Abstract
Known in over 150 species, cytoplasmic male sterility is encoded by aberrant mitochondrial genes that prevent pollen development. The RNA- or protein-level expression of most of the mitochondrial genes encoding cytoplasmic male sterility is altered in the presence of one or more nuclear genes called restorers of fertility that suppress the male-sterile phenotype. Cytoplasmic male sterility/restorer systems have been proven to be an invaluable tool in the production of hybrid seeds. Despite their importance for both the production of major crops such as rice and sunflower and the study of organelle/nuclear interactions in plants, none of the nuclear fertility-restorer genes that reduce the expression of aberrant mitochondrial proteins have previously been cloned. Here we report the isolation of a gene directly involved in the control of the expression of a cytoplasmic male sterility-encoding gene. The Petunia restorer of fertility gene product is a mitochondrially targeted protein that is almost entirely composed of 14 repeats of the 35-aa pentatricopeptide repeat motif. In a nonrestoring genotype we identified a homologous gene that exhibits a deletion in the promoter region and is expressed in roots but not in floral buds.
Cytoplasmic male sterility (CMS), a maternally inherited condition in which a plant is unable to produce functional pollen, has been observed in numerous species (1). Mitochondrial defects have been shown to be responsible for all of the CMS characterized so far. The regions whose expression is associated with CMS contain unusual ORFs that are often chimeric in structure and frequently cotranscribed with conventional mitochondrial genes (2).
Nuclear genes called restorers of fertility (Rf) have the ability to suppress the male-sterile phenotype and, hence, restore the production of pollen to plants carrying the deleterious mitochondrial genome. CMS/Rf systems greatly facilitate hybrid seed production by eliminating the need for tedious hand emasculation and ensuring that each seed is a result of cross-pollination. The Rf allele from the pollen parent restores fertility and seed production in the heterotic hybrid progeny. Apart from its commercial exploitation, CMS offers one of the few opportunities to examine the regulation of mitochondrial gene expression by a nuclear gene in multicellular organisms.
Despite the diversity of mitochondrial mutations causing CMS, there are a number of striking similarities in the mechanism of restoration. In many instances, the transcript pattern of the CMS gene is altered in the presence of the Rf gene (3–7). Fertility restoration has been shown to correlate with an enhanced processing of the transcript from the ORFs associated with the CMS in Sorghum (6) and in Brassica napus (8). There is evidence that the nonrestoring alleles at the Rf loci of these two species are involved in the processing of other mitochondrial transcripts not associated with CMS (9–11).
The maize Rf2 gene, which acts in conjunction with the Rf1 gene to restore fertility to T-cytoplasm maize, is an unusual restorer gene and is the only one previously cloned (12). Rather than affecting the expression of the CMS protein, URF13, Rf2 is an aldehyde dehydrogenase (13). It has been proposed that Rf2 acts by compensating for a metabolic defect caused by the low levels of the URF13 protein that remain despite the presence of Rf1, the gene that reduces the amount of the toxic protein (14) and also alters the T-urf13 transcript profile (3, 15).
In Petunia, a single dominant nuclear gene termed Rf confers fertility to lines carrying the only known CMS cytoplasm in this genus (16). CMS in Petunia is encoded by an abnormal gene termed pcf, a chimeric mitochondrial ORF composed of portions of the coding region of ATP synthase subunit 9 and cytochrome oxidase subunit 2 fused to an ORF of unknown origin (17). In lines containing the Rf allele, the pcf transcript profile is altered and the amount of the PCF protein is greatly reduced (5, 18). The action of Petunia Rf is, thus, analogous to that of maize Rf1, not to maize Rf2.
In an effort to better understand the molecular mechanism of fertility restoration, we decided to follow a positional cloning strategy to identify the Rf gene in the 1200-Mb Petunia genome. An amplified fragment length polymorphism (AFLP) marker that cosegregated with Rf in a fine-mapping population was cloned, and used to screen a Petunia binary bacterial artificial chromosome (BIBAC) library that we constructed from a restorer line. As a result, a 37.5-kb BIBAC clone that cosegregated with Rf was identified (19). Shotgun sequencing of this clone enabled us to identify ORFs as potential candidates for the Rf gene.
In this study, we present evidence that the Petunia Rf locus is composed of duplicated genes containing a pentatricopeptide repeat (PPR) motif. This motif has been recently described and is found in one of the largest plant gene families (20). One of the two PPR genes found in the Rf locus, denoted Rf-PPR592, encodes a 592-aa protein and is able to restore fertility when transferred to rf/rf CMS plants. Isolation of a recessive and nonfunctional homolog rf-PPR592 from a CMS plant indicates a deletion in the promoter area as the likely cause of its inability to restore fertility.
Materials and Methods
Shotgun Sequencing.
