Abstract
A large fraction of the information content of metazoan genomes resides in the transcriptional and posttranscriptional cis-regulatory elements that collectively provide the blueprint for using the protein-coding capacity of the DNA, thus guiding the development and physiology of the entire organism. As successive whole-genome sequencing projects—–including those of mice and humans—are completed, we have full access to the regulatory genome of yet another species. But our ability to decipher the cis-regulatory code, and hence to link genes into regulatory networks on a global scale, is currently very limited. Here we describe SCORE (Site Clustering Over Random Expectation), a computational method for identifying transcriptional cis-regulatory modules based on the fact that they often contain, in statistically improbable concentrations, multiple binding sites for the same transcription factor. We have carried out a Drosophila genomewide inventory of predicted binding sites for the Notch-regulated transcription factor Suppressor of Hairless [Su(H)] and found that the fly genome contains highly nonrandom clusterings of Su(H) sites over a broad range of sequence intervals. We found that the most statistically significant clusters are very heavily enriched in both known and logical targets of Su(H) binding and regulation. The utility of the SCORE approach was validated by in vivo experiments showing that proper expression of the novel gene Him in adult muscle precursor cells depends both on Su(H) gene activity and sequences that include a previously unstudied cluster of four Su(H) sites, indicating that Him is a likely direct target of Su(H).
Realizing the full promise of whole-genome sequencing projects depends on our ability to read and understand the tremendous informational richness contained therein. Computational methods for predicting novel protein coding genes in whole-genome sequence data are quite advanced, and various strategies for recognizing transcription units that generate noncoding RNAs are also available. However, these techniques address only part of the informational content of the genome. The complex blueprint that controls the utilization of the coding information in DNA is contained in the huge number of cis-regulatory elements, both transcriptional and posttranscriptional, that surround and invade the transcribed part of the genome. But it is clear that we are in our infancy in learning how to read the regulatory genome and thus decipher the blueprint.
Here we describe SCORE (Site Clustering Over Random Expectation), a computational method for identifying potential cis-regulatory modules and the target genes they serve. Transcriptional enhancer elements are generally quite compact, and they frequently include closely spaced binding sites for the same or multiple transcription factors (1). SCORE is designed to detect and statistically evaluate these structural features in whole-genome sequence data, and thus to reveal previously unrecognized enhancers. A conceptually similar method has been described recently by Markstein et al. (2).
Suppressor of Hairless [Su(H)] is the key transducing transcription factor for the Notch cell–cell signaling pathway (3–5), which is involved in a large variety of cell fate specification and patterning events during bilaterian development (6). The sequence specificity of DNA binding by Su(H) is well defined (4, 7, 8), and multiple direct targets of regulation by this factor and the Notch pathway have been identified (4, 8–11). That defined cis-regulatory modules associated with these targets frequently include multiple high-affinity binding sites for Su(H) (8–10) suggested that this factor might be favorable for evaluating the SCORE technique.
Materials and Methods
Whole-Genome Inventory of Consensus Sequence Matches.
Perl scripts (available on request) were written to search the Drosophila genome for matches to binding site consensus sequences. Release 2 (October, 2000) chromosome arm sequences and gene annotation data were downloaded from the Berkeley Drosophila Genome Project web site (http://www.bdgp.org). The positions of consensus sequence matches relative to known or predicted genes are calculated by the script, and the identity of the gene nearest the match is reported.
Clustering Analysis.
Binding site clustering was assessed by tallying the number of additional sites lying to the right of each binding site, within a specified range of window sizes. Each successive binding site along the sequence was similarly treated as the left end of a new cluster set, and the tallying process was repeated. This method results in an inventory consisting of overlapping, but unique, clusters.
Monte Carlo Simulations.
In anticipation of applying SCORE to clusters of binding sites for multiple factors, we chose Monte Carlo simulations as the means of estimating random clustering probabilities. We have verified that our simulations of single-factor binding site clustering conform to the Poisson distribution, as expected. Data sets of random site positions were generated by scattering a fixed number of sites (equal to the total number in the genome) randomly in a space equal to the size of the genome. One thousand of these randomized inventories were then analyzed by using the clustering algorithm just described, to estimate the probability P that a given cluster frequency observed in the genome (or greater) could arise by chance. Purity values for each cluster bin, expressed as percentages, were calculated as the observed frequency minus the mean random frequency, divided by the observed frequency. To classify bins as pure or enriched we used cutoffs of P < 0.005 and purities of >99% and >50%, respectively.
