Abstract
Cold, hyperosmolarity, and abscisic acid (ABA) signaling induce RD29A expression, which is an indicator of the plant stress adaptation response. Two nonallelic Arabidopsis thaliana (ecotype C24) T-DNA insertional mutations, cpl1 and cpl3, were identified based on hyperinduction of RD29A expression that was monitored by using the luciferase (LUC) reporter gene (RD29A:LUC) imaging system. Genetic linkage analysis and complementation data established that the recessive cpl1 and cpl3 mutations are caused by T-DNA insertions in AtCPL1 (Arabidopsis C-terminal domain phosphatase-like) and AtCPL3, respectively. Gel assays using recombinant AtCPL1 and AtCPL3 detected innate phosphatase activity like other members of the phylogenetically conserved family that dephosphorylate the C-terminal domain of RNA polymerase II (RNAP II). cpl1 mutation causes RD29A:LUC hyperexpression and transcript accumulation in response to cold, ABA, and NaCl treatments, whereas the cpl3 mutation mediates hyperresponsiveness only to ABA. Northern analysis confirmed that LUC transcript accumulation also occurs in response to these stimuli. cpl1 plants accumulate biomass more rapidly and exhibit delayed flowering relative to wild type whereas cpl3 plants grow more slowly and flower earlier than wild-type plants. Hence AtCPL1 and AtCPL3 are negative regulators of stress responsive gene transcription and modulators of growth and development. These results suggest that C-terminal domain phosphatase regulation of RNAP II phosphorylation status is a focal control point of complex processes like plant stress responses and development. AtCPL family members apparently have both unique and overlapping transcriptional regulatory functions that differentiate the signal output that determines the plant response.
Plants tolerate environmental stress because of numerous physiological adaptations, which have been attributed to the function of various determinant genes (1). In Arabidopsis thaliana, transcription of RD (Responsive to Dehydration) (2) and COR (Cold Responsive) (3) genes is activated by cold or hyperosmotic stress. The plant hormone abscisic acid (ABA) activates transcription of some RD and COR genes through an interaction involving the cis element ABRE (ABA-responsive element) and basic leucine zipper transcription factors such as ABFs/AREBs (4, 5). However, expression of some RD or COR genes is activated by low temperature or desiccation, independent of ABA, by the interaction of CBF/DREB DNA-binding proteins with another cis element, DRE (6, 7). Overexpression of CBF/DREB in transgenic Arabidopsis plants induces ectopic expression of RD/COR genes and confers desiccation and cold tolerance (7, 8).
Recent genetic dissection of cold-, hyperosmolarity-, and ABA-induced signaling that regulates gene expression and adaptation indicates that the cascade signature is modulated by numerous positive and negative regulators (9–14). Signaling control of plant gene expression is known now to include components that function at various stages in mRNA metabolism (15). Gene expression regulation that is a consequence of controlling transcript processing and elongation is an emerging topic in biology (16).
RNA polymerase II (RNAP II) is a core component of the transcription complex. RNAP II catalyzes mRNA synthesis and is also involved in the regulation of various mRNA maturation processes such as capping, splicing, and polyadenylation (see review ref. 15). The largest subunit of RNAP II contains a C-terminal domain (CTD) that consists of an evolutionally conserved heptapeptide consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which is the pivotal phosphorylation target (17). In Arabidopsis, 41 repeats of the consensus sequence are present in RNAP II CTD (18). Evidence in nonplant systems indicates that RNAP II function is specified by the phosphorylation status of the CTD. RNAP II with an unphosphorylated CTD participates in the formation of the preinitiation complex (19). During the transcription cycle, the CTD is differentially phosphorylated, predominantly at Ser-2 or Ser-5. Ser-5 is phosphorylated by the TFIIH (Kin28) at promoters of Saccharomyces cerevisiae, which facilitates recruitment of capping enzyme (20). Ctk1 phosphorylates CTD at Ser-2 to facilitate transcription elongation. Ctk1 appears to be a yeast ortholog of mammalian P-TEFb that hyperphosphorylates CTD as the early elongation complex transitions into a productive elongation complex (16).
