpain substrate (Rhodamine-110). (B) Activity assay of Cn in vitro. Cell extracts prepared from Jurkat cells exposed to A23187 were assayed for Cn activity. The Cn activity determined from four independent experiments is indicated as picomoles of phosphate per minute per microgram of protein. (C) Calpain-mediated activation of Cn. Jurkat cells were exposed for 4 h to A23187 in the presence or absence of calpeptin (40 μg/ml) or zLLY (20 μM), and cell extracts were then assayed for the Cn activity. (D) Calpain-mediated activation of Cn in vivo. Jurkat cells were transfected with pNF-AT-luc and pβactgal as an internal control. After 48 h, cells were treated with A23187 for 4 h in the presence or absence of calpeptin (40 μg/ml), zLLY (20 μM), or FK506 (3 μM), and luciferase activity was then measured.
Cn Is Activated in a Calpain-Dependent Manner to Mediate Calcium Ionophore-Induced Cell Death.
Cain/cabin1 is an endogenous inhibitor of Cn (15, 16). We therefore measured Cn activity during A23187-induced cell death (Fig. 3B). Determination of Cn activity in vitro showed that Cn activity was induced in Jurkat cells exposed to A23187 and undergoing cell death. Cn activity was maximal at 30 min after treatment with calcium ionophore and remained high in dying cells (Fig. 3B). When Jurkat cells were preincubated with the calpain inhibitors, calpeptin and zLLY, Cn activation evoked by A23187 was suppressed to the control level, indicating that Cn is activated in a calpain-dependent manner (Fig. 3C). We further examined in vivo activation of Cn by measuring the NF-AT activity by using the reporter gene under the control of the NF-AT-binding site derived from the IL-2 promoter (Fig. 3D) (16). Treatment of Jurkat cells with A23187 alone induced NF-AT activity about 2-fold; the A23187-induced NF-AT activation was diminished by calpain inhibitors, calpeptin and zLLY, and Cn inhibitor, FK506, respectively. These results indicate that Cn is activated in Jurkat cells undergoing apoptosis.
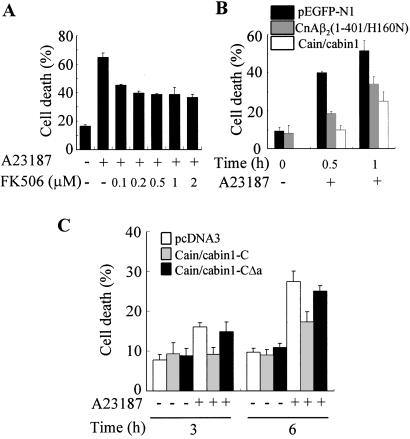
We then examined the contribution of Cn activation to A23187-induced cell death. FK506 markedly reduced A23187-induced cell death (65–44% at 0.1 μM) (Fig. 4A). In addition, transient expression of CnAβ2(1–401/H160N), a catalytically inactive mutant of Cn A, or cain/cabin1 in Jurkat cells also reduced substantially the calcium ionophore-induced cell death (Fig. 4B). Further cell death analysis by using Cn deletion mutants showed that expression of cain/cabin1-C, the C terminus of cain/cabin1 containing Cn-binding domain, significantly suppressed A23187-induced cell death (Fig. 4C). On the contrary, cain/cabin1-CΔa lacking a part of the Cn-binding domain in the C terminus did not inhibit A23187-induced cell death. These results indicate that Cn activation mediates A23187-induced cell death and that the Cn-binding domain of cain/cabin1 modulates the calcium-triggered cell death.
Figure 4.
Attenuation of A23187-induced cell death by treatment with FK506 and expression of either cain/cabin1, an inactive Cn mutant [CnAβ2(1–401/H160N)], or Cn deletions. (A) The effects of FK506 on A23187-induced cell death. Jurkat cells were exposed to A23187 (1 μM) for 4 h in the presence of the indicated amounts of FK506, and cell viability (percent of cell death) was determined. (B) Jurkat cells were transiently cotransfected with pEGFP-N1 and either pcDNA3, pCnAβ2(1–401/H160N), or pCain/cabin1. One day later, cells were exposed to A23187 for the indicated times, and cell viability was determined on the basis of the morphology of GFP-positive cells under a fluorescence microscope. (C) Effect of cain/cabin1 deletions, cain/cabin1-C and cain/cabin1-CΔa (see diagram in Fig. 5A), on A23187-induced cell death. Jurkat cells were transfected for 24 h with pEGFP-N1 and either pcDNA3-HA, pHA-cain/cabin1-C, or pHA-cain/cabin1-CΔa and then exposed to A23187 for the indicated times. The data shown are the means ± SD from at least three independent experiments.