Plasmid Construction.
Enhancer/promoter reporter constructs expressing nuclear green fluorescent protein were prepared by using the pStinger transformation vector (12). The 3′ terminus of both fragments representing the Him gene (2.2 and 4.0 kb) was the nucleotide just 5′ to the translation initiation codon; both constructs thus included the Him promoter and the entire 5′ untranslated region.
Fly Genotype.
Su(H)SF8/Su(H)AR9 was used as a Su(H) null genotype.
In Situ Hybridization.
Digoxygenin-labeled antisense RNA probes representing the Him gene were transcribed from a 2.4-kb genomic DNA fragment that contains the full extent of the predicted gene (13). In situ hybridization was performed as described (14).
Results
Global Inventory of High-Affinity Su(H) Binding Sites in the Drosophila Genome.
The first step in our computational approach to identifying novel targets of Su(H) and the Notch pathway was to search the complete Drosophila genome sequence (13) for matches to a consensus definition (YGTGDGAA) of high-affinity binding sites for the Su(H) protein. This consensus is derived from a combination of known target sites in Drosophila and information from a random binding site selection analysis using the mouse ortholog of Su(H) (4, 7, 8). After eliminating the lower-affinity sequence TGTGTGAA, the net consensus consisted of the five octamers CGTGGGAA, CGTGAGAA, CGTGTGAA, TGTGGGAA, and TGTGAGAA. A total of 15,659 perfect matches to these five sequences was found in the global genome inventory, compared with the statistical expectation (based on mononucleotide frequencies) of 16,989. Thus, the real genome has a substantial (≈8%) deficit of these high-affinity Su(H) binding site octamers. Of the observed 15,659 total sites, 9,886 were categorized as occurring in intergenic regions, 2,078 in predicted exons, 429 in putative 5′ and 3′ untranslated regions, and 3,266 in predicted introns. After removal of putatively exonic consensus matches, it was found that 5,592 unique genes are associated by position with at least one predicted Su(H) binding site.
Inventory of Su(H) Binding Site Clusters.
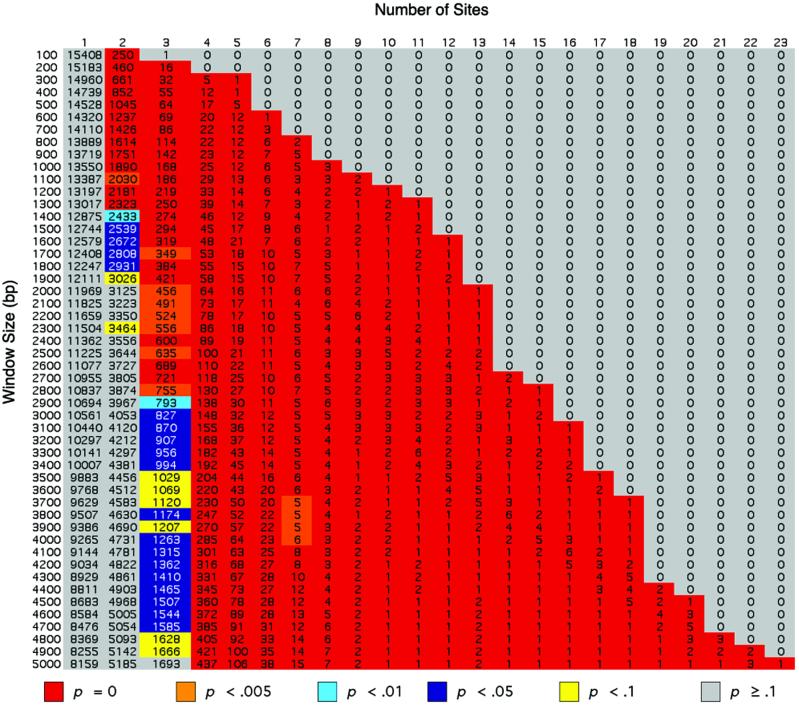
Transcriptional enhancer modules that are responsive to Notch signaling activity frequently are characterized by the presence of multiple high-affinity Su(H) binding sites in a relatively small sequence interval (<1 kb) (4, 8–11). This finding suggests the utility of identifying unusual Su(H) binding site concentrations in the genome as a means of recognizing possible novel target enhancers. For this purpose we developed the cluster detection algorithm described in Materials and Methods. The search window size was varied by increments of 100 bp over the range of 100 to 5,000 bp to cover a large variety of enhancer module sizes and complexities. The complete matrix of Su(H) binding site cluster frequencies over this sequence interval range is presented in Fig. 1. We will refer to each position in the matrix as a cluster bin, wherein the frequency (number) of a particular size of cluster (x number of sites in y bp) is recorded.