Although several kinases phosphorylate the CTD of RNAP II, the function of only one CTD phosphatase (yeast FCP1 and its human homolog hFCP1) has been reported (21). The essential yeast gene product FCP1 (TFIIF-associating CTD phosphatase) dephosphorylates CTD at Ser-2 (19). FCP1 apparently associates with RNAP II through interaction with the RNAP74 subunit of the TFIIF complex (21, 22), controls the phosphorylation status of Ser-2 during elongation, and promotes RNAP II recycling presumably by dephosphorylating the CTD at the termination step (19, 23). TFIIF is an integral component of the transcription preinitiation complex that interacts with RNAP II to facilitate message chain elongation (24–28).
Both positive and negative regulation of transcription by FCP1 has been reported. Inhibition of human (h)FCP1 in T cells by HIV-1 Tat protein is necessary for proliferation of the retrovirus HIV-1 (29). This finding indicates that hFCP1 antagonizes the capacity of the Tat protein to recruit the CTD kinase to RNAP II, which is necessary for hyperphosphorylation and chain elongation of the HIV-1 genome. Heat shock inactivates hFCP1, which leads to hyperphosphorylation of RNAP II CTD that is presumed to promote heat shock gene transcription (30). Transcript levels are reduced substantially when cells of the yeast fcp1 mutant are exposed to restrictive high temperature (22). These results indicate that the FCP1 phosphatase is a critical regulator of the phosphorylation status of the CTD, the biological consequences of which are only now beginning to emerge. It is now known that FCP1 functions in the dephosphorylation of Ser-2 in the CTD, but another phosphatase(s) dephosphorylates Ser-5 (19).
Screening of an Arabidopsis T-DNA mutant population for hyperactivation of the RD29A promoter after cold or ABA treatment resulted in the identification of two monogenic recessive hos (high expression of osmotically regulated genes) mutations, cpl1 and cpl3. Interestingly, cpl1 and cpl3 mutations are caused by T-DNA insertions in different members of the Arabidopsis CTD phosphatase-like (AtCPL) gene family and exhibit distinct RD29A activation, growth, and flowering time phenotypes. AtCPL1 and AtCPL3 phosphatases apparently are global regulatory entities that coordinate gene expression most likely by modulating the phosphorylation of RNAP II.
Materials and Methods
Plant Materials and Growth Conditions.
A T-DNA population in the A. thaliana C24 line that is homozygous for the chimeric RD29A:LUC reporter gene (31) was obtained after floral transformation of inflorescences with Agrobacterium GV3101 (pMP90RK) carrying the binary vector pSKI015 (provided by D. Weigel, The Salk Institute, La Jolla, CA). T1 plants were selected based on Liberty (30 mg/liter) resistance (32) and combined into 10-plant pools. Plants were grown either in a controlled room with 16 h of light at 22°C and 8 h of darkness at 18°C or in a greenhouse.
In vitro grown seedlings to be used for luciferase (LUC) imaging were obtained after seeds were surface-sterilized (1% sodium hypochlorite and 0.05% Tween 20 for 20 min), stratified for 2–4 days at 4°C, and sowed onto medium (pH 5.7) containing Murashige and Skoog (MS) salts, 30 g/liter sucrose, and 6 g/liter agar (31). For hygromycin selection, seeds were plated onto medium containing 1/4 × MS salts, 30 mg/liter hygromycin B, 100 mg/liter Timentin, and 8 g/liter agar.
LUC Imaging.
About 200 T2 seedlings from each 10-plant pool were screened for constitutive or cold-modulated (0°C, 2 days) or ABA-modulated (100 μM, 4 h) RD29A:LUC expression as described by Ishitani et al. (31). Mutants were also evaluated after NaCl (300 mM, 4 h) treatment. Image acquisition (5 min) with a charge-coupled device (CCD) system and processing with winview software (Roper Scientific, Trenton, NJ) were performed as described (31).
Molecular Analysis.
