Abstract
A patient with a typical form of chronic myeloid leukemia was found to carry a large deletion on the derivative chromosome 9q+ and an unusual BCR-ABL transcript characterized by the insertion, between BCR exon 14 and ABL exon 2, of 126 bp derived from a region located on chromosome 9, 1.4 Mb 5′ to ABL. This sequence was contained in the bacterial artificial chromosome RP11-65J3, which in fluorescence in situ hybridization experiments on normal metaphases was found to detect, in addition to the predicted clear signal at 9q34, a faint but distinct signal at 22q11.2, where the BCR gene is located, suggesting the presence of a large region of homology between the two chromosomal regions. Indeed, blast analysis of the RP11-65J3 sequence against the entire human genome revealed the presence of a stretch of homology, about 76 kb long, located approximately 150 kb 3′ to the BCR gene, and containing the 126-bp insertion sequence. Evolutionary studies using fluorescence in situ hybridization identified the region as a duplicon, which transposed from the region orthologous to human 9q34 to chromosome 22 after the divergence of orangutan from the human-chimpanzee-gorilla common ancestor about 14 million years ago. Recent sequence analyses have disclosed an unpredicted extensive segmental duplication of our genome, and the impact of duplicons in triggering genomic disorders is becoming more and more apparent. The discovery of a large duplicon relatively close to the ABL and BCR genes and the finding that the 126-bp insertion is very close to the duplicon at 9q34 open the question of the possible involvement of the duplicon in the formation of the Philadelphia chromosome translocation.
The Philadelphia chromosome (Ph), the most frequent cytogenetic abnormality present in human leukemias, is a derivative chromosome 22 arising as a consequence of a reciprocal translocation between the long arms of chromosomes 9 and 22 (1, 2). The spectrum of leukemia phenotypes associated with the presence of a Ph is extremely wide, ranging from chronic myeloid leukemia (CML) to acute lymphoid leukemia and acute myeloid leukemia (3, 4). The presence of a Ph always parallels the presence of a rearrangement between the BCR and the ABL genes, which presents a certain degree of molecular variability, giving rise to different types of BCR/ABL transcripts and proteins that show a preferential, although not exclusive, association with certain leukemia phenotypes (3, 4).
Besides the molecular variability, a cytogenetic variability is also present in Ph-positive leukemias. It has long been known that 5–6% of patients with Ph-positive leukemias show variant Ph translocations involving other chromosomes in addition to 9 and 22 (5), and that more than half of the cases apparently negative for the presence of a classical Ph show a BCR-ABL rearrangement arising from a transposition of the ABL gene to chromosome 22 or of the BCR gene to chromosome 9 (6).
Recently, another cytogenetic variability of the CML patients possibly bearing important prognostic implications has been pointed out by Sinclair et al. (7). Fluorescence in situ hybridization (FISH) showed that a substantial minority of patients (about 15–20%) bear large deletions adjacent to the translocation breakpoint on the derivative chromosome 9 [(der(9)]. The same abnormalities have been described to occur also in Ph-positive acute lymphoid leukemia (8). The deletions span up to several megabases and have variable breakpoints. In CML, these deletions have been found to be associated with poor prognosis in terms of overall survival, and multivariate analysis demonstrates that their prognostic importance is independent of other parameters (7–9). At the moment, although we know that the Ph translocation causes the formation of a fusion oncogene essential for leukemogenesis, the molecular processes underlying the formation of the Ph translocation remain elusive. The analysis of the presence of sequences potentially relevant to chromosomal translocations as Alu repeated elements, Chi-like octamers, Ig heptamers, a-protein binding sites surrounding the breakpoint positions on both derivative chromosome 9q+ and 22q− in a number of cloned BCR-ABL rearrangements, has been inconclusive (10). This is intriguing in light of the fact that very low amounts of BCR-ABL fusion transcripts may be found in a percentage of adult people who will never develop a leukemia phenotype (11, 12), suggesting that the mechanisms underlying the Ph translocation may produce many more exchanges between chromosomes 9 and 22 than those that will eventually become clinically relevant.