Figure 1.
Global inventory of Su(H) binding site clusters in the Drosophila genome. Each position (bin) in the matrix shows the whole-genome frequency of a particular type of Su(H) binding site cluster (x sites in a window of y base pairs). The probability of obtaining by chance the observed real-genome frequency (or greater) in each bin is indicated by the color of the bin, according to the key.
Monte Carlo Simulation of Random Site Clustering.
Any sufficiently large collection of sequence features will exhibit a certain degree of clustering even if they are distributed in the genome randomly. We used a Monte Carlo approach to simulate this background of randomly occurring Su(H) binding site clusters, to evaluate the statistical significance of the cluster frequencies observed in the real genome. One round of simulation is carried out as follows. A number of sites equal to the number of putative Su(H) sites in the Drosophila genome (15,659) is randomly positioned in a genome space of the same size. Clustering of these site positions is then inventoried by the same algorithm used for cluster analysis of real genome data. Cluster frequencies for a large number of such random simulations are accumulated. These data are analyzed to determine for each cluster bin what fraction of random data sets show a cluster frequency equal to or greater than the frequency observed in the real genome. This process yields an estimated probability P that the observed real-genome cluster frequency is caused by chance.
When the cluster frequencies for Su(H) binding sites in the Drosophila genome were compared with those obtained in 1,000 random simulations, a large domain of bins with P = 0 was observed (Fig. 1), revealing that the genome has a surprisingly high degree of extremely improbable clustering of Su(H) sites. This broad P = 0 “valley” is flanked on both left and right by very steep “walls” rising to domains of bins with much higher P values (Fig. 1). On the left (Fig. 1), at all window sizes, the frequencies of clusters containing one site were reproduced in a high proportion of random simulations. Similarly, at the larger window sizes (>2,000 bp), the frequencies of clusters containing two sites were also reproduced regularly in random data sets (Fig. 1). On the right end of the frequency matrix (Fig. 1), there is an abrupt transition from very low P values to values of P ≥ 0.1. The latter represent cluster bins with a frequency of 0 in the real genome (Fig. 1) and 0 in the random data, resulting in P = 1.
We further confirmed the extreme statistical unlikelihood of the Su(H) binding site clustering in the fly genome by a second method: carrying out probability analysis on randomly generated genome sequences. Using the mononucleotide base frequencies in the total fly genome sequence (A = T = 0.288; C = G = 0.212), we constructed 50 random sequence versions of the genome that are identical in size to the real genome. Each of these random genomes was inventoried to determine its total number of Su(H) sites, and this fixed number was then used in 1,000 random clustering simulations by the Monte Carlo method described above, to obtain P values for each cluster bin in each random genome. We found that none of the 50 random genomes displayed probability plots (see Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org) even remotely resembling that of the real genome (Figs. 1 and 4A). As expected, we did observe occasional Su(H) binding site densities in the random genomes with values of P < 0.005 (Fig. 4C). However, probability plots like the one shown in Fig. 4B were far more typical. We conclude that the Drosophila genome exhibits highly nonrandom concentrations of putative high-affinity Su(H) binding sites over a broad range of sequence intervals (Figs. 1 and 4A).
Mining Pure Bins of Su(H) Binding Site Clusters.
When considering how best to make use of global data on clustering of transcription factor binding sites or other cis-regulatory elements in the genome, it is perhaps useful to distinguish two special categories of bins in the cluster frequency matrix. The first of these we term “pure” bins, defined as those with a nonzero frequency in the real genome data, but with a zero or very near-zero frequency in the random data sets resulting from the Monte Carlo simulations. These bins represent site densities that are so unlikely to have arisen by chance that they may be considered pure—i.e., statistically unexpected features of the real genome, uncontaminated by random clusters. A second and much more common category consists of “enriched” bins—those in which the expected random frequency is substantial, but in which the cluster frequency for the real genome is nonetheless considerably higher.