Genomic sequence flanking the T-DNA insertion was determined by using the thermal asymmetric interlaced PCR procedure of Liu et al. (33) with primers corresponding to nested regions internal to the left border and degenerate primers as listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. For reverse transcriptase–PCR analysis, 5 μg of total RNA from wild-type, cpl1, or cpl3 seedlings was used as template for reverse transcription using Superscript II (Invitrogen). Specific AtCPL fragments (AtCPL1: 0.5 kbp and AtCPL3: 0.7 kbp) were amplified from a fraction (1/100) of the reverse transcriptase reaction product by PCR [94°C 5 min (94°C 30 sec, 60°C 1 min, 72°C 2 min) × 30 and 72°C 5 min] using ExTaq DNA polymerase (Takara Shuzo, Kyoto, Japan) and the appropriate primers (CPL1F and CPL1R or CPL3F and CPL3R, Table 1). For Northern blot analysis, seedlings growing in vitro were stress-treated similarly to those used for LUC analysis. Total RNA was isolated with TRIzol (Invitrogen). Ten micrograms of total RNA was size-fractioned by electrophoresis and then blotted onto nylon membrane. Digoxygenin-labeled probe was prepared by PCR, hybridized to the blotted RNA, and detected according to the manufacturer's protocol (Roche).
Nuclear Run-On Analysis.
One-week-old wild-type (C24) and cpl1 seedlings grown in vitro were cold-treated (0°C, 15 h). Nuclei were isolated, and the reaction was performed as described by Cushman (34). Labeled transcripts were purified with TRIzol (Invitrogen) according to the manufacturer's protocol. To prepare immobilized probes, PCR products of cDNAs encoding RD29A (GenBank accession no. L22567: positions 79–640 and 1339–2096), pyrroline-5-carboxylate synthase (P5CS) (GenBank accession no. X86777: positions 73–671 and 1424–2142), proline oxidase (POX) (GenBank accession no. D83025: position 113–627), tubulin (GenBank accession no. AF367301: position 76–1551), LUC (GenBank accession no. U03687: position 1430–2230), and genomic fragment of 18S RNA (GenBank accession no.X52322: position 4135–4927) were produced by using cDNA or genomic fragments that have been cloned into pBluescript as templates. Three hundred nanograms of purified PCR product was slot-blotted onto nitrocellulose membrane, hybridized with purified run-on product for 48 h, and washed twice for 15 min with 6 × SSC, 0.1% SDS at room temperature and then twice for 15 min with 0.1 × SSC, 0.1% SDS at 55°C. The blot was placed onto an imaging plate overnight and analyzed by using the Typhoon 8600 PhosphorImager (Molecular Dynamics).
Genetic Complementation Analysis.
The complementation constructs for expressing AtCPL1 or AtCPL3 were produced by joining PCR fragments of the AtCPL promoter region and the cDNA encoding AtCPL. The PCR products were first cloned into pBluescript derivatives, and the sequence was confirmed. The AtCPL1 cassette consists of the promoter (bacterial artificial chromosome F17L22, position 42149–43915) joined to the cDNA corresponding to entire coding sequence (F17L22.130) at a unique NheI site and placed between the SalI and SmaI sites upstream of the Nos terminator in pBIB-HYG binary vector (35) to produce pBCPL1. Similarly, pBCPL3 contains a cassette that encompasses the AtCPL3 promoter region to the middle of the fifth exon (F4P9 position 110851–113732) and a cDNA fragment (from SacI at 1311 to the stop codon of AF486633) joined by using a unique SacI site and placed between HindIII and XbaI sites in pBIB-HYG. The resulting plasmids pBCPL1 and pBCPL3 were introduced into Agrobacterium GV3101 and used for transformation of cpl1 and cpl3 mutant plants, respectively. T1 plants were selected by using hygromycin.
Phosphatase Assay.
cDNA fragments encoding AtCPL1 and AtCPL3 were amplified by Pfx polymerase with primers containing attB1 (5′ primers) and attB2 (3′ primers) sequence (B1C15 and B2C13, B1C35 and B2C33, Table 1) and recombined into pDONR201 and then recombined into pDEST17 according to the manufacturer's protocol (Invitrogen) to produce pDCPL1 and pDCPL3. pDCPL1 was transformed into Escherichia coli BL21SI and pDCPL3 into BL21-star (Invitrogen). For recombinant protein synthesis, BL21SI cells without and with pDCPL1 plasmid were induced for 10 h by adding 0.3 M NaCl into the medium. pDCPL3 was introduced into BL21 star because of low AtCPL3 expression in BL21SI, and cells were induced with 1 mM isopropyl β-d-thiogalactoside for 10 h. The recombinant AtCPL1 and AtCPL3 proteins were recovered in the insoluble fraction after sonication of bacterial cells in 50 mM Tris (pH 7.5) buffer and extensive washes with 1% Triton X-100. AtCPL proteins were prepared in nonreducing SDS/PAGE loading buffer and size-fractioned in a 6.25% SDS/PAGE gel (36). After eletrophoresis, the gel was incubated in 2.5% Triton X-100 for 30 min and then phosphatase assay buffer (50 mM Tris⋅HCl, pH 7.9/10% glycerol/10 mM MgCl2/10 mM potassium acetate/1 mM ZnCl2) for 30 min. The equilibrated gel was then sealed in a plastic bag with 10 ml of phosphatase buffer containing 33 μM CDP-star, a phosphate ester substrate (37). The chemiluminescent signal was collected by using the CCD imaging system for 60 min at room temperature.