Calpain Cleaves the Cn-Binding Domain of Cain/Cabin1.
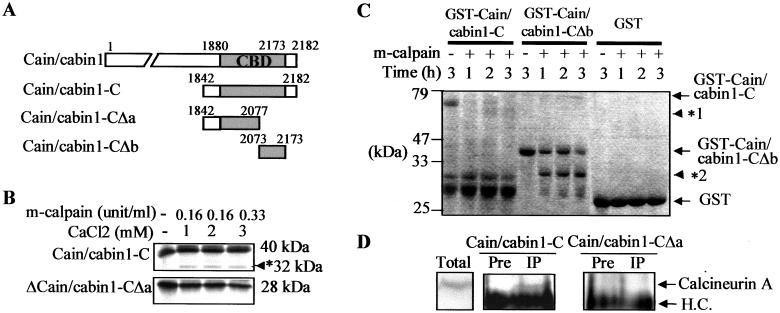
We determined the calpain cleavage site in the C terminus of cain/cabin1 by performing in vitro cleavage assays with deletion mutants (Fig. 5A). When a cain/cabin1-C mutant containing an intact Cn-binding domain (1880–2173) and cain/cabin1-CΔa (1842–2077) mutant containing half of the Cn-binding domain of cain/cabin1 were incubated with calpain, cain/cabin1-CΔa was resistant to calpain (Fig. 5B). By contrast, the cain/cabin1-CΔb mutant (2073–2173) containing the other half of Cn-binding domain, but not GST, was cleaved by calpain in vitro to a similar extent as cain/cabin1-C (Fig. 5C). Because GST is fused to the N terminus of cain/cabin1-CΔb, we propose that calpain-mediated cleavage occurs near the N terminus of cain/cabin1-CΔb construct.
Figure 5.
In vitro mapping of calpain-sensitive region in the C terminus of cain/cabin1 and intracellular interaction with Cn A. (A) Schematic diagram of cain/cabin1 and its deletions, cain/cabin1-C, cain/cabin1-CΔa, and cain/cabin1-CΔb. Gray box indicates the Cn-binding domain (CBD). (B) Lack of calpain-mediated cleavage in the cain/cabin1-CΔa. Cain/cabin1-C and cain/cabin1-CΔa were labeled with [35S]methionine and subjected to in vitro cleavage assays. Asterisk indicates a cleavage product of cain/cabin1-C. (C) Purified GST, GST-cain/cabin1-C, and GST-cain/cabin1-CΔb fusion proteins were incubated with calpain (0.005 unit) at 30°C for the indicated times and then visualized by Coomassie blue staining after SDS/PAGE. Asterisks indicate the cleavage product of respective protein. (D) Intracellular binding assays. HEK293 cells were transiently cotransfected with pHA-CnA, pHA-CnB, and either pHA-cain/cabin1-C or pHA-cain/cabin1-CΔa. One day later, proteins of HEK293 cell extracts were immunoprecipitated with preimmune serum (Pre) or anti-cain/cabin1 antibody (IP), and immunoblotted with anti-Cn A antibody. Cn A and heavy chain (H.C.) of Ig are indicated with arrows in the immunoprecipitates (Middle) and in total cell extracts (Left).
Through immunoprecipitation assays in the transfected cells, we have confirmed that, in contrast to cain/cabin1-C, calpain-resistant cain/cabin1-CΔa could not form complexes with Cn A (Fig. 5D). Together with the observation that cain/cabin1-CΔa mutant could not suppress A23187-induced apoptosis as shown in Fig. 4C, these results imply that Cn binding of cain/cabin1 through the Cn-binding domain modulates the calcium-triggered cell death and thus calpain-mediated cleavage in the Cn-binding domain of cain/cabin1 prevents cain/cabin1 from binding to and inhibiting Cn.
Cain/Cabin1 Binds to Cn B-Binding Domain of Cn A.