Here we report on a patient bearing a classical form of CML, in which reverse transcriptase–PCR (RT-PCR) revealed 126 bp inserted between the ABL and BCR genes. blast search identified the insertion as derived from the genomic sequence AL353803 [bacterial artificial chromosome (BAC) RP11-65J3] mapping approximately at 1.4 Mb 5′ to the ABL gene. Surprisingly, we found that the RP11-65J3 contains a large region, 48 kb 3′ to the 126 bp, duplicated on chromosome 22, contained within the sequence AP000345, 150 kb 3′ of BCR. After these results, we decided to further investigate the nature and the evolution of this duplicon and its potential implication on 9/22 translocation.
Materials and Methods
Patient.
A 50-year-old male patient was diagnosed with chronic-phase Ph-positive CML in November 1999. His peripheral blood showed: WBC, 75.6 × 109/liter; platelet count, 842 × 109/liter; hemoglobin level, 14.7 g/dl. Liver and spleen were not enlarged. Bone marrow examination showed a marked hypercellularity with an increased number of megakaryocytes, myeloid hyperplasia with basophilia, and 1% myeloblasts. Cytogenetic analysis revealed the Ph in all 20 metaphases examined without other abnormalities. Hydroxyurea and IFN-α were started. Currently, the patient shows a complete hematological remission but only a minor cytogenetic response.
Cytogenetic Analysis, RT-PCR Analysis, and Sequencing.
For cytogenetic analyses, cells from bone marrow were cultured in McCoy's medium supplemented with 20% FCS and antibiotics for 48 h. Chromosome preparations were stained with 5% Giemsa solution according to standard procedures.
The RT-PCR procedure was performed with the primers and the protocol established by the European BIOMED 1 Concerted Action (13). The purified PCR products were sequenced by using the Thermo Sequenase Cy5.5 dye terminator cycle sequencing kit (Amersham Pharmacia).
FISH.
FISH hybridizations were performed essentially as described by Lichter et al. (14). Interspecies FISH was performed at low stringency (15). Digital images were obtained by using a Leica DMRXA epifluorescence microscope equipped with a cooled charge-coupled device camera (Princeton Instruments, Princeton, NJ). Cy3 (Perkin–Elmer) and 4′,6′,diamidino-phenylindole dihydroclhoride (DAPI) fluorescence signals, detected by using specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed with Adobe photoshop software.
BAC and P1 artificial chromosome probes, belonging to the Roswell Park Cancer Institute and P. de Jong libraries, were obtained from the Sanger Centre (http://www.sanger.ac.uk) or directly from P. de Jong, Children's Hospital Oakland Research Institute, Oakland, CA (http://www.chori.org/bacpac/). CalTech human BAC library A clones (CTA) were from the Sanger Centre. The precise position of probes on the sequence of chromosome 22 was established by blast search of the end sequences of the clone against the complete sequence of this chromosome. The analysis of the human chromosome 22 and chromosome 9 genomic sequences were based mostly on the University of California Santa Cruz Human Genome Project Working Draft [December 22, 2001 assembly (hg10), available at http://genome.cse.ucsc.edu/]. pipmaker analysis was performed at the web site http://nog.cse.psu.edu/cgi-bin/pipmaker?advanced, according to the authors' instructions (16).
Evolutionary Studies.
Evolutionary studies were performed on great apes (Pan troglodytes, PTR; gorilla, Gorilla gorilla, GGO; orang, Pongo pygmaeus), and representative species of Old World monkeys (macaque, Macaca mulatta) and New World monkeys (squirrel monkey, Saimiri sciureus; dusky titi Callicebus molloch, CMO). The evolutionary history of chromosome 9 has been described (15).