Results
Isolation of cpl Mutants.
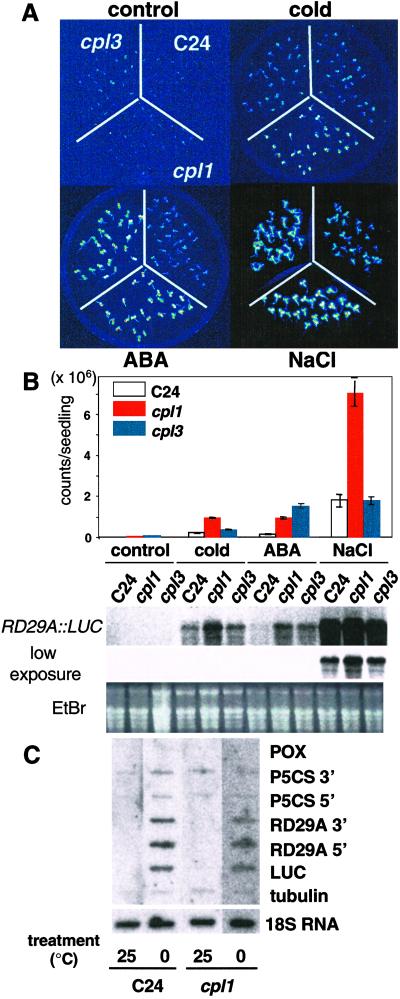
A screen of 6,000 T2 lines by CCD imaging identified two mutations that exhibit a hos phenotype based on RD29A:LUC expression after cold and/or ABA treatment of seedlings (31). These mutations were designated as cpl1 and cpl3 (see below for explanation). Hyperinduction of LUC occurred in response to cold, ABA, or salt in cpl1 seedlings but only in response to ABA in cpl3 seedlings (Fig. 1). Northern blot analysis determined that the RD29A:LUC transcript increases concomitantly with luminescence. Only a small increase in the expression level of endogenous RD29A was detected (data not shown). Although a modest increase in LUC, RD29A, and CBF3 (encodes upstream transcription factor) transcript accumulation was observed when cpl1 seedlings were cold-treated (Fig. 1B, data not shown), nuclear run-on analysis did not differentiate expression between wild-type and cpl1 seedlings after cold treatment (Fig. 1C). These results suggest that CPL1 functions downstream of transcription initiation, i.e., in transcription elongation, RNA processing, or degradation. cpl1 plants grow more vigorously and flower later than wild type whereas cpl3 plants exhibit reduced fresh weight gain and flower early (Fig. 2). cpl1 and cpl3 seedlings have longer hypocotyls than wild type when grown under light. Root growth in medium with NaCl (0–180 mM), freezing tolerance before and after cold acclimatization, and germination in the presence of ABA were not affected in cpl1 and cpl3 plants when compared with wild type (data not shown).
Fig 1.
A. thaliana cpl1 and cpl3 cause differential RD29A:LUC hyperexpression in response to cold, ABA, and NaCl. (A) Ten-day-old cpl1 and cpl3 T4 and wild-type (C24) seedlings were analyzed after no treatment (control) and after cold (0°C, 2 days), ABA (100 μM, 4 h), and NaCl (300 mM, 4 h) treatment. (B) Luminescence intensity of 8–20 plants per treatment was determined by quantitative analysis of the image recorded with the CCD camera. Error bar represents standard error of the mean. Steady-state transcript level of the LUC transgene was determined after plants were treated as indicated above except for the cold (0°C, 24 h) treatment. The RNA gel blot was hybridized with a digoxygenin-labeled probe corresponding to the entire LUC coding region. (C) Nuclear run-on transcription analysis of cold-treated seedlings. Eight-day-old seedlings growing in vitro were incubated either in 25°C or 0°C for 15 h before isolation of nuclei. Nuclear isolation, run-on reaction, and hybridization were performed as described by Cushman (34). Transcripts hybridized to immobilized probe were detected by Typhoon phosphorimager after 16 h of exposure.