Because of difficulties in contig assembly caused by the presence of repeated sequences, BamHI subclones rather than the entire BIBAC clone were used as the starting material for shotgun sequencing. DNA was sonicated into 1- to 3-kb fragments, which were gel purified (Geneclean Spin Kit, Bio 101), end-repaired with T4 DNA polymerase (GIBCO/BRL) in the presence of all four dNTPs, and ligated at a mass ratio of 3 inserts to 1 vector into the SmaI site of the pTrueBlue vector (Genomics One, Buffalo, NY). The ligation product was introduced into Electromax DH10B Escherichia coli cells (GIBCO/BRL), and DNA obtained from the white colonies by minipreparation was sequenced with the T7 primer in the Cornell BioResource Center. The sequences were assembled into contigs with Sequencher (Gene Codes, Ann Arbor, MI).
Sequence Analysis.
ORFs from BIBAC SB5, their promoter region, and poly(A) signals were predicted by using genscan (21) (http://genes.mit.edu/GENSCAN.html) with the Arabidopsis parameter matrix. Duplicated blocks in the Rf locus were determined by aligning the genomic sequence against itself by using the dot-plot feature from the megalign program (DNAstar, Madison, WI) with a 90% match. The presence of a transit peptide in the ORFs was determined by using predotar version 0.5 (http://www.inra.fr/Internet/Produits/Predotar/), targetp (refs. 22 and 23; http://www.cbs.dtu.dk/services/TargetP/), and mitoprot (ref. 24; http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter). The length of the transit peptide was predicted by targetp and mitoprot.
PPR motifs were identified in Rf-PPR592 and Rf-PPR591 by the meme software (ref. 25; http://meme.sdsc.edu/meme/website/meme.html). The parameters for motif searching were set as minimum width = 35, maximum width = 35. The PPR consensus motif computed from the comparison of 1,303 motifs has been described previously (20).
Analysis of a Nonrestoring Homolog.
The sequence of rf-PPR592 was obtained by amplifying genomic DNA of an rf/rf line with the PfuTurbo Hotstart DNA polymerase (Stratagene) and primers 1 (5′-TGCACAGTGTTATATTTACATACCC-3′) and 2 (5′-TTTATGATACATGGATTTCAACGAC-3′). The PCR product was cloned into the pCR-Blunt II-TOPO vector (Invitrogen). Because of the primer design, the rf-PPR592 sequence lacks 35 nt available for Rf-PPR592.
Similarity blocks between rf-PPR592, Rf-PPR592, and Rf-PPR591 were determined by comparing the aligned sequences with sequencher. Percent similarity was computed by using the megalign program.
Expression of Rf-PPR592 in Floral Buds.
The reverse primer R3 used for reverse transcription (RT)-PCR lies in the 3′ untranslated region of the Rf-PPR592 gene at position +430 to +454. The forward primer used for the PCR lies in the coding sequence and is specific to the rf or Rf allele, F2S or F2, respectively, because of DNA polymorphisms between rf and Rf in this area. Primer pairs F2SR3 amplify a 1333-bp product and F2R3 amplify a 1507-bp product. The RT reaction was performed with Superscript II RNase H− reverse transcriptase (GIBCO), and the PCR was performed with the PfuTurbo Hotstart DNA polymerase. R3, 5′-TGAAAATGACAATCGTAACAGAAAA-3′; F2, 5′-AACATTCCTCCAGACATTATTACA-3′; F2S, 5′-GACGCTGAGGAAATAATGAGATAC-3′.
Transgenic Experiments.
A sequence encoding the N-terminal 44 aa of Rf-PPR592 was inserted 5′ to the green fluorescent protein (GFP) sequence in the pOL vector (26) to use in transient assay of protein localization. As a control, a vector carrying GFP fused with a known mitochondrial coxIV transit peptide (27) was also used in the transient assays. DNAs of GFP constructs were bombarded into onion epidermal cells as described (28).
For the stable transformation experiments, genomic DNA from the Rf-PPR592 gene was amplified from the SB5 BIBAC clone with the PfuTurbo Hotstart DNA polymerase and the primers F11-XbaI (5′-TCTAGAAAAAATGAAGGGGGAATCAAT-3′) and R11-EcoRI (5′-GAATTCACTTTGCTCTCACGATAAACTAAGA-3′) (underlined are the restriction sites added to the 5′ end of the primers for further use in the cloning of the PCR product). The PCR product was first cloned into the pCR-Blunt II-TOPO vector, and its sequence was checked to be free of possible mutations generated by the polymerase. The PCR product was then released from the pCR-Blunt II-TOPO vector by digestion with XbaI and EcoRI, gel purified, and cloned into XbaI/EcoRI-digested binary vector pGPTVKan (29). Petunia transformation and regeneration were performed as described by Horsch et al. (30). Transformants were selected on 300 mg/liter kanamycin, 100 mg/liter ticarcillin/clavulanic acid (15:1, Duchefa Biochemie, Harlem, The Netherlands). Shoots were rooted on N13 medium (31) before transfer to soil.
DNA Blots and Immunoblots.
DNA extractions and Southern blotting were performed as described (19). Floral bud protein was prepared as described for cell culture protein (32). After separation by SDS/PAGE (15%), immunoblots on Hybond-P poly(vinylidene difluoride) membranes (PVDF; Amersham Pharmacia) were prepared as previously described (33) and probed with a 1:5000 dilution of the anti-PCF antibody (34).