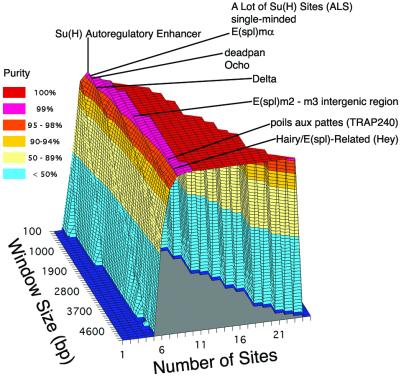
To quantify the differences among bins in this regard, we used a parameter we refer to as purity. We first computed the mean frequency of randomly expected clusters in each bin from 1,000 Monte Carlo permutations. Then, for each bin, we took the difference between the real-genome frequency and the average random frequency, to measure the excess number of clusters of a given size in the real genome over that expected by chance. This nonrandom excess value was divided by the real-genome frequency in the bin, yielding an estimate of purity (expressed as a percentage). Fig. 2 shows a plot of purity across the whole cluster matrix.
Figure 2.
Purity of Su(H) binding site cluster bins in the fly genome. Purity value for each cluster bin (expressed as a percentage) was calculated as described in Materials and Methods; values are plotted on the z axis and coded by the colors shown in the key. The identity of the gene nearest each cluster region found in bins of P = 0 and purity ≥99% (see Table 1) is indicated at the first bin in which the region appears.
We first examined the contents of bins that met the criteria of having a purity of ≥99% and a probability value of P < 0.005. Only 10 distinct genomic regions are identified by these bins. The first bin in the matrix where each region was found is marked in Fig. 2; the specifications of each region, including the identity of the nearest gene, are given in Table 1. We found that this short list is very heavily biased toward both known and logical targets of Su(H) binding and regulation. Of the 10 genes, five [Su(H), singleminded, Ocho, E(spl)ma, and E(spl)m2/E(spl)m3)] have been shown previously by various experimental criteria (see Table 1) to be subject to transcriptional activation by Su(H) (9, 10, 15–19). Like E(spl)m3, two other genes (Hey and deadpan) encode basic helix–loop–helix (bHLH) repressor proteins. Most genes of this class in Drosophila are directly regulated by Su(H) (4, 8, 11, 20); moreover, the mouse Hey1 gene has already been shown to be directly activated by the mouse ortholog of Su(H) (21). Interestingly, Delta, which encodes a major ligand for the Notch receptor, also appears on this high-purity list, although a direct role for Su(H) in regulating Delta expression has not previously been suggested. Finally, we found that one major concentration of Su(H) sites contributes to many bins and is responsible for the large expanse of 100% pure, P = 0 regions of the cluster matrix. This highly unusual genome segment, which we call the A Lot of Su(H) Sites (ALS) region, contains 25 predicted high-affinity Su(H) sites within 5.3 kb. Overall, it is clear that the very high-purity, low-probability bins are extremely selective detectors of bona fide Su(H) targets.
Table 1.
Su(H) binding site clusters identified by SCORE
| Evidence* | Name of nearest gene/element | Symbol or CG number | Position of closest site | Bin† |
|---|---|---|---|---|
| Pure | ||||
| a, c, d | Suppressor of Hairless | Su(H) | (+)105 | 5/300 |
| A Lot of Su(H) Sites | ALS | (+)464 | 5/500 | |
| a, b, c, d | single-minded | sim | (−)2058 | 5/500 |
| a, b, c | E(spl)mα | mα | (−)410 | 5/500 |
| deadpan | dpn | (+)1990 | 5/600 | |
| a, c | Ocho | Ocho | (−)205 | 5/600 |
| Delta | Dl | (−)3874 | 5/900 | |
| a, b, c | E(spl)m2/m3 | HLHm3 | (−)939 | 7/2,400 |
| poils aux pattes | pap | Intron 2 | 8/4,300 | |
| Hairy/E(spl)-Related | Hey | Intron 2 | 8/4,700 | |
| Enriched | ||||
| CG4057 | CG4057 | 5′ UTR | 4/1,400 | |
| numb | numb | Intron 1 | 4/1,400 | |
| neuralized | neur | Intron 1 | 4/1,400 | |
| Allostatin Receptor 2 | AR-2 | Intron 1 | 4/1,400 | |
| CG1136 | CG1136 | Intron 1 | 4/1,400 | |
| √ | derailed | drl | Intron 1 | 4/1,600 |
| Cyp313a3 | CG10093 | Intron 1 | 5/2,800 | |
| nebula | nla | Intron 1 | 5/3,400 | |
| headcase | hdc | Intron 2 | 4/1,400 | |
| arrest | aret | Intron 3 | 4/1,500 | |