Fig 2.
cpl1 and cpl3 mutations differentially affect biomass accumulation and flowering time and increase hypocotyl length. (A) Photograph illustrates morphological differences of 5-week-old cpl1, cpl3, and wild type (C24). (Magnification: ×0.2.) (B) Fresh weight of the seventh rosette leaf was determined as an indicator of biomass. (C) Flowering time is expressed as the number of leaves per plant at the onset of anthesis. (D) Hypocotyl length of 5-day-old seedlings was measured. For B–D, data are averages of 40–50 plants ± the standard error of the mean.
CPL Loci Encode FCP-Like Phosphatases.
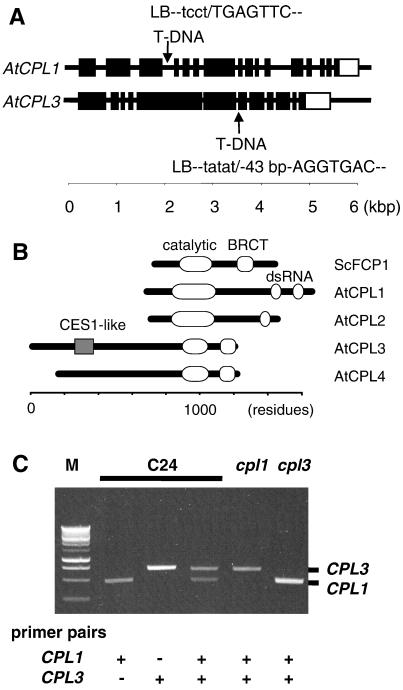
Thermal asymmetric interlaced-PCR analysis (33) located a T-DNA insertion in the fourth intron of AtCPL1 (chromosome IV, position 45179 of bacterial artificial chromosome F17L22) and in the seventh exon of AtCPL3 (chromosome II, position 120888 of bacterial artificial chromosome F4P9), in the genome of cpl1 and cpl3 plants, respectively (Fig. 3A). AtCPL1 and AtCPL3 encode proteins with high sequence similarity to the evolutionarily conserved CTD phosphatases (21) as shown in Fig. 3B and Fig. 6, which is published as supporting information on the PNAS web site. Four genes encoding CTD phosphatase family members can be identified in the Arabidopsis genome (www.tair.org), and the corresponding proteins are now designated as CPL phosphatases (Fig. 3B). The existence of AtCPL1-4 expressed sequence tag sequences (GenBank accession nos. T44887, AV530190, F15563, and BE529486) indicate that all are functional genes. AtCPL3 transcript was undetectable in cpl3 plants, whereas a very low abundance of AtCPL1 transcript was detectable in cpl1 plants (Fig. 3C).
Fig 3.
T-DNA insertional mutations (cpl1 and cpl3) abrogate the expression of AtCPL1 and AtCPL3, respectively, which are members of a four-gene family in A. thaliana. (A) Schematic illustration indicates the location of the T-DNA insertions in AtCPL1 and AcCPL3. Exons (▪) were deduced from the cDNA sequence corresponding to AtCPL1 and AtCPL3. The open boxes indicate the 3′ untranslated region. Sequences at the T-DNA left border (lowercase)-genome (uppercase) junctions were determined by thermal asymmetric interlaced-PCR analysis. The cpl3 contains a 43-bp insertion of unknown origin between the T-DNA left border and genomic sequence. (B) Schematic of AtCPLs 1–4 (GenBank accession nos. CAB36811.1, CAB69855.1, AF486633, and AAD25584.1, respectively) domain structures compared with the prototypical CTD phosphatase yeast FCP1 (ScFCP1, GenBank accession no. NP014004). Identified are the phosphatase catalytic domain (19), the BRCT domain for proposed RAP74 interaction (19), the CES1 (capping enzyme suppressor) (41)-like region, and the dsRNA-binding domain (43). (C) Transcript abundance in wild-type (C24), cpl1, or cpl3 plants determined by reverse transcriptase–PCR using gene-specific primers for AtCPL1 (CPL1) and AtCPL3 (CPL3) (see Table 1). M, Promega 1-kbp DNA ladder.