Results
Identification of Two PPR-Containing ORFs as Potential Candidates for the Rf Gene.
We previously reported the isolation of a 37.5-kb BIBAC clone, SB5, that cosegregates with the Rf gene (19). SB5 is part of a contig that was constructed by screening a Petunia BIBAC library with a marker, EACA/MCTC, tightly linked to Rf. No recombination was identified between EACA/MCTC and Rf after examining 1,078 meiotic events. The genetic delimitation of the Rf locus was achieved only partially on the BIBAC contig. One extremity of the contig was separated from Rf by the occurrence of four recombination events, whereas no crossing-over was found between Rf and the other extremity (19). Because of the possibility that Rf might lie further away in the area not covered by the contig, we decided to initiate a walk by screening the BIBAC library with a probe lying on the extremity that cosegregates with Rf. Unfortunately, the only hits were clones already isolated in the contig, demonstrating the presence of a gap in the Petunia BIBAC library.
Before increasing the redundancy of our library to find new clones covering the gap, we decided to determine whether the Rf gene might lie in the SB5 clone. Because the BIBAC vector is a binary vector allowing Agrobacterium-mediated plant transformation (35), we attempted to use SB5 to restore fertility to CMS plants. Unfortunately, although SB5 is stable in E. coli, it undergoes multiple rearrangements when introduced into A. tumefaciens, thus precluding its use in transgenic experiments (data not shown). Randomly chosen clones of various sizes did not show this instability in A. tumefaciens, pointing to special features in the sequence of the SB5 insert.
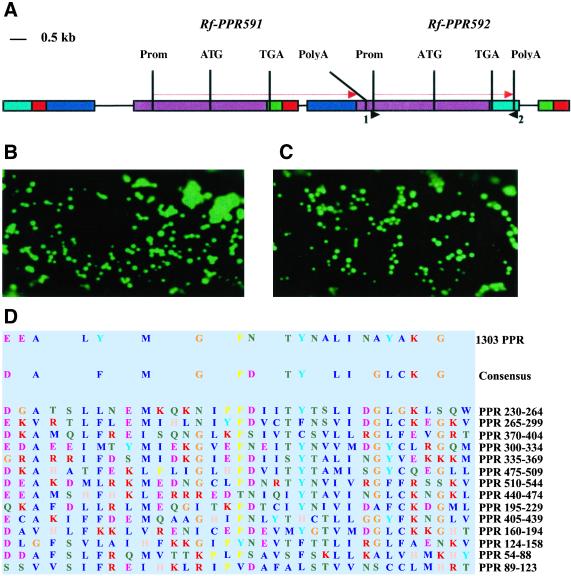
To address whether Rf might lie in the SB5 clone, we carried out shotgun sequencing of the entire clone and examined the predicted ORFs for candidate Rf genes. Because the Rf gene is expected to be targeted to mitochondria where it can act upon the pcf gene to prevent its expression, we searched for an ORF predicted to carry mitochondrial transit sequences. Two ORFs with putative mitochondrial targeting signals were identified. The two ORFs are adjacent to each other and appear to have originated from duplications in the promoter and coding region, but carry divergent 3′ flanking regions (Fig. 1A). The ORFs are 92% identical at the nucleotide level, and the predicted proteins are 93% similar, with C termini that differ completely in their final 12 aa. Both ORFs carry PPR motifs; one encodes 591 aa and the other encodes 592 aa, and are therefore named Rf-PPR591 and Rf-PPR592. A third PPR-containing ORF might lie in the vicinity of the two PPR-containing ORFs shown in Fig. 1A. On the left extremity lies a genomic block (shown in blue) that shares high similarity with the end of the coding sequence of Rf-PPR592 and its terminator region. Because this extremity expands into the gap of the BIBAC library, determining whether there is a third PPR motif protein-encoding gene in this area will require the screening of a new library.
Fig 1.
The Rf locus contains two tandem mitochondrially targeted PPR motif genes. (A) Genomic organization of the region containing the Rf-PPR592 and Rf-PPR591 genes. Duplicated blocks are indicated by similar colors. Arrows indicate the direction of transcription. 1 and 2 show locations of the primers used to amplify the rf-PPR592 gene from a CMS plant. (B) A single onion epidermal cell expressing a known mitochondrially targeted GFP after DNA bombardment. (C) A single onion epidermal cell transiently expressing 44 N-terminal amino acids of Rf-PPR592 fused to GFP. (D) Comparison of PPR motifs found in Rf-PPR592 with the MEME-derived consensus from 1,303 PPR motifs. The 14 PPR repeats are sorted by decreasing statistical significance, with PPR 230–264 showing the highest match to the consensus motif that is generated by retaining only the amino acids that occur at least in 6 of the 14 repeats. The color code is taken from the MEME output.