| frizzled | fz | Intron 4 | 5/3,100 | |
| CG13489 | CG13489 | (−)132 | 4/1,400 | |
| CG5103 | CG5103 | (−)191 | 4/1,400 | |
| a, b, c, d | E(spl)m5 | HLHm5 | (−)222 | 4/1,400 |
| a, b, c, d | E(spl)mγ | HLHmγ | (−)276 | 4/1,400 |
| CG13636 | CG13636 | (+)284 | 6/5,000 | |
| a | E(spl)m7 | HLHm7 | (−)662 | 4/1,400 |
| √ | peanut | pnut | (−)806 | 4/1,400 |
| 48 related 2 | Fer2 | (−)943 | 4/1,400 | |
| a, b, d | HES-related | Her | (−)966 | 4/1,400 |
| CG13936 | CG13936 | (+)1376 | 4/1,400 | |
| CG15217 | CG15217 | (−)1384 | 4/1,400 | |
| tailless | tll | (−)1474 | 6/4,700 | |
| CG10450 | CG10450 | (+)2250 | 4/1,400 | |
| CG14760 | CG14760 | (−)3425 | 4/1,600 | |
| no hitter | nht | (+)3956 | 4/1,400 | |
| √ | CG12689 | CG12689 | (−)5405 | 4/1,400 |
| CG5643 | CG5643 | (−)5408 | 4/1,400 | |
| CG7370 | CG7370 | (+)6113 | 4/1,400 | |
| CG14598 | CG14598 | (+)9525 | 4/1,500 | |
| CG4683 | CG4683 | (+)12027 | 5/2,800 | |
| CG9650 | CG9650 | (−)12645 | 4/1,600 | |
| CG17668 | CG17668 | (+)15683 | 4/1,600 | |
| CG4814 | CG4814 | (−)20617 | 4/1,400 | |
| CG9598 | CG9598 | (−)39861 | 4/1,400 | |
| grim | grim | (−)43077 | 4/1,400 |
Su(H) binding site clusters residing in pure and enriched bins of the cluster frequency matrix (Figs. 1 and 2; see text) are listed according to the identity of the nearest gene. Genes and binding site clusters shown in bold are discussed in the text. The location of the binding site closest to the listed gene is indicated.
Letters in this column indicate the nature of experimental evidence supporting the physiological relevance of the Su(H) sites in the identified cluster (see text for details and references). a: Wild-type expression pattern; b: expression in a Su(H) mutant background; c: in vitro DNA-binding assays; and d: binding site-dependent enhancer/promoter activity in vivo. A check (√) indicates that a genomic DNA fragment containing or overlapping the cluster has been demonstrated to exhibit enhancer activity in vivo that recapitulates some aspect of the normal expression pattern of a nearby gene.
This column denotes for each cluster region the first bin in the cluster frequency matrix that identifies the cluster and meets the criteria for pure or enriched bins (see text).
Mining Enriched Bins of Su(H) Binding Site Clusters.
Pure bins of Su(H) binding site clusters are the easiest and most obvious to mine for potential target enhancers and the associated genes. However, enriched bins of only moderate purity are also expected to yield valuable candidate targets. To avoid skewing of purity and probability values by contributions from clusters that are also found in pure bins, we removed these regions from the cluster frequency matrix before choosing enriched bins to mine. To this modified data set we applied the same probability and purity analysis procedures described above. In deciding on a definition of enriched bins that would permit efficient mining, we sought to balance frequency and purity, so that a substantial number of candidate clusters could be evaluated, but without undue contamination by randomly expected clusters. We selected a purity cutoff of 50%, and, as before, a probability cutoff of P < 0.005. The specifications of the 36 distinct genomic regions meeting these two criteria are listed in Table 1. As with the pure bins, this list was found to include several binding site clusters [near E(spl)mγ, E(spl)m5, and E(spl)m7] that have previously been demonstrated to be functional targets of transcriptional regulation by Su(H) and the Notch pathway (4, 8, 11, 20). Additionally, two other genes that appear on the enriched bin list, numb and neuralized, are closely associated with the function of the Notch pathway. It is highly unlikely that these Notch pathway components appear on our list by chance, and further investigation of the function of their associated Su(H) binding site clusters is needed.