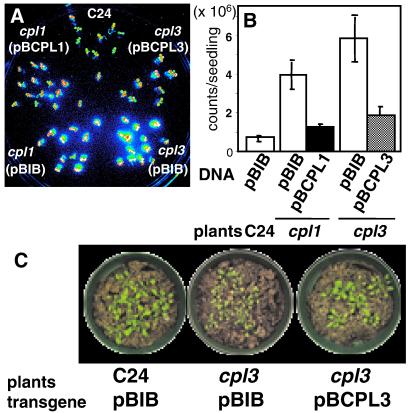
Genetic cosegregation analysis of progeny from the backcross to C24(RD29A:LUC) established a linkage between phenotype and T-DNA insertion in each AtCPL gene. All 54 (C24 × cpl1) and 64 (C24 × cpl3) F1 plants were similar to wild type, indicating that both mutations are recessive. All 48 hos plants selected from F2 progeny by CCD imaging contained a homozygous T-DNA insertion. These results indicate that a tight linkage existed between each mutated cpl locus and the hos phenotype (data not shown). Expression of AtCPL1 and AtCPL3 cDNAs driven by the native promoter suppressed the mutant phenotype of cpl1 and cpl3, respectively (Fig. 4). Furthermore, Xiong, et al. (38) indicate that several mutant alleles of CPL1/FRY2 exhibit phenotypes similar to cpl1. These results confirm that AtCPL1 and AtCPL3 are negative regulators of RD29A expression.
Fig 4.
AtCPL expression functionally complements the hos (high expression of osmotically regulated genes) phenotype of cpl seedlings. Illustrated are wild-type (C24), cpl1, or cpl3 T1 seedlings transformed with vector (pBIB: empty bars in B) or AtCPL1 (pBCPL1: solid bar in B), or AtCPL3 (pBCPL3: hatched bar in B). T1 transformants were transferred to hygromycin-free Murashige and Skoog medium before image acquisition. LUC luminescence image (A) was captured after ABA treatment (100 μM, 3 h) and quantified (B). Bars represent standard error of the mean. Subsequently cpl3 transformants were transferred to soil (C). (Magnifications: ×0.16.)
Structure and Enzymatic Activity of AtCPL1 and AtCPL3.
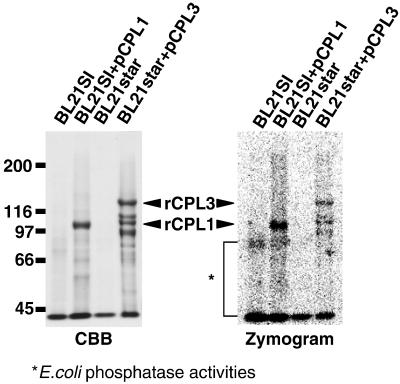
Fig. 3B illustrates the domains in AtCPLs that can be deduced from cDNA and genomic nucleotide sequence data. AtCPL1 and AtCPL3 encode CPLs of 995 and 1,241 aa, respectively. When AtCPL1 and AtCPL3 were expressed in E. coli as (His)6-tagged recombinant proteins, the proteins migrated on SDS/PAGE gels with Mr of 100 kDa and 130 kDa, respectively. Both AtCPL proteins contain the highly conserved phosphatase motif ΨΨΨDXDX(T/V)ΨΨ (39). Recombinant AtCPL1 and AtCPL3 exhibit phosphatase activity that is detected based on hydrolysis of artificial phosphatase substrate CDP-star (Fig. 5). We identified two consensus double-stranded RNA (dsRNA)-binding motifs (40) in the C terminus of AtCPL1. This motif was also found in Arabidopsis HYL1 that regulates plant ABA sensitivity (41). The AtCPL1 dsRNA-binding domain is most homologous to that in TRBP (TAR-RNA binding protein) that binds to the TAR-RNA in the long terminal repeat of the HIV genome (42). Similarly, AtCPL2 contains one dsRNA-binding domain. In contrast, C termini of AtCPL3 and AtCPL4 contain a motif with marked similarity to BRCA C-terminal (BRCT) domain, which is found in many proteins involved in DNA damage responsive cell cycle checkpoint (43). The BRCT domain is also in human and yeast FCP1, making AtCPL3 and AtCPL4 strong candidates for FCP1 orthologs.