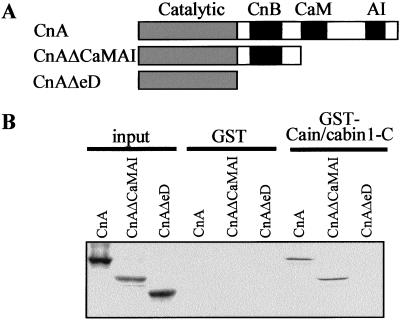
The cain/cabin1-binding region in Cn A, which interacted with Cn B and calmodulin (25, 26), was investigated by in vitro binding assays (Fig. 6). Purified GST-cain/cabin1-C, which contains the Cn-binding domain, bound to full-length Cn A and to a deletion mutant lacking the calmodulin-binding domain (CnAΔCaMAI), but not to a further deletion mutant lacking the Cn B-binding domain (CnAΔeD) (Fig. 6B). These results suggest that cain/cabin1 interacts with Cn A through its Cn B-binding domain.
Figure 6.
In vitro binding assay identifying the cain/cabin1-binding domain in the Cn A subunit. (A) Schematic diagram showing deletion mutants of Cn A (CnA): The CnAΔCaMAI mutant lacks the calmodulin (CaM)-binding and autoinhibitory (AI) domains and the CnAΔeD mutant lacks Cn B (CnB)-binding region and contains only the catalytic domain. (B) Purified GST and GST-cain/cabin1-C were subjected to in vitro binding assays by using [35S]methionine-labeled full-length or deletions of Cn A. Interacting proteins bound to GST-cain/cabin1-C were pulled down with GST-Sepharose, separated by SDS/PAGE, and exposed to x-ray film for autoradiography.
Discussion
Calpain is a critical mediator of cell death triggered by calcium signals and the ability of calpain to cleave a growing number of substrates suggests a potentially important role for this enzyme in the regulation of cell death. Thus, we have isolated cain/cabin1 as a new calpain substrate by a systemic and functional screening strategy. Although numerous other cysteine and serine proteases in cells exist, we have provided several lines of evidence demonstrating that calpain is likely to cleave cain/cabin1. (i) Degradation of cain/cabin1 was observed by incubation of cell extracts with purified m-calpain and its cleavage was suppressed by calpain inhibitors. (ii) In vitro cleavage assays showed that cain/cabin1 was cleaved by calpain. (iii) Time-response studies in A23187-treated cells revealed a close relationship between calpain activation, degradation of cain/cabin1, and activation of Cn. (iv) Cain/cabin1 cleavage and Cn activation during A23187-induced cell death were simultaneously suppressed by calpain inhibitors, but not by a caspase inhibitor. Although caspase was activated at a later stage of cell death, cain/cabin1 was not cleaved by caspase (data not shown).
To examine whether the cleavage of cain/cabin1 caused or resulted from cell death, we monitored cain/cabin1 cleavage throughout a time course relative to the progression of cell death. When calpain and Cn were activated with the appearance of dead cells, the degradation of full-length cain/cabin1 occurred at an early time (30 min). In addition, the cleavage-resistant truncated cain/cabin1 could not bind to Cn A and the cleavage occurred in the Cn-binding domain, implying that calpain-mediated cleavage of cain/cabin1 may result in a loss of its ability to bind to and inhibit Cn. Furthermore, overexpression of cain/cabin1 suppressed A23187-induced cell death as efficiently as an inactive Cn mutant [CnAβ2(1–401/H160N)], indicating that inactivation of cain/cabin1, presumably by proteolysis, is crucial for A23187-induced cell death. The data presented here provide evidence for calpain-dependent activation of Cn in A23187-induced cell death, suggesting a potential means by which calcium signaling in pathogenic cell death may be manipulated.
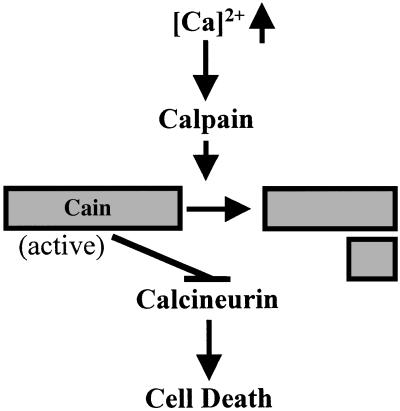
Cain/cabin1 may bind to Cn through its Cn B-binding domain to inhibit Cn activity. Cn (PP2B) consists of a catalytic (Cn A) and a regulatory subunit (Cn B) in which Cn B interacts with Cn A through the CnB-binding domain (1, 3, 25). At the present time, although the stoichiometry of Cn complexes is not yet clear in living and dying cells, cain/cabin1 binds to the CnB-binding domain of Cn A and may compete with Cn B to modulate Cn activity. We propose that under apoptotic stimuli, cain/cabin1 is cleaved by calcium-activated calpain and the cleavage of cain/cabin1 may allow cain/cabin1 to be inactivated and dissociated from the Cn complex, leading to the activation of Cn (Fig. 7).