We were also very interested to note that several clusters on the enriched bin list (Table 1) overlap DNA segments with defined transcriptional enhancer activities that mimic the expression patterns of the adjacent genes. A fragment overlapping three of the four Su(H) binding sites in the cluster between deadpan and peanut has been demonstrated to exhibit peripheral nervous system-specific enhancer activity that recapitulates a normal aspect of deadpan expression (22). Three of four Su(H) sites in the cluster nearest CG12689 overlap an enhancer fragment associated with the cut gene that drives expression in adult external sensory organ lineages (23). Finally, a cluster of four Su(H) sites in the first intron of derailed is contained in an enhancer fragment that drives expression in the three epidermal attachment cells for larval muscles 21–23 and in the bipolar glia (24). In all three cases, it is plausible that direct regulation by Su(H) could be a component of the enhancer activity.
Functional Analysis of the Su(H) Binding Site Cluster Adjacent to Her.
By definition, enriched bins of moderate purity such as we have just considered (Table 1) are expected to be contaminated by false positives, i.e., binding site clusters that appear in the genome by chance and do not represent bona fide enhancer elements. Such contaminated bins may perhaps be mined most effectively through the use of secondary criteria to select candidate clusters for further study. These might include known patterns of expression of nearby genes, if consistent with regulation by the transcription factor of interest, microarray data indicating the possibility of such regulation, or simple gene identity. This last criterion brought to our attention the Su(H) binding site cluster near the gene Hes-related (Her) (Table 1; refs. 13 and 25), which like many known targets of direct activation by Su(H) (see above), encodes a bHLH repressor protein. We selected this cluster for experimental analysis to determine whether it identifies a novel target of Su(H) control (Fig. 3).
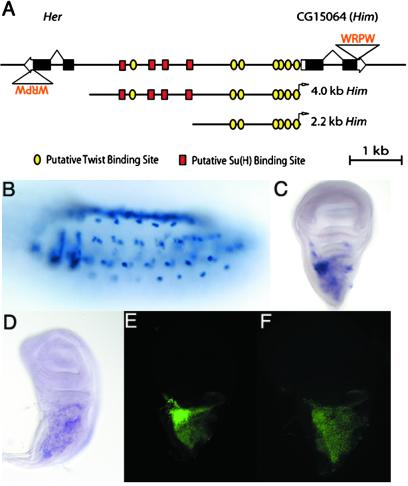
Figure 3.
Sequences including a cluster of four Su(H) binding sites between Her and Him are required for normal expression of Him. (A) Schematic of Her and Him genes and the intergenic region, showing two enhancer/promoter fragments used to construct Him reporter genes. (B–D) Pattern of Him transcript accumulation in stage 15 embryo (B) and late third-instar wing imaginal disc (C); disc expression is severely reduced in a Su(H) null background (D). (E and F) Expression of green fluorescent protein reporter constructs containing the Him promoter and upstream region (see A; see text for details). (E) A 4.0-kb fragment that includes the Su(H) binding site cluster. (F) A 2.2-kb fragment lacking the Su(H) sites but still including six Twist binding sites.
By in situ hybridization to whole embryos and late third-instar imaginal discs, Her transcripts appear to accumulate at low levels in a broad or ubiquitous pattern (data not shown; ref. 25), inconsistent with specific regulation by Notch signaling and Su(H). We further observed that green fluorescent protein reporter constructs containing the Her promoter and upstream region, and either including or not including the four Su(H) sites, exhibit no detectable activity in vivo, suggesting that the Su(H) binding site cluster is not relevant to Her's pattern of expression (data not shown). These results raised the possibility that the Su(H) site cluster might instead be involved in the regulation of CG15064, a predicted gene that lies ≈4.3 kb upstream of Her and is transcribed in the opposite direction (Fig. 3A). We named this gene Him for its proximity to Her. The predicted protein product of Him is itself of interest: the C-terminal four amino acids are WRPW (Fig. 3A), a motif that recruits the corepressor Groucho to a wide variety of bHLH and other repressors (26). Unlike Her, the Him protein is not predicted to include a bHLH domain, suggesting that it may instead be a repressor of a different class, or possibly an adaptor protein that functions as an intermediary between Groucho and a DNA-binding repressor. We found that Him transcripts accumulate in adult muscle precursor cells in both stage 15–17 embryos and imaginal discs of late third-instar larvae (Fig. 3 B and C), in a pattern that strongly resembles the expression of a known Notch/Su(H)-responsive gene, E(spl)m6 (17). In particular, Him transcripts appear in a subset of the adepithelial cell population of the third-instar wing disc (Fig. 3C); these cells give rise to the adult thoracic musculature (27). We first verified that Him transcript accumulation in the wing disc depends on Su(H) function (Fig. 3D). We then observed that a reporter construct that includes the Him promoter and 3.9 kb of upstream sequence, including the Su(H) binding site cluster that appears on our enriched bin list (Table 1), recapitulates the expression pattern of Him in the wing disc (Fig. 3 C and E). This construct also includes seven putative binding sites (CACATG; ref. 28) for the mesodermal bHLH activator Twist (Fig. 3A), which is expressed at a largely uniform level in all adepithelial cells (29). By contrast, a truncated Him promoter construct that includes 2.1 kb of upstream sequence and lacks the entirety of the Su(H) site cluster (but retains six of the seven Twist sites; Fig. 3A) exhibits a greatly reduced level of expression in the region of the adepithelial cell population that strongly expresses both E(spl)m6 and Him in a Su(H)-dependent manner (Fig. 3F). Our results strongly suggest that the expression of Him is directly regulated by Su(H) via the binding site cluster revealed by our SCORE analysis.