According to cleavage prediction programs, both putative proteins exhibit 28-residue mitochondrial transit peptides. Predicted transit peptides of Rf-PPR592 and Rf-PPR591 differ by only one substitution. To determine whether the predicted transit peptide could target a passenger protein to mitochondria, 44 codons from the 5′ end of the Rf-PPR592 coding region were inserted 5′ to the coding region of an enhanced GFP. DNAs of this construct and of one known to target GFP to mitochondria were bombarded into onion epidermal cells. Both GFPs appear to be localized to the same type of organelle in the single cells shown in Fig. 1 B and C. Because the predicted transit peptides of Rf-PPR592 and Rf-PPR591 differ by only one amino acid, we expect that not only Rf-PPR592 but also Rf-PPR591 would be mitochondrially localized.
Most of the predicted mature protein (87%) of Rf-PPR592 consists of 14 PPRs (Fig. 1D). These repeats extend from the amino acid in position 54 to the amino acid in position 544 and are organized in two sets of tandem repeats, one set containing 3 PPRs from amino acid 54 to amino acid 158, the other set containing 11 PPRs from amino acid 160 to amino acid 544. Because the Rf-PPR591 and Rf-PPR592 proteins are 93% similar and differ mainly in the last 12 C-terminal amino acids, their organization with respect to PPRs is identical. There is a very good agreement between the consensus motif derived from the 14 PPRs found in Rf-PPR592 (hereafter designated 14 PPR consensus) and the consensus motif derived from 1,303 PPRs (hereafter designated 1303 PPR consensus) reported previously (20) (Fig. 1D). Whenever a discrepancy occurs between the consensus motif of the 14 PPRs in Rf-PPR592 and the 1303 PPR consensus, the difference usually is a conservative substitution. For instance, the aspartic acid in the first position of the 14 PPR consensus is replaced by a glutamic acid in the 1303 PPR consensus. Moreover, when the most frequent amino acid in the 14 PPR consensus at a given position differs from the corresponding amino acid found in the 1303 PPR consensus, the amino acid in the 1303 consensus is generally the second most frequent in the 14 PPR consensus (glutamic acid at position 1, asparagine at position 18, alanine at position 28, tyrosine at position 29; Fig. 1D).
A Deletion in the Promoter of rf-PPR592 Prevents Its Expression in CMS Floral Buds.
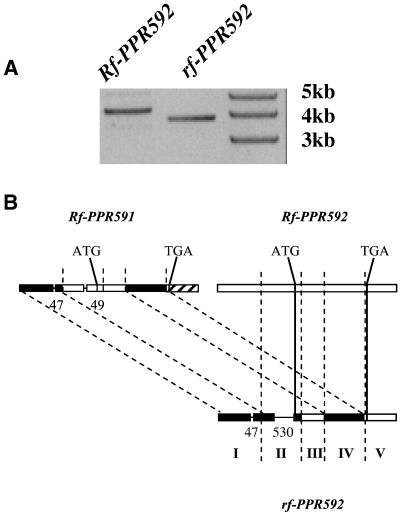
If one of the candidate ORFs, Rf-PPR591 or Rf-PPR592, is the Rf gene, we might expect some sequence polymorphism between the allele of these ORFs found in a restorer line (Rf/Rf) and the allele found in a CMS plant (rf/rf). Presumably some difference in the sequences of the dominant Rf allele vs. the recessive nonrestoring allele rf must reflect their opposite restoring ability. PCR primers flanking Rf-PPR591 and Rf-PPR592 were used to amplify the corresponding sequences of a Petunia hybrida rf/rf plant where rf was inherited from a P. hybrida line called 2423. A PCR product was obtained only with a primer specific to the 3′ flanking region of Rf-PPR592, not with a primer specific to the 3′ flanking region of Rf-PPR591 (data not shown). The rf-PPR592 PCR product shows a reduction in size of about 500nt compared with the Rf-PPR592 PCR product amplified from the genomic DNA of an Rf/Rf line (Fig. 2A). Using the same primers, a PCR product similar in size to rf-PPR592 was amplified from another nonrestoring P. hybrida line as well as from a nonrestoring Petunia parodii line (data not shown). The rf-PPR592 PCR product amplified from the P. hybrida 2423 sequence was cloned and sequenced, revealing a gene 97% identical to Rf-PPR591 and 94% identical to Rf-PPR592 in the coding region, with the predicted proteins 98% and 94% similar, respectively. Comparison of the similarities of regions of the three different PPR genes reveals that the 5′ promoter region of rf-PPR592 is most similar to Rf-PPR591, whereas the 3′ flanking region of rf-PPR592 is most similar to Rf-PPR592. The genomic structure of rf-PPR592 is consistent with the past occurrence of recombination between two genes similar to Rf-PPR591 and Rf-PPR592 (Fig. 2B). Because PCR amplification could have resulted in an artificial recombination between Rf-PPR591 and Rf-PPR592 due to their high similarity, we resequenced the Rf-PPR592 PCR product as a control experiment. The sequences of three rf-PPR592 and Rf-PPR592 clones were determined. No evidence of recombination was found in any of the sequenced Rf-PPR592 clones, thus precluding PCR amplification as the source of the genetic mosaic found in the rf-PPR592 ORF.
Fig 2.
