Dear editor,
We read with great interest the manuscript by Contier et al. published in Critical Care [1]. This is a valuable study that highlights the use of rapid molecular pathogen detection in immunocompromised patients—a population in urgent need of early and accurate diagnostic strategies. However, we would like to raise several concerns for consideration by the authors and the readership of Critical Care.
First, the Pneumonia multiplex PCR assay employed in the study detects only 18 bacterial species. Although multiplex PCR offers advantages in terms of speed and sensitivity, its limited pathogen coverage is a significant drawback, particularly in immunocompromised individuals, whose infectious profiles often differ markedly from those of the general population.
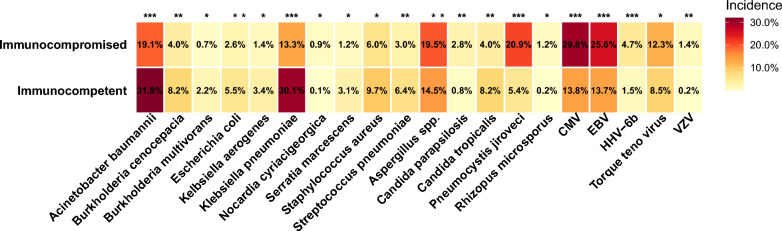
In this context, we would like to share insights from our own multicenter retrospective study using metagenomic sequencing. This cohort, previously described in a series of publications, included bronchoalveolar lavage fluid samples from both immunocompromised and immunocompetent patients [2–9]. Our findings revealed substantial differences in pathogen distribution: immunocompromised patients exhibited significantly higher rates of CMV, EBV, Pneumocystis jirovecii, and Aspergillus spp., while typical bacterial pathogens such as Acinetobacter baumannii, Burkholderia cenocepacia, and Klebsiella pneumoniae were less frequently detected (Fig. 1). This suggests that the diagnostic yield of Pneumonia multiplex PCR is likely to be significantly lower in immunocompromised patients compared to immunocompetent individuals.
Fig. 1.
Differences in pathogen isolation by clinical metagenomics between immunocompromised and immunocompetent patients. As this is an exploratory analysis, no multiple testing correction was applied. *, p < 0.05; **, p < 0.01; ***, p < 0.001
Second, co-detection of multiple pathogens is more common in immunocompromised patients, with clinically relevant co-infections frequently involving viruses and fungi rather than bacteria. This further limit the diagnostic utility of a bacteria-restricted panel in this vulnerable population.
In summary, while the study by Jérémy et al. offers meaningful insights for clinical practice, we emphasize the importance of tailoring diagnostic approaches to the pathogen landscape specific to different patient populations. Broader-spectrum detection tools, particularly those inclusive of viral and fungal pathogens, may be more appropriate for immunocompromised individuals.
Acknowledgements
None.
Author contributions
WXZ, LTH, HZS participated in the discussion and wrote the manuscript.
Funding
This work was supported by the "Pioneer" and "Leading Goose" R&D Program of Zhejiang (grant no. 2025C02090), the National Natural Science Foundation of China (grant no. 82202356) and the Zhejiang Provincial Natural Science Fund (grant no. LTGY24H190001).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
None.
Consent for publication
All authors agreed to publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lingtong Huang, Email: lingtonghuang@zju.edu.cn.
Huanzhang Shao, Email: shaohuanzhang001@126.com.
References:
- 1.Contier J, Platon L, Benchabane N, Tchakerian S, Herman F, Mollevi C, et al. Diagnostic performance of pneumonia multiplex PCR in critically ill immunocompromised patients. Crit Care. 2025;29:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J, Zhong L, Shao H, Wang Q, Dai M, Shen P, et al. Incidence and clinical features of HHV-7 detection in lower respiratory tract in patients with severe pneumonia: a multicenter, retrospective study. Crit Care. 2023;27:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, Huang X, Gu S, Cai H, Wang M, Wang H, et al. Landscape of fungal detection in the lungs of patients with severe pneumonia in the ICU, a multicenter study based on clinical metagenomics. J Infect. 2024;89:106195. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y, Huang X, Zhou H, Wang M, Wang S, Ren X, et al. Clinical Characteristics and Prognosis of Patients With Severe Pneumonia With Pneumocystis jirovecii Colonization: A Multicenter. Retrosp Study Chest. 2025;167:54–66. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Ren X, Huang X, Jin Y, Wang M, Zhong L, et al. Epidemiological and clinical characteristics of ammonia-producing microorganisms in the lungs of patients with severe pneumonia: a multicentre cohort study. J Transl Med. 2024;22:1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Lin B, He G, He C, Li K, Huang Y, et al. Association between caspofungin treatment and mortality in non-HIV Pneumocystis jirovecii pneumonia: a multicenter, retrospective study. J Infect. 2025;91:106548. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Zhuang Y, Huang X, Papazian L, Cai H, Shao H, et al. The landscape of lower respiratory tract herpesviruses in severe pneumonia patients: a multicenter, retrospective study with prospective validation. Crit Care. 2025;29:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Ren X, Wang Q, Shen S, Li Y, Qian X, et al. A predictive model for pulmonary aspergillosis in ICU patients: a multicenter retrospective cohort study. Infect Drug Resist. 2025;18:441–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Jin Y, Zhang W, Ye L, Shao H. Identifying subgroup of severe community-acquired pneumonia based on clinical metagenomics, a multicenter retrospective cohort study. Front Cell Infect Microbiol. 2024;14:1516620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.