Results
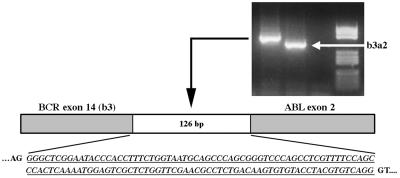
Bone marrow cytogenetic analysis of the patient did not reveal any other abnormality besides the presence of a common t(9;22)(q34;q11) translocation. By contrast the diagnostic RT-PCR analysis, aimed at detecting the hybrid BCR-ABL transcript, revealed the presence of an amplification band longer than the one expected from the fusion between BCR exon 14 and ABL exon 2 (e14a2 fusion) (Fig. 1). Direct sequencing of the band excised from the gel showed an insertion of 126 bp positioned exactly between the BCR exon 14 and ABL exon 2 boundaries and able to maintain the correct reading frame of the ABL portion.
Figure 1.
RT-PCR analysis showing the unusual BCR-ABL transcript present in the patient characterized by the insertion of 126 bp (italics underlined) between BCR exon 14 and ABL exon 2.
blast search (17) revealed that the 126-bp fragment was contained within the genomic sequence AL353803, from BAC RP11-65J3, chromosome 9, and mapping approximately 1,400 kb centromeric to the ABL gene (http://www.ensembl.org). The fragment was preceded and followed by typical splicing recognition signals, suggesting that it corresponded to an exon. Furthermore, the sequence was identical to several expressed sequence tags (GenBank accession numbers BI518252, BI517686, BF699626, H43169, H22380, BF680532, and H30889), which belong to the UniGene cluster Hs.349544. This cluster contains expressed sequence tag clones derived from a variety of tissues and cells, including brain, retina, prostate, fetal liver, spleen, fibroblasts, kidney, and colon. Altogether, these blast findings confirm that the inserted fragment is indeed part of an uncharacterized transcript with a wide pattern of expression. It must be noted that the 126-bp inserted fragment partially recognized in blast analysis two additional sequences located on human chromosome 5 (90% identity) and on chromosome 12 (86% identity). The latter sequences are neither preceded nor followed by canonical splicing sites and are not represented in expressed sequence tag databases, which suggests that they are not transcribed.
PCR experiments performed on the patient's genomic DNA, using a forward primer derived from BCR exon 14 and a reverse primer from the 126-bp inserted sequence, produced an amplification product of 1.5 kb. Its sequencing revealed the exact genomic breakpoint between chromosome 22 and chromosome 9 on the Ph, which was located 155 bp downstream to BCR exon 14, within intron 14 of the gene, and 886 bp upstream of the 126-bp inserted exon on chromosome 9 (data not shown).
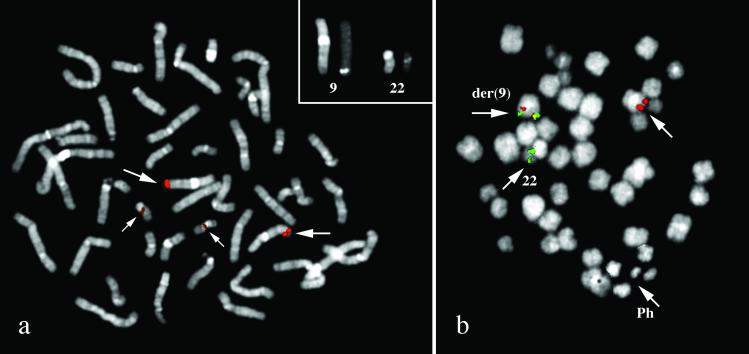
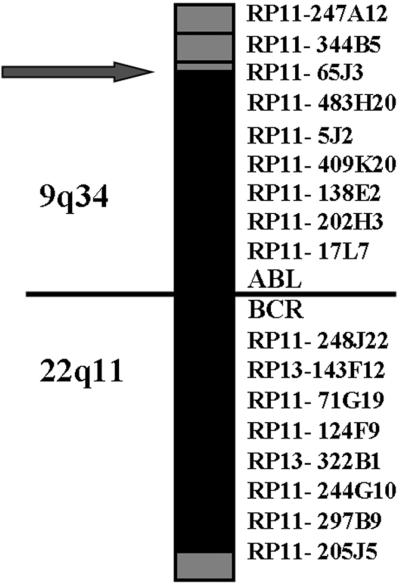