Fig 5.
AtCPL are phosphatases. AtCPL1 and AtCPL3 cDNAs were placed under the control of the T7 promoter in the vectors pCPL1 and pCPL3 and expressed in E. coli BL21SI and BL21 star cells to produce rCPL1 and rCPL3, respectively. Protein was isolated from cells without and with pCPL1 or pCPL3 plasmid, normalized based on the culture volume (0.25 ml culture per lane), and then fractionated by SDS/PAGE (6.25% gel). Phosphatase activity was visualized by CCD imaging, after impregnating the gel with CDP star substrate (Zymogram). Protein bands were visualized by Coomassie brilliant blue R-25 (CBB) staining after image acquisition. Innate phosphatase activity of bacterial cells is indicated by *. Zymogram indicates the presence of low Mr degradation product of rCPL3 in bacterial cells.
Discussion
Fine Tuning of Gene Expression at the Level of Transcription Elongation.
Plant stress adaptation and development requires the coordination of diverse cellular process including positive and negative regulation of gene expression. Constitutive activation of the stress response by overexpression of DREB/CBF transcription factors facilitates stress adaptation but is disadvantageous to plants growing in the absence of the stress (7, 8). As environmental conditions can change rapidly, the ability to modulate gene expression after transcriptional activation is presumably an adaptive capacity of plants. We have determined that two Arabidopsis loci encoding FCP-like phosphatases (AtCPLs) function genetically as negative regulators of RD29A expression whose up-regulation is linked to cold and osmotic stress adaptation (7, 8). cpl1 and cpl3 mutations also alter growth and flowering time. Nuclear run-on analysis indicated that the transcription initiation rate of several cold-inducible genes and LUC was not affected by the cpl1 mutation, even though the steady-state LUC transcript level was clearly increased in response to cold stress. This finding is consistent with the hypothesis that cpl mutations affect transcription elongation; a difference would not be detected by nuclear run-on analysis. However, it is not possible to exclude equivocally that the undetectable increase in transcription initiation rate can contribute to the difference in steady-state transcript level.
The fry2 mutations reported by Xiong et al. (38) are allelic to cpl1. fry2–1 is a nonsense mutation caused by an internal stop codon in CPL1/FRY2 coding sequence whereas cpl1 is an allele that results in reduced CPL1/FRY2 expression. Stress-induced gene expression in fry2–1 is much higher than in cpl1. This allelic difference also may explain why it is not possible to establish that a cold stress-sensitive phenotype is associated with cpl1 mutation even though fry2–1 mutants displayed cold stress sensitivity. However, further analysis is required to dissect additional diverse phenotypes of different cpl1 alleles.
Two Arabidopsis CTD Phosphatase Subfamilies and Different Phenotypes.
Neither the cpl1 nor the cpl3 mutation, unlike the fcp1 mutation, is lethal and each causes some unique phenotypes. Therefore, AtCPL gene products apparently have an overlapping function that may be associated with the duplicate genes that exist in the two AtCPL subfamilies (see below). Each may also regulate the expression of specific gene sets when plants respond to environmental changes or proceed through development.
The phosphorylation status of RNAP II CTD coordinates transcription and RNA processing, such as mRNA capping, splicing, and polyadenylation (15). Most of the RNAP II CTD kinases, Ctk1, Kin28, and Srb10, belong to the same cyclin-dependent kinase superfamily (19). However, FCP1 is the only characterized CTD phosphatase. It has been established recently that yeast FCP1 is a Ser-2-specific CTD phosphatase but an, as yet, unidentified enzyme(s) is responsible for Ser-5 dephosphorylation. Perhaps other members of the FCP1 family are to be identified in yeast. The human FCP1 produces two alternatively spliced products, FCP1a and FCP1b, although the biochemical function of FCP1b has not been elucidated (43).
Several genes in the Arabidopsis genome have been annotated as encoding CTD phosphatases but most are composed of about 300 aa and are likely “DDDD” nonspecific acid phosphatase superfamily members (39). AtCPLs 1–4 encode peptides that contain not only the catalytic FCP CTD phosphatase domain (21) but additional domains that are associated with proteins that function in the transcription elongation complex (Figs. 3 and 6). Although these four AtCPLs are likely CTD phosphatases, the presence of distinct domains within the proteins facilitates categorization into two groups based on predicted mode of interaction with RNAP II. The BRCA1 C-terminal (BRCT) domain (43) in AtCPL3 and AtCPL4 resembles that in the prototypical CTD phosphatases such as FCP1 of S. cerevisiae or hFCP. The BRCT domain in FCP1 proteins interact with RAP74 subunit of TFIIF of the RNAP II complex during transcription elongation (21, 22).