Genetic structure of the rf-PPR592 gene. (A) Comparison of Rf-PPR592 and rf-PPR592 reveals a size polymorphism. The first lane was loaded with the Rf-PPR592 PCR amplicon obtained from a restorer line (Rf/Rf), the adjacent lane was loaded with the rf-PPR592 PCR amplicon obtained with the same primer pair from a CMS line (rf/rf). (B) Comparison of Rf-PPR592, Rf-PPR591, and rf-PPR592 reveals five similarity blocks. For each block, (I to V), the two blocks that exhibit the greatest similarity are shown with the same shading. Overall all three sequences are greater than 90% identical at the nucleotide level except in block V, where Rf-PPR591 exhibits only 23% identity to the other two genes. The locations of 47- and 49-nt deletions in Rf-PPR591 and 47- and 530-nt deletions in rf-PPR592 with respect to the Rf-PPR592 sequence in blocks I and II are shown as lines.
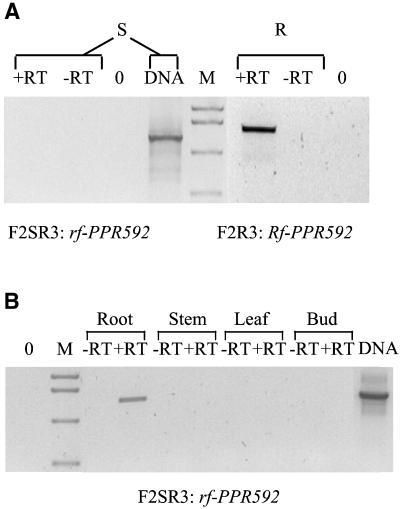
rf-PPR592 carries a 530-nt deletion from −556 to −27 relative to the start codon of Rf-PPR592. This deletion is responsible for the observed difference in the sizes of the respective amplicons. Rf-PPR591 has a 49-nt gap within the same region, from −273 to −224 relative to the start codon of Rf-PPR592 (Fig. 2B). RT-PCR experiments were performed to determine whether both Rf-PPR592 and rf-PPR592 are expressed in Petunia floral buds. An Rf-PPR592 transcript was detected in floral buds in lines carrying the Rf allele, but no transcripts of rf-PPR592 were detected in a homozygous nonrestoring rf/rf line (Fig. 3A). The absence of the upstream 530-nt region in rf-PPR592 is likely to prevent the expression of PPR592 in the floral buds of nonrestoring lines.
Fig 3.
Expression pattern of rf-PPR592 and Rf-PPR592. (A) Examination of floral bud RNA for expression of rf-PPR592 and Rf-PPR592. RT-PCR of floral bud RNA of a CMS plant (S) with primers specific to rf-PPR592, and RT-PCR of floral bud RNA of an RfRf (nontransgenic) fertile plant with primers specific for Rf-PPR592 (R). DNA, positive control for the amplification where the substrate is leaf DNA from a CMS plant; M, mass markers; 0, no template added, negative control. (B) Examination of different tissues for expression of rf-PPR592. RT-PCR of RNA from different tissues of a CMS plant with primers specific to rf-PPR592. DNA, M, and 0 same as in A.
Since rf-PPR592 encodes a protein that is very similar to the one encoded by Rf-PPR592, a survey of its expression was conducted in tissues other than the floral buds. From all of the tissues analyzed, an rf-PPR592 transcript was detected only in roots of a nonrestoring rf/rf line (Fig. 3B).
Rf-PPR592 Is Able to Restore Fertility to CMS Plants.
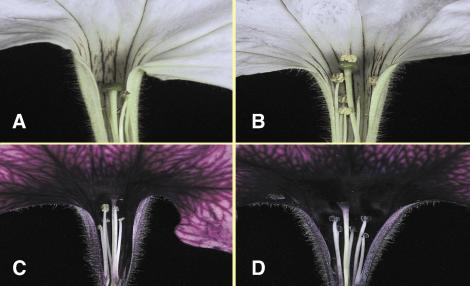
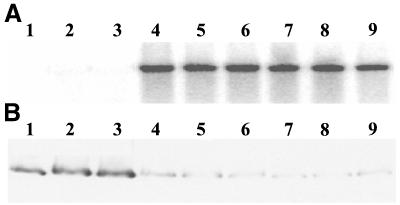
To determine whether Rf-PPR592 could restore fertility to rf/rf CMS lines, a 4.6-kb fragment carrying the entire coding region was introduced into the binary vector pGPTVKan. This fragment carries 2007 nt upstream of the start codon and 861 nt downstream of the stop codon. The pGPTVKan-4.6 kb Rf-PPR592 vector was transferred into A. tumefaciens strain LBA4404, which was used to transform a P. parodii rf/rf CMS line (Fig. 4A) and a P. hybrida rf/rf CMS line (Fig. 4C). More than two dozen independent transformants were obtained and grown to flowering. Fertile transformants were observed after transformation of both lines (Fig. 4 B and D). Among these were several fertile transformants carrying a single copy of the introduced Rf-PPR592 genomic DNA. Flowers of one of the P. parodii primary transformant plants were selfed, and a population of 40 T1 progeny was grown to flowering. DNA blot hybridization revealed that the fertile phenotype cosegregated with the Rf-PPR592 transgene (Fig. 5A). The T1 progeny were also surveyed for the presence of the CMS-associated 19.5-kDa PCF protein. The 19.5-kDa protein was found to be decreased about 10-fold in fertile progeny restored by Rf-PPR592 relative to sterile progeny and the parental CMS line (Fig. 5B). Thus, Rf-PPR592 is capable of restoring fertility by decreasing the amount of the PCF protein.