To further investigate the involvement of the BAC RP11-65J3 in the rearrangement we used this probe in FISH experiments. Before using it in the patient, we validated the probe on normal metaphases. Surprisingly, the probe, in addition to the expected signal at 9q34, also gave a faint but distinct signal at 22q11.2 where the BCR gene is located (Fig. 2a), suggesting the presence of a duplicon. In the patient, by contrast, the BAC probe gave on der(9) a signal of lesser intensity in respect to the normal 9 and did not yield any signal on the Ph (Fig. 2b). This finding suggested a deletion of most of the sequences of the BAC RP11-65J3, expected to be translocated on der(22), and that the genomic sequences from RP11-65J3 inserted on der(22) (see above) were too short to give an appreciable signal. The D-FISH probe (Vysis, Downer's Grove, IL), designed to detect the rearrangements on both derivative chromosomes, showed the presence of a fusion signal on the Ph but not on der(9), further suggesting a deletion similar to those described by Sinclair et al. (7). The exact extension of the deletion was investigated by FISH experiments using two appropriate panels of BAC/P1 artificial chromosome probes encompassing the regions centromeric to the ABL gene and telomeric to the BCR gene, respectively. A summary of the results is reported in Fig. 3. The deletion of the sequences 5′ to ABL spanned the entire segment of 1,400 kb between RP11-65J3 and the ABL gene, whereas the deletion of sequences 3′ to BCR extended approximately for 1.4 Mb.
Figure 2.
(a) FISH analysis with BAC RP11-65J3 on a normal metaphase. In addition to the region at 9q34, the probe is enlightening also a region at 22q11.2 where the BCR gene is located, suggesting the presence of a region of homology between the two chromosomes. (b) FISH analysis with BAC RP11-65J3 on a patient's metaphase. The BAC RP11-65J3 (red) yielded a signal of lesser intensity on the derivative chromosome 9q+, and no signal on the derivative chromosome 22q−. CTA-221G9 probe (mapping at 22q11.2, distal to 3′BCR, in green) is translocated on der(9). (Magnification: ×1,500.)
Figure 3.
Map of the deletions on the derivative chromosome 9q+. BAC and P1 artificial chromosome probes corresponding to the regions centromeric to the ABL gene and telomeric to the BCR gene. Gray boxes indicate region retained; black boxes region deleted.
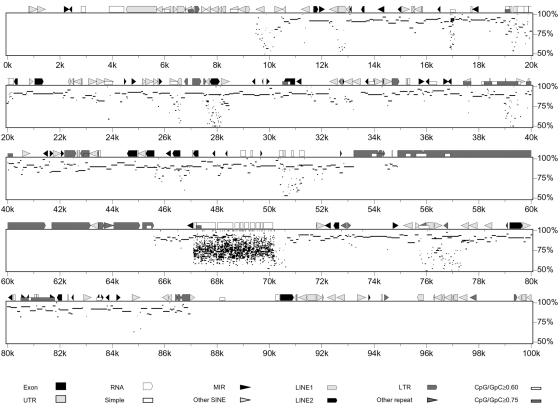
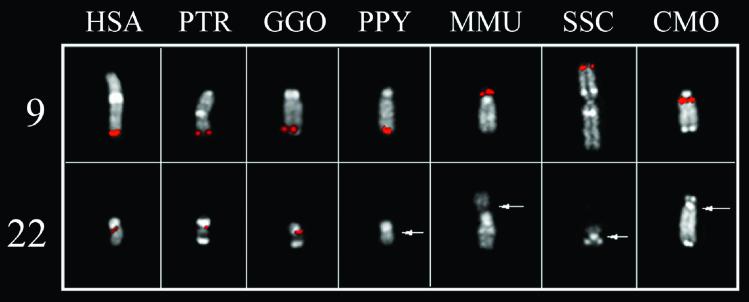
The AL353803 sequence (BAC RP11-65J3) was used to search for homologies against the entire human genome, by using blast software. A long stretch of homology was found on chromosome 22 sequence AP000345. A similar analysis was performed by using the entire AP000345 sequence as query. The region of similarity on chromosome 9 extended beyond AL353803, to the adjacent sequences, namely AC073075, derived from clone RP11-253N4, and AL391056 from clone RP11-492E3. The graphic representation of the homology between chromosome 22 and chromosome 9, obtained with pipmaker software, is reported in Fig. 4, where AP000345 is the reference sequence. It must be noted that the chromosome 9 sequence is still unfinished, with