AtCPL1 and AtCPL2 are the only known examples of peptides containing both dsRNA-binding and FCP-like phosphatase domains. Consequently, there is no precedence on which to base prediction about the biological substrate(s), although the presence of the FCP homology domain strongly implicates the CTD of RNAP II as a substrate for catalysis. The dsRNA-binding domain of AtCPL1 is homologous to human TRBP that was isolated based on molecular interaction with TAR RNA (40). TAR RNA plays a critical role in proliferation of HIV-1, as it is the interaction site for Tat protein complexed with CTD kinase that hyperphosphorylates RNAP II, which is transcribing the HIV-1 genome (45). Furthermore, a putative RNA-binding protein RD is an essential subunit of NELF (negative elongation factor), which represses transcription elongation (46) and is a target of hepatitis delta antigen (HDAg) that stimulates the transcription elongation by RNAP II (47). It is proposed that the RD subunit stabilizes interaction of NELF and RNAP II by binding to a nascent transcript (46). By analogy, AtCPL1 may bind to the 5′ region of elongating mRNA, a process that positions its phosphatase domain in close proximity to the CTD of RNAP II. The sequence specificity of the dsRNA-binding domains in AtCPL1 and AtCPL2 may direct targeting to specific RNA substrates (48). Specific protein–protein interaction with regions in the N termini of AtCPL3 and AtCPL4 could facilitate the targeting of each to unique transcript species. The presence of an AtCPL3 region that is homologous to CES1 (capping enzyme suppressor) in yeast (49) is unique among known CTD phosphatases and may imply that AtCPL3 may be directly involved in capping process. Further experimentation is required for unequivocal identification of the biological target(s).
RNA Metabolism and Signaling.
In Arabidopsis, several genes encoding proteins related to RNA metabolism are involved in osmotic stress and ABA signaling (50). Deficiency of mRNA cap-binding protein ABH1, Sm-like protein SAD1, or dsRNA-binding protein HYL1 causes hypersensitivity to ABA (9, 10, 41). Control of transcription is achieved by a combination of positive and negative regulatory factors that facilitate cycling of RNAP II through the active and inactive states (51). It is conceivable that modulation of the phosphorylation status of the CTD by different phosphatase family members is a focal regulatory process in the coordination of transcription in plants. As stress-induced regulation of RNAP II functions in plants is largely unexplored, identification of downstream regulons controlled by AtCPL family members and the signal perception mechanism that activates AtCPLs are likely the next focus for dissection of this global regulatory mechanism.
Supplementary Material
Acknowledgments
We thank Professor Detlef Weigel (The Salk Institute) for providing pSKI015 plasmid. Bacterial artificial chromosome clones were provided by the Arabidopsis Biological Resource Center, Columbus, OH. We also thank Jean Clithero and Tara Ware for technical assistance. Research was supported, in part, by National Science Foundation Plant Genome Award DBI-9813360, National Science Foundation Grant IBN-9808398, and U.S. Department of Agriculture National Research Initiative Grant 2000-00664. This is journal article no. 16593 of the Purdue University Agricultural Experimental Station.
Abbreviations
ABA, abscisic acid
CTD, C-terminal domain
CPL, CTD phosphatase-like
LUC, luciferase
RNAP II, RNA polymerase II
CCD, charge-coupled device
dsRNA, double-stranded RNA
AtCPL, Arabidopsis CPL
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF486633).
References
- 1.Hasegawa P. M., Bressan, R. A., Zhu, J.-K. & Bohnert, H. J. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 463-499. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi-Shinozaki K., Koizumi, K., Urao, S. & Shinozaki, K. (1992) Plant Cell Physiol. 33 217-224. [Google Scholar]
- 3.Hajela R. K., Horvath, D. P., Gilmour, S. J. & Thomashow, M. F. (1990) Plant Physiol. 93 1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi H., Hong, J., Ha, J., Kang, J. & Kim, S. Y. (1999) J. Biol. Chem. 275