Fig 4.
Restoration of fertility to CMS Petunia lines by transformation with a 4.6-kb genomic sequence carrying Rf-PPR592. (A) Flower of P. parodii CMS line 3688. (B) Regenerant carrying Rf-PPR592. (C) P. hybrida CMS line 2423. (D) Regenerant carrying Rf-PPR592.
Fig 5.
Cosegregation of the Rf-PPR592 transgene, restoration of fertility, and reduction of PCF. (A) DNA blot hybridized with an npt II transgene-specific probe. Lane 1, P. parodii CMS line 3688; lanes 2-and 3, sterile T1 progeny of transformed P. parodii; lanes 4–9, fertile T1 progeny. (B) Immunoblot of floral bud proteins probed with anti-PCF antibody. Lanes as in A.
Discussion
Petunia Rf Is a PPR-Containing Gene.
In this study we demonstrated that Rf-PPR592, a gene encoding a 592-aa protein containing 14 PPRs, was able to restore fertility to CMS plants. The PPR motif, a degenerate 35-aa repeat, has been found in a very large gene family in the Arabidopsis genome (20). The repeats are organized in tandem arrays with the number of motifs per peptide ranging from 2 to 26. About two-thirds of these Arabidopsis PPR proteins are predicted to be targeted to either mitochondria or chloroplasts (20). Although distinct from the tetratricopeptide repeat (TPR), a motif that is likely to be involved in protein binding, the PPR motif shares with the former a predicted spatial structure consisting of two α-helices (20, 36). Tandem PPRs are thought to form a superhelix with a central spiral groove that presumably serves as the ligand-binding surface in a similar way as the one predicted for the tandem TPRs (20). However, unlike in the TPR motif, the side chains lining the central groove of the PPR are almost exclusively hydrophilic, suggesting that some or all of the PPR motifs are RNA-binding rather than protein-binding motifs. This hypothesis is supported by the involvement in RNA metabolism and/or translation of the very few PPR motif-containing proteins characterized so far: maize chloroplast CRP1, involved in chloroplast petD RNA processing and petD and petA translation (37), Chlamydomonas MCA1, required for the accumulation of the chloroplast petA transcript (38), yeast PET309, required for the stability and translation of the coxI mitochondrial mRNA (39), and Drosophila BSF, which binds to and stabilizes the bicoid mRNA (40). That Petunia Rf belongs to this family is consistent with its similarity of action to crp1, mca1, and pet309. Mutations in these three genes result in lack of accumulation of a particular transcript and reduced abundance of an organelle protein. Likewise, in Petunia restored plants, among the population of pcf transcripts with different 5′ termini, the ones with termini at −121 exhibit reduced abundance and the amount of the PCF protein is greatly reduced (5, 18). However, the alleles of the other PPR genes that are known to reduce RNA and/or protein accumulation are recessive, whereas the Petunia Rf allele is dominant. Rf genes from other species have been shown to alter the RNA transcript profile of the CMS-associated genes (3, 4, 6, 7). In some cases, restoration has been shown to result from enhanced processing of the CMS-associated transcripts (6, 8). Taken together, these observations suggest that Rfs in other species could also be PPR-containing genes like the Petunia Rf.
The Petunia Rf Locus Contains at Least Two Duplicated PPR-Containing Genes.
The data presented in this study show that a pair of duplicated PPR-containing genes, denoted Rf-PPR591 and Rf-PPR592, lie in the Petunia Rf locus. A third related PPR gene might lie in the area not covered by the SB5 BIBAC clone as suggested by the high similarity between the sequence available at the end of the clone and the sequence present at the end of the coding sequence of Rf-PPR592 and in its terminator region.
In Brassica napus, the restorer locus has been shown to affect the transcripts of several mitochondrial genes, two of them being associated with the nap and pol CMS (9, 10). At the same locus have been mapped Rfp, the restorer gene to the pol CMS, that modifies the transcripts of the pol CMS-associated orf224/atp6 mitochondrial DNA region, Rfn, the restorer gene to the nap CMS that modifies the transcripts of the nap CMS-associated orf222/nad5c/orf139 mitochondrial DNA region, and Mmt (modifier of mitochondrial transcripts), a gene that modifies the transcripts of the nad4 gene and another gene possibly involved in cytochrome c biogenesis (10). The resolution of the genetic mapping in these studies did not allow the authors to address whether the three genes represent different alleles of a single gene or whether the restorer locus might contain multiple, related, tightly linked genes. A similar situation occurs in Sorghum, where at the Rf3 locus, one of the two restorers to A3 CMS, has been mapped a gene that regulates the transcript-processing activity of A3 CMS-associated orf107 and the Mmt1 gene that enhances the transcript processing of urf209 (11). As in Brassica napus, either a multiallelic model or tightly linked genes could account for this result.