some gaps still present, and therefore we cannot exclude that the extent of sequence homology on this chromosome may differ from the above reported value. The 126-bp insertion is within the duplication, at about 35 kb from its centromeric border. The duplication hypothesis was investigated by tracking the evolutionary history of the sequence. FISH experiments using BAC RP11-65J3 as probe were performed on metaphase spreads of great apes and representatives of Old and New World monkeys (PTR, GGO, P. pygmaeus, M. mulatta, S. sciureus, and C. molloch). Hybridization signals were found in all species at regions orthologous to human 9q34. On the contrary, cross-hybridization signals on regions orthologous to chromosome 22 were detected only in Homo sapiens (HSA), PTR, and GGO (Fig. 5). Additional experiments were designed to clarify this point. Fragments generated by long PCR and derived from the duplicated region of chromosome 22, whose sequence is complete, were pooled to reach almost contiguous DNA stretches of about 20 kb and used in FISH experiments on the same primate species. Each pool detected main signals at phylogenetic 22q11.2 and cross-hybridization signals at phylogenetic 9q34 (data not shown). By contrast, the pool of long PCR products from sequences close to, but external to, the duplicated region did not yield any signal on phylogenetic chromosome 9 (data not shown). These overall results strongly suggest that the transposition event occurred after orang divergence from HSA-PTR-GGO ancestor, that is, about 14 million years ago (18).
Figure 4.
Results of pipmaker analysis. The sequence AP000345 (chromosome 22) has been plotted against a chromosome 9 composite sequence derived from the University of California Santa Cruz Human Genome Project web site (http://genome.cse.ucsc.edu/) and spanning the genomic clones RP11-65J3, RP11-253N4, and RP11-492E3. Note the long stretch of high homology from 10 kb through 87 kb. The apparent lack of homology between 55 and 65 kb is caused by the presence of long-terminal repeats that have therefore been masked in the course of the pipmaker analysis.
Figure 5.
FISH experiments with the RP11-65J3 probe on metaphases of great apes and Old and New World monkeys. Hybridization signals on regions hortologous to human 9q34 are evident in all species. On the contrary, cross-hybridization signals in regions hortologous to chromosome 22 are observed only in HSA, PTR, and GOO. Chromosome 22 in M. mulatta (MMU) and C. molloch (CMO), as well as chromosome 9 in S. sciureus (SSC), are part of a bigger chromosome.
Discussion
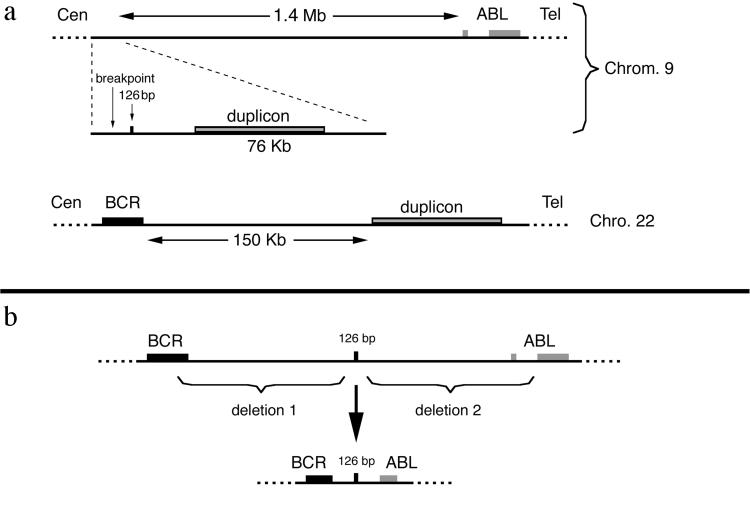
In this article we report on a CML patient showing an insertion of 126 bp in the BCR-ABL hybrid transcript typical of this disease, deriving from sequences upstream of the ABL locus. This finding led us to discover the existence of a 76-kb duplicon located 1,400 kb 5′ to ABL and 150 kb 3′ to BCR.