Figure 7.
Schematic diagram of the proposed model. Cain/cabin1 is cleaved by calpain to be inactivated, and Cn is subsequently activated to mediate calcium-ionophore-induced cell death.
Cn is likely to be downstream of the inositol-1,4,5-triphosphate receptor in the cell death pathway (27). Cn plays a critical role in calcium-mediated cell death by triggering mitochondrial targeting and dephosphorylation of BAD, a proapoptotic member of the Bcl-2 family, thus enhancing BAD heterodimerization with Bcl-XL (19, 28). In contrast, Bcl-2 seems to regulate Cn-mediated cell death by binding and sequestering Cn, thereby blocking NF-AT signaling (29, 30). Recently, Cn signals were reported to be integrated by NF-AT to mediate T cell apoptosis through Nur77 (31–33). We also speculate that other downstream effectors of calpain may contribute to the calcium-induced cell death, including caspase-12 and cdk5 (12, 13), because, in contrast to calpain, inhibition of Cn partially blocks the calcium-induced cell death.
Although calcium effects are so widespread that it has been difficult to define specific mechanisms, intracellular increase of calcium elicits diverse pathological effects through cell death in cerebral ischemia and other neural insults including glutamate, kainite, maitotoxin, and brain trauma, and neurodegenerative processes such as Alzheimer's disease (2, 5, 6, 35–38). A wide range of stimuli eliciting apoptotic cell death acts through elevations in intracellular calcium and activations of calpain. Accordingly, cain/cabin1-mediated apoptosis described here may be relevant to multiple forms of cell death.
Acknowledgments
We thank Drs. N. Spoerel and J. Bandyopabhyay for critical reading of this manuscript and technical support. This work was supported by the grant from the National Research Laboratory Program (to Y.J.) and the Life Phenomena and Function Research Group Program (2000) of the Korean Ministry of Science and Technology.
Abbreviations
- HA
hemagglutinin
- Cn
calcineurin
- GST
glutathione S-transferase
- NF-AT
nuclear factor of activated T cells
References
- 1.Croall D E, DeMartino G N. Physiol Rev. 1991;71:813–847. doi: 10.1152/physrev.1991.71.3.813. [DOI] [PubMed] [Google Scholar]
- 2.Saido T C, Sorimachi H, Suzuki K. FASEB J. 1994;8:814–822. [PubMed] [Google Scholar]
- 3.Huang Y, Wang K K. Trends Mol Med. 2001;7:355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- 4.Margaret K, Squier T, John C. J Immunol. 1997;158:3690–3697. [PubMed] [Google Scholar]
- 5.Saito K, Elce J S, Hamos J E, Nixon R A. Proc Natl Acad Sci USA. 1993;90:2628–2632. doi: 10.1073/pnas.90.7.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuji T, Shimohama S, Kimura J, Shimizu K. Neurosci Lett. 1998;248:109–112. doi: 10.1016/s0304-3940(98)00348-6. [DOI] [PubMed] [Google Scholar]
- 7.Shields D C, Ray S K, Gantt-Wilford G, Banik N L. J Neurosci Res. 1998;53:482–489. doi: 10.1002/(SICI)1097-4547(19980815)53:4<482::AID-JNR10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Markgraf C G, Velayo N L, Johnson M P, McCarty D R, Medhi S, Koehl J R, Chmielewski P A, Linnik M D. Stroke. 1998;29:152–158. doi: 10.1161/01.str.29.1.152. [DOI] [PubMed] [Google Scholar]
- 9.Li P A, Howlett W, He Q P, Miyashita H, Siddiqui M, Shuaib A. Neurosci Lett. 1998;247:17–20. doi: 10.1016/s0304-3940(98)00266-3. [DOI] [PubMed] [Google Scholar]
- 10.Yokota M, Tani E, Tsubuki S, Yamaura I, Nakagaki I, Hori S, Saido T C. Brain Res. 1999;819:8–14. doi: 10.1016/s0006-8993(98)01334-1. [DOI] [PubMed] [Google Scholar]