It will be worthwhile to determine whether Rf-PPR591 affects the profile of mitochondrial transcripts other than pcf in transgenic plants. If so, it would strengthen the hypothesis that Rf alleles arise as modifications, perhaps through duplication, of existing alleles that control mitochondrial gene expression. According to this theory, once CMS occurs in a plant species, there may be strong selective pressure for the plant to overcome it by recruiting preexisting activities and redirecting them to down-regulate the expression of CMS-encoding genes. Conceivably, recombination among closely related PPR-containing genes could have led to the appearance of the Rf-PPR592 gene.
The Nonrestoring rf-PPR592 Exhibits a Promoter Deletion and Likely Arose from a Recombination Between Two Genes Similar to Rf-PPR591 and Rf-PPR592.
A deletion of 530 nt in the promoter area of the rf-PPR592 gene is the likely cause of its nonexpression in the floral buds of CMS plants. That the rf-PPR592 gene, which encodes a protein 98% similar to Rf-PPR591 and 94% similar to Rf-PPR592, has not yet accumulated missense mutations suggests either a recent deletion in the promoter or a functional expression in plant organs other than the floral buds. This latter possibility was supported by the finding of an rf-PPR592 transcript in the roots of homozygous nonrestoring rf/rf line.
Sequence inspection demonstrates that a recombination event between two genes similar to Rf-PPR591 and Rf-PPR592 can explain the formation of rf-PPR592. Perhaps once Rf-PPR592 was generated and happened to prevent the expression of pcf, its maintenance required the presence of the CMS-associated gene. The absence of the CMS-associated gene in new nucleocytoplasmic combinations might have resulted in recombination between Rf-PPR591 and Rf-PPR592 because of their high similarity. In Brassica and related genera, Rfn is found only in association with the nap cytoplasm, suggesting that the evolutionary appearance of the nap cytoplasm and the attending male sterility may have provided the selective pressure for the origin, and possibly the continued presence, of Rfn in B. napus (10). Sampling of more rf-PPR592 genes from different Petunia species should help us to understand the evolution of CMS and fertility restoration in this genus.
Conclusions
The cloning of a gene that can restore fertility to male-sterile Petunia lines will facilitate elucidation of the mechanism by which expression of the CMS-associated mitochondrial gene is suppressed. The reduced amount of the PCF protein could be due to a reduction in the abundance of one of the Petunia CMS-associated transcripts, which was reported previously (5), or to a translation defect that destabilizes the transcript. In yeast, mutation in a transcript-specific translation factor destabilizes the particular transcript with which the factor normally interacts (41).
A number of fertility restorer genes in other species are known to alter transcript profiles and mitochondrial gene product accumulation (7, 9, 42, 43). In addition to the molecular phenotype of restoration, the Petunia Rf locus and Rf loci from other species may be similar in genomic organization (10, 11). The identification of Petunia Rf as a PPR family member suggests that searching for PPR motif genes near known restorer loci should be a useful strategy to identify candidate restorer genes in other species. Further studies of Rf-PPR592 and other PPR motif-containing genes in plants, fungi, and animals will be required to determine whether the motif has a direct role in RNA–protein and/or protein–protein interactions.
Acknowledgments
We thank S. Izhar for Petunia lines and N. Peeters for the pOL vector. This work is supported by a grant from the U.S. Department of Agriculture National Research Initiative Plant Genome Program to M.R.H. A.A.A. held a Rockefeller Foundation Advanced Training Fellowship.