The evolutionary history of the duplicon was investigated by comparative FISH studies in great apes and selected Old World and New World primate species, using appropriate probes. The results clearly indicated that the transposition took place at the time of divergence of the HSA-PTR-GGO common ancestor from orang, which occurred approximately 14 million years ago (18). Duplicons are chromosome-specific low-copy repeats, ranging from a few kb to 200 kb and representing about 5% of the human genome (19). The divergence process between duplicated segments can generate new genes over evolutionary time, and we have just described an extraordinary example (20). On the other hand, the chance of misalignment during meiosis can result in “genomic disorders” (21). The role of duplicons in triggering genomic disorders is becoming more and more evident (22, 23). The recent examples reported by Giglio et al. (24) is paradigmatic in this respect. A series of 8p constitutional chromosomal changes, usually considered as occurring de novo, have been shown, by contrast, to be the direct consequence of a specific genomic arrangement in which the duplicons, represented by olfactory receptor gene clusters at 8p, play a crucial role. More recently, we have documented that heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation, shading new light on interchromosomal relationships (25).
What about recurrent chromosomal changes frequently occurring in leukemias, and in tumors in general? Are duplicons involved in triggering mitotic changes in somatic cells? Our findings of a large duplicon near regions involved in a common translocation in human leukemias opens the question of the potential involvement of duplicons in triggering specific somatic exchanges. At the moment, however, if and in which way this duplicon could act in favoring the genesis of the Ph translocation is not clear.
We can speculate that the duplicon could play a role in the initial step of the process, simply by drawing together the specific chromosomal regions and favoring an exchange between them. In most CML cases, this exchange could result directly in the formation of a functional BCR-ABL rearrangement on the derivative chromosome 22, able to lead to the synthesis of a sufficient amount of BCR-ABL protein to confer a growth advantage to the cells, finally ending in the Ph-positive clone expansion.
In a minority of CML cases, however, the genesis of the Ph and that of the consequent BCR-ABL rearrangement appears to be the result of a more complex and maybe multistep event. Indeed, the occurrence of the so-called variant translocations, which involves other chromosomes in addition to chromosomes 9 and 22, supports this hypothesis. Also the finding of CML cases showing the presence of aberrant BCR-ABL transcripts showing the presence of sequences of unknown origin interposed between BCR and ABL (26–28) or unexpected duplications of BCR sequences present in both the BCR-ABL and the reciprocal ABL-BCR transcripts (26, 29–31) suggest that the formation of a BCR-ABL rearrangement may be achieved through the occurrence of a series of events, possibly driven by the selective growth advantage that the synthesis of a BCR-ABL protein may give.
In this context, the genesis of the deletions on the derivative chromosome 9q+ described by Sinclair et al. (7) is not clear at the moment. In particular, we do not know whether they simply are the result of a more complex mechanism in the genesis of the Ph and the consequent BCR-ABL rearrangement or whether they represent secondary events able to confer an additional functional advantage to the cells. Indeed, the two hypotheses may not be mutually exclusive.