- 11.Toda G, Matsushita S, Kuramoto K, Oda S, Ezaki H, Hattori A, Kawashima S. Jpn Heart J. 1989;30:375–386. doi: 10.1536/ihj.30.375. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa T, Yuan J. J Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M S, Kwon Y T, Li M, Peng J, Friedlander R M, Tsai L H. Nature (London) 2000;1405:360–364. doi: 10.1038/35012636. [DOI] [PubMed] [Google Scholar]
- 14.Kusakawa G I, Saito T, Onuki R, Ishguro K, Kishimoto T, Hisanaga S-I. J Biol Chem. 2000;275:17166–17172. doi: 10.1074/jbc.M907757199. [DOI] [PubMed] [Google Scholar]
- 15.Lai M M, Burnett P E, Wolosker H, Blackshaw S, Snyder S H. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Youn H D, Loh C, Stolow M, He W, Liu J O. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- 17.Lai M M, Luo H R, Burnett P E, Hong J J, Snyder S H. J Biol Chem. 2000;275:34017–34020. doi: 10.1074/jbc.C000429200. [DOI] [PubMed] [Google Scholar]
- 18.Shibasaki F, McKeon F. J Cell Biol. 1995;131:735–743. doi: 10.1083/jcb.131.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H G, Pathan N, Ethell I M, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke T F, Reed J C. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 20.Youn H D, Sun L, Prywes R, Liu J O. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 21.Kurz J E, Sheets D, Parson J T, Rana A, Delorenzo R J, Churn S B. J Neurochem. 2001;78:304–315. doi: 10.1046/j.1471-4159.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- 22.Jung Y K, Yuan J. Oncogene. 1997;14:1207–1214. doi: 10.1038/sj.onc.1200943. [DOI] [PubMed] [Google Scholar]
- 23.Eastman A. Methods Cell Biol. 1995;46:41–55. doi: 10.1016/s0091-679x(08)61923-8. [DOI] [PubMed] [Google Scholar]
- 24.Chung C W, Song Y H, Kim I K, Yoon W J, Ryu B R, Jo D G, Woo H N, Kwon Y K, Kim H H, Gwag B J, et al. Neurobiol Dis. 2001;8:162–172. doi: 10.1006/nbdi.2000.0335. [DOI] [PubMed] [Google Scholar]
- 25.Yoshizawa T, Sorimachi H, Tomioka S, Ishiura S, Suzuki K. Biochem Biophys Res Commun. 1995;208:376–383. doi: 10.1006/bbrc.1995.1348. [DOI] [PubMed] [Google Scholar]
- 26.Klee C B, Ren H, Wang X. J Biol Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 27.Jayaraman T, Marks A R. J Biol Chem. 2000;275:6417–6420. doi: 10.1074/jbc.275.9.6417. [DOI] [PubMed] [Google Scholar]
- 28.Springer J E, Azbill R D, Nottingham S A, Kennedy S E. J Neurosci. 2000;20:7246–7251. doi: 10.1523/JNEUROSCI.20-19-07246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibasaki F, Kondo E, Akagi T, McKeon F. Nature (London) 1997;386:728–731. doi: 10.1038/386728a0. [DOI] [PubMed] [Google Scholar]
- 30.Asai A, Qiu J-h, Narita Y, Chi S, Saito N, Shinoura N, Hamada H, Kuchino Y, Kirino T. J Biol Chem. 1999;274:34450–34458. doi: 10.1074/jbc.274.48.34450. [DOI] [PubMed] [Google Scholar]
- 31.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 32.Youn H D, Liu J O. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 33.Youn H D, Chatila T A, Liu J O. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z Y, Thieme-Sefler A M, Maclean D, McNamara D J, Dobrusin E M, Sawyer T K, Dixon J E. Proc Natl Acad Sci USA. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K S, Frank S, Vanderklish P, Arai A, Lynch G. Proc Natl Acad Sci USA. 1991;88:7233–7237. doi: 10.1073/pnas.88.16.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hajimohammadreza I, Raser K J, Nath R, Nadimpalli R, Scott M, Wang K K W. J Neurochem. 1997;69:1006–1013. doi: 10.1046/j.1471-4159.1997.69031006.x. [DOI] [PubMed] [Google Scholar]
- 37.Saatman K E, Murai H, Bartus R T, Smith D H, Hayward N J, Perri B R, McIntosh T K. Proc Natl Acad Sci USA. 1996;93:3428–3433. doi: 10.1073/pnas.93.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J. Nature (London) 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]