Abbreviations
CMS, cytoplasmic male sterility
BIBAC, binary bacterial artificial chromosome
PPR, pentatricopeptide repeat
RT, reverse transcription
GFP, green fluorescent protein
References
- 1.Laser K. D. & Lersten, N. R. (1972) Bot. Rev. 38, 425-454. [Google Scholar]
- 2.Schnable P. S. & Wise, R. P. (1998) Trends Plant Sci. 3, 175-180. [Google Scholar]
- 3.Wise R. P., Dill, C. L. & Schnable, P. S. (1996) Genetics 143, 1383-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh M. & Brown, G. G. (1991) Plant Cell 3, 1349-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruitt K. D. & Hanson, M. R. (1991) Mol. Gen. Genet. 227, 348-355. [DOI] [PubMed] [Google Scholar]
- 6.Tang H. V., Pring, D. R., Shaw, L. C., Salazar, R. A., Muza, F. R., Yan, B. & Schertz, K. F. (1996) Plant J. 10, 123-133. [DOI] [PubMed] [Google Scholar]
- 7.Moneger F., Smart, C. J. & Leaver, C. J. (1994) EMBO J. 13, 8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menassa R., L'Homme, Y. & Brown, G. G. (1999) Plant J. 17, 491-499. [DOI] [PubMed] [Google Scholar]
- 9.Singh M., Hamel, N., Menassa, R., Li, X. Q., Young, B., Jean, M., Landry, B. S. & Brown, G. G. (1996) Genetics 143, 505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X. Q., Jean, M., Landry, B. S. & Brown, G. G. (1998) Proc. Natl. Acad. Sci. USA 95, 10032-10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H. V., Chang, R. & Pring, D. R. (1998) Genetics 150, 383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui X., Wise, R. P. & Schnable, P. S. (1996) Science 272, 1334-1336. [DOI] [PubMed] [Google Scholar]
- 13.Liu F., Cui, X., Horner, H. T., Weiner, H. & Schnable, P. S. (2001) Plant Cell 13, 1063-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewey R. E., Timothy, D. H. & Levings, C. S., III (1987) Proc. Natl. Acad. Sci. USA 84, 5374-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennell J. C., Wise, R. P. & Pring, D. R. (1987) Mol. Gen. Genet. 210, 399-406. [Google Scholar]
- 16.Izhar S. (1978) J. Hered. 69, 22-26. [Google Scholar]
- 17.Young E. G. & Hanson, M. R. (1987) Cell 50, 41-49. [DOI] [PubMed] [Google Scholar]
- 18.Nivison H. T. & Hanson, M. R. (1989) Plant Cell 1, 1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentolila S. & Hanson, M. R. (2001) Mol. Genet. Genomics 266, 223-230. [DOI] [PubMed] [Google Scholar]
- 20.Small I. D. & Peeters, N. (2000) Trends Biochem. Sci. 25, 46-47. [DOI] [PubMed] [Google Scholar]
- 21.Burge C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 22.Emanuelsson O., Nielsen, H., Brunak, S. & von Heijne, G. (2000) J. Mol. Biol. 300, 1005-1016. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Prot. Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 24.Scharfe C., Zaccaria, P., Hoertnagel, K., Jaksch, M., Klopstock, T., Dembrowski, M., Lill, R., Prokisch, H., Gerbitz, K. D., Neupert, W., et al. (2000) Nucleic Acids Res. 28, 155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey T. L. & Elkan, C. (1994) in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, eds. Altman, R., Brutlag, D., Karp, P., Lathrop, R. & Searls, D. (Am. Assoc. Artificial Intelligence Press, Menlo Park, CA), pp. 28–36.
- 26.Peeters N. M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A. & Small, I. (2000) J. Mol. Evol. 50, 413-423. [DOI] [PubMed] [Google Scholar]
- 27.Akashi K., Grandjean, O. & Small, I. (1998) FEBS Lett. 431, 39-44. [DOI] [PubMed] [Google Scholar]
- 28.Scott A., Wyatt, S., Tsou, P. L., Robertson, D. & Allen, N. S. (1999) BioTechniques 26, 1125., 1128–1132. [DOI] [PubMed] [Google Scholar]
- 29.Becker D., Kemper, E., Schell, J. & Masterson, R. (1992) Plant Mol. Biol. 20, 1195-1197. [DOI] [PubMed] [Google Scholar]
- 30.Horsch R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229-1231.17757866 [Google Scholar]
- 31.O'Connell M. A. & Hanson, M. R. (1985) Theor. Appl. Genet. 70, 1-12. [DOI] [PubMed] [Google Scholar]
- 32.Kohler R. H., Zipfel, W. R., Webb, W. W. & Hanson, M. R. (1997) Plant J. 11, 613-621. [DOI] [PubMed] [Google Scholar]
- 33.Reed M. L., Wilson, S. K., Sutton, C. A. & Hanson, M. R. (2001) Plant J. 27, 257-265. [DOI] [PubMed] [Google Scholar]
- 34.Nivison H. T., Sutton, C. A., Wilson, R. K. & Hanson, M. R. (1994) Plant J. 5, 613-623. [DOI] [PubMed] [Google Scholar]
- 35.Hamilton C. M. (1997) Gene 200, 107-116. [DOI] [PubMed] [Google Scholar]
- 36.Das A., Cohen, P. & Barford, D. (1998) EMBO J. 17, 1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisk D. G., Walker, M. B. & Barkan, A. (1999) EMBO J. 18, 2621-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lown F. J., Watson, A. T. & Purton, S. (2001) Biochem. Soc. Trans. 29, 452-455. [DOI] [PubMed] [Google Scholar]
- 39.Manthey G. M. & McEwen, J. E. (1995) EMBO J. 14, 4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancebo R., Zhou, X., Shillinglaw, W., Henzel, W. & Macdonald, P. M. (2001) Mol. Cell. Biol. 21, 3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poutre C. G. & Fox, T. D. (1987) Genetics 115, 637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewey R. E., Levings, C. S., III & Timothy, D. H. (1986) Cell 44, 439-449. [DOI] [PubMed] [Google Scholar]
- 43.Wise R. P., Gobelman, W. K., Pei, D., Dill, C. L. & Schnable, P. S. (1999) J. Hered. 90, 380-385. [DOI] [PubMed] [Google Scholar]