The CML case reported in this article in our opinion could provide some insights on this matter. This patient, besides having the abnormal BCR-ABL transcript, is also characterized by the presence of large deletions of sequences located 5′ to the ABL gene on chromosome 9 and 3′ to the BCR gene on chromosome 22 and expected to be on the derivative chromosome 9 as a consequence of the t(9;22) translocation. The finding in this case that a small piece of the segment where the deletion begins on chromosome 9 is found on the Ph between the BCR and the ABL sequences suggests that the original breakpoint position on chromosome 9 was indeed in the segment AL353803 (and therefore very 5′ to the ABL gene) (Fig. 6a) and that the sequences between the AL353803 segment and the ABL gene were lost after their translocation to chromosome 22. Moreover, the concomitant presence of a large deletion 3′ to the BCR gene suggests that a very complex process took place in this case. It is not easy to perceive how these complex rearrangements could have occurred all at the same time. In our opinion, a multistep process is more likely. As a first step, we hypothesize a t(9;22) translocation involving sequences located very 3′ to the BCR gene on chromosome 22 and very 5′ to ABL on chromosome 9 and giving rise to an ancestral Ph in which the BCR and ABL genes were quite faraway and, therefore, not able to produce a functional fusion protein. Subsequent steps characterized by deletions on this ancestral Ph could have brought the BCR and ABL in the final type of rearrangement, able to give rise to hybrid transcripts and functional fusion proteins and selected by growth advantage. According to this model, at least in some instances, the cases characterized by the presence of deletions on the derivative chromosome 9q+ can actually represent cases in which the deletions took place on the Ph in a second time and after the original translocation. This hypothetical model has been illustrated in the scheme of Fig. 6b. Some of the CML cases reported in the literature in which, as in our case, sequences of unknown origin have been found interposed between the BCR and ABL sequences in the fusion transcripts (26–28) could fit with this model. Indeed, in one of these cases described only in an abstract,** the “alien” intervening sequences were found to derive from a gene originally located very 3′ to the BCR gene on chromosome 22.
Figure 6.
(a) Maps of the position of the 76-kp duplicated region on chromosome 9 and on chromosome 22 with respect to the ABL and BCR genes. (b) Scheme of the possible events giving rise to the BCR-ABL rearrangement present in the patient.
The t(9;22) translocations are probably more frequent than we suspect, as suggested by the finding that small amounts of BCR-ABL transcripts can be found in a high percentage (approximately 30%) of adult normal subjects (11, 12). The presence of the identified duplicon in this region could represent a molecular basis to explain the frequency of this event. However, only in very few occasions are the t(9;22) translocations able to give rise to a functional BCR-ABL rearrangement and, as a consequence, to produce the appearance of a CML phenotype. The case reported in this article suggests that in some cases, this also can result through subsequent deletions, which bring together in a functional rearrangement originally distant BCR and ABL genes.
In this view, the formation of a BCR-ABL rearrangement as a consequence of the Ph translocation is a very rare and accidental occurrence in the context of a quite frequent event triggered by the plasticity of the human genome. Indeed, some studies have hypothesized that some sequence repeats, Alu sequences in particular, can be directly involved in mitotic exchanges (9). We think that these hypotheses complement our view and that improvements in our understanding of mitotic exchanges will probably help us in elucidating the mitotic role of duplicons. In conclusion, the findings we report draw attention to the potential role of duplicons in mediating mitotic exchanges underlying the formation of translocations that characterize human tumors.
Acknowledgments
The financial support of Associazione Italiana per la Ricerca sul Cancro (Italy), Associazione Italiana Leucemie, Ministero della Istruzione della Universitá e della Ricerca, Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare, and Grant FIS 01/0662 from Fondo de Investigacion Sanitaria (Spain) is gratefully acknowledged.
Abbreviations
- Ph
Philadelphia chromosome
- CML
chronic myeloid leukemia
- FISH
fluorescence in situ hybridization
- RT-PCR
reverse transcriptase–PCR
- BAC
bacterial artificial chromosome
- PTR
Pan troglodytes
- GGO
Gorilla gorilla
- HSA
Homo sapiens
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Reichert, A., Emig, M., Reiter, A., Schultheis, B., Saussele, S., Schoch, S., Duyster, J., Melo, J. V., Cross, N. C. P., Hehlmann, R., et al. (1998) Blood 92, Suppl. 1, 4065 (abstr.).
References
- 1.Nowell P C, Hungerford D A. Science. 1960;32:1497–1501. [Google Scholar]
- 2.Rowley J D. Nature (London) 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Melo J V. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- 4.Saglio G, Pane F, Martinelli G, Guerrasio A. Leuk Lymphoma. 1997;26:281–286. doi: 10.3109/10428199709051777. [DOI] [PubMed] [Google Scholar]
- 5.Mitelman F. Leuk Lymphoma. 1993;11, Suppl. 1:11–15. doi: 10.3109/10428199309047856. [DOI] [PubMed] [Google Scholar]
- 6.Morris C M, Heisterkamp N, Kennedy M A, Fitzgerald P H, Groffen J. Blood. 1990;76:1812–1818. [PubMed] [Google Scholar]
- 7.Sinclair P B, Nacheva E P, Leversha M, Telford N, Chang J, Reid A, Bench A, Champion K, Huntly B. Blood. 2000;95:738–743. [PubMed] [Google Scholar]
- 8.Kolomietz E, Al-Maghrabi J, Brennan S, Karaskova J, Minkin S, Lipton J, Squire J A. Blood. 2001;97:3581–3588. doi: 10.1182/blood.v97.11.3581. [DOI] [PubMed] [Google Scholar]
- 9.Huntly B J, Reid A G, Bench A J, Campbell L J, Telford N, Shepherd P, Szer J, Prince H M, Turner P, Grace C, et al. Blood. 2001;98:1732–1738. doi: 10.1182/blood.v98.6.1732. [DOI] [PubMed] [Google Scholar]
- 10.Chissoe S L, Bodenteich A, Wang Y F, Wang Y P, Burian D, Clifton S W, Crabtree J, Freeman A, Iyer K, Jian L, et al. Genomics. 1995;27:67–82. doi: 10.1006/geno.1995.1008. [DOI] [PubMed] [Google Scholar]
- 11.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]
- 12.Bose S, Deininger M, Gora-Tybor J, Goldman J M, Melo J V. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 13.van Dongen J J, Macintyre E A, Gabert J A, Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G, Griesinger F, et al. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 14.Lichter P, Tang Chang C-J, Call K, Hermanson G, Evans G A, Housman D, Ward D C. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 15.Montefalcone G, Tempesta S, Rocchi M, Archidiacono N. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey J A, Yavor A M, Massa H F, Trask B J, Eichler E E. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson M E, Viggiano L, Bailey J A, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler E E. Nature (London) 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 21.Stankiewicz P, Lupski J R. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 22.Ji Y, Eichler E E, Schwartz S, Nicholls R D. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- 23. Osborne, L. R., Li, M., Pober, B., Chitayat, D., Bodurtha, J., Mandel, A., Costa, T., Grebe, T., Cox, S., Tsui, L. C., et al. (2001) Nat Genet. 321–325. [DOI] [PMC free article] [PubMed]
- 24.Giglio S, Broman K W, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, et al. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giglio, S., Calvari V., Gregato G., Gimelli G., Camanini S., Giorda R., Ragusa A., Guerneri S., Selicorni A., Stumm, M., et al. (2002) Am. J. Hum. Genet., in press. [DOI] [PMC free article] [PubMed]
- 26.Melo J V. Baillieres Clin Haematol. 1997;10:203–222. doi: 10.1016/s0950-3536(97)80003-0. [DOI] [PubMed] [Google Scholar]
- 27.Branford S, Rudzki Z, Hughes T P. Br J Haematol. 2000;109:635–637. doi: 10.1046/j.1365-2141.2000.02042.x. [DOI] [PubMed] [Google Scholar]
- 28.Roman J, Parziale A, Gottardi E, De Micheli D, Cilloni D, Tiribelli M, Gonzalez M G, del Carmen Rodriguez M, Torres A, Saglio G, et al. Br J Haematol. 2000;111:644–646. doi: 10.1046/j.1365-2141.2000.02394.x. [DOI] [PubMed] [Google Scholar]
- 29.Litz C E, McClure J S, Copenhaver C M, Brunning R D. Blood. 1993;81:1567–1572. [PubMed] [Google Scholar]
- 30.Melo J V, Gordon D E, Cross N C, Goldman J M. Blood. 1993;81:158–165. [PubMed] [Google Scholar]
- 31.Zhang J G, Goldman J M, Cross N C. Br J Haematol. 1995;90:138–146. doi: 10.1111/j.1365-2141.1995.tb03392.x. [DOI] [PubMed] [Google Scholar]