Discussion
We have described here a computational approach to the identification of transcriptional cis-regulatory modules and the associated genes in whole-genome sequence data. This method is based on the now well-established observation that bona fide enhancer elements in bilaterian genomes are usually discrete modules of compact size that frequently contain multiple binding sites for the same transcription factor (1). By identifying statistically nonrandom densities of putative high-affinity binding sites for the Notch-regulated transcription factor Su(H), we were able to recognize in the full Drosophila genome (13) a substantial number of both known and highly probable cis-regulatory modules that use this factor, along with the genes they serve. Our experimental analysis of one such novel target enhancer, including four Su(H) sites and associated with the gene Him, fully supports the conclusion that it is a site of direct activation by Su(H).
In designing the SCORE approach to binding site cluster analysis, we sought to incorporate several features that facilitate efficient mining of the genome. First, obtaining an unbiased inventory of site clusters over a wide range of sequence intervals (Fig. 1) permits a detailed examination of site density as a global genomic character. Second, the strong statistical foundation afforded by the Monte Carlo simulations of random binding site clustering allows a rigorous evaluation of the significance of all cluster frequencies observed in the genome (Figs. 1 and 4). Finally, the concept of bin purity (Fig. 2) offers an intuitive, yet quantitative, measure of the degree to which cluster frequencies in the genome exceed the random background, greatly assisting in the choice of favorable bins to mine for potential regulatory targets.
Several parameters contribute to the success of cluster analysis when applied to any given transcription factor, including the quality of the binding site definition, the number of sites in the genome, and whether the factor uses site clustering to any degree to evoke its regulatory response. Su(H) may be particularly favorable in this regard. Nevertheless, we have had considerable success in applying SCORE analysis to Drosophila DNA-binding proteins other than Su(H), including the “proneural” bHLH activators encoded by the achaete-scute complex. Enriched bins (purity ≥50%, P < 0.005) from the proneural activator inventory identify binding site clusters located near logical targets of direct regulation by these factors. Genes such as DTRAF1 (30) and worniu (31), both expressed in the developing nervous system, are associated with strong concentrations of proneural protein binding sites. We have also found that site cluster analysis can be applied successfully to cis-regulatory elements involved in posttranscriptional regulation, such as the negatively acting 3′ untranslated region motifs our laboratory has characterized previously (32–34). These observations suggest that SCORE will be of quite general utility in mining genome sequence data for potential targets of multiple types of regulation.
A clear limitation of the SCORE method when applied to a single transcription factor is that it will generally fail to draw attention to enhancer modules and target genes that use only a single binding site for that factor, or that include more than one site with a statistically random spacing. However, this difficulty can be overcome at least in part by conducting a SCORE analysis with binding sites for more than one factor. The contribution of a single Su(H) site may become significant if this site is part of a statistically unusual cluster of sites for multiple factors. Our knowledge of frequently used combinations of transcription factors, and the expression specificities they control, is growing rapidly (1), making multifactor SCORE increasingly valuable and feasible. For example, our survey of statistically improbable clusters that include binding sites for both proneural bHLH activators (RCAGSTG) and bHLH repressors (CACGYG) has identified a potential cis-regulatory module in an intron of nervy, which encodes a transcription factor that is the fly homolog of the human oncogenic protein ETO. nervy is expressed in both neuroblasts and sensory organ precursors,† a specificity fully consistent with direct regulation by a combination of proneural activators and Notch-regulated bHLH repressors. Significant success in multifactor clustering analysis has also been reported recently by Berman et al. (35).
The data presented in this article demonstrate that the fly genome exhibits widespread and highly significant clustering of binding sites for the transcription factor Su(H) and indicate that cluster analysis can be a sensitive detector of cis-regulatory modules and the associated target genes. As high-quality definitions of transcription factor binding sites and other cis-regulatory sequence elements become increasingly available, SCORE and other similar techniques will no doubt prove increasingly valuable as tools for reading the regulatory genome.
Supplementary Material
Acknowledgments
We are grateful to Elizabeth Blankenhorn for suggesting the Monte Carlo simulations. We thank Scott Barolo and Matt Ronshaugen for critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM46993 and GM62279 to J.W.P.
Abbreviations
- SCORE
Site Clustering Over Random Expectation
- Su(H)
Suppressor of Hairless
- bHLH
basic helix–loop–helix
Footnotes
Wildonger, J. & Mann, R. S., 41st Annual Drosophila Research Conference, March 22–26, 2000, Pittsburgh, p. 605A (abstr.).
References
- 1.Davidson E H. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic; 2001. [Google Scholar]
- 2.Markstein M, Markstein P, Markstein V, Levine M S. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweisguth F, Posakony J W. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A M, Posakony J W. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 5.Fortini M E, Artavanis-Tsakonas S. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 7.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nellesen D T, Lai E C, Posakony J W. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 9.Barolo S, Walker R G, Polyanovsky A D, Freschi G, Keil T, Posakony J W. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- 10.Morel V, Schweisguth F. Genes Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- 11.Lecourtois M, Schweisguth F. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 12.Barolo S, Carver L A, Posakony J W. BioTechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 13.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2196. [Google Scholar]
- 14.Sturtevant M A, Roark M, Bier E. Genes Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- 15.Jennings B, Preiss A, Delidakis C, Bray S. Development (Cambridge, UK) 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 16.Lai E C, Bodner R, Kavaler J, Freschi G, Posakony J W. Development (Cambridge, UK) 2000;127:291–306. doi: 10.1242/dev.127.2.291. [DOI] [PubMed] [Google Scholar]
- 17.Lai E C, Bodner R, Posakony J W. Development (Cambridge, UK) 2000;127:3441–3455. doi: 10.1242/dev.127.16.3441. [DOI] [PubMed] [Google Scholar]
- 18.Wech I, Bray S, Delidakis C, Preiss A. Dev Genes Evol. 1999;209:370–375. doi: 10.1007/s004270050266. [DOI] [PubMed] [Google Scholar]
- 19.Knust E, Tietze K, Campos-Ortega J A. EMBO J. 1987;6:4113–4123. doi: 10.1002/j.1460-2075.1987.tb02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eastman D S, Slee R, Skoufos E, Bangalore L, Bray S, Delidakis C. Mol Cell Biol. 1997;17:5620–5628. doi: 10.1128/mcb.17.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier M M, Gessler M. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 22.Emery J F, Bier E. Development (Cambridge, UK) 1995;121:3549–3560. doi: 10.1242/dev.121.11.3549. [DOI] [PubMed] [Google Scholar]
- 23.Jack J, DeLotto Y. Genetics. 1995;139:1689–1700. doi: 10.1093/genetics/139.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonkowsky J L, Thomas J B. Mech Dev. 1999;82:181–184. doi: 10.1016/s0925-4773(99)00007-6. [DOI] [PubMed] [Google Scholar]
- 25.Moore A W, Barbel S, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Courey A J. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, VijayRaghavan K. BioEssays. 1999;21:486–498. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Cripps R M, Black B L, Zhao B, Lien C L, Schulz R A, Olson E N. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bate M, Rushton E, Currie D A. Development (Cambridge, UK) 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 30.Preiss A, Johannes B, Nagel A C, Maier D, Peters N, Wajant H. Mech Dev. 2001;100:109–113. doi: 10.1016/s0925-4773(00)00506-2. [DOI] [PubMed] [Google Scholar]
- 31.Ashraf S I, Hu X, Roote J, Ip Y T. EMBO J. 1999;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai E C, Burks C, Posakony J W. Development (Cambridge, UK) 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- 33.Lai E C, Posakony J W. Development (Cambridge, UK) 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- 34.Lai E C, Posakony J W. Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- 35.Berman B P, Nibu Y, Pfeiffer B D, Tomancak P, Celniker S E, Levine M, Rubin G M, Eisen M B. Proc Natl Acad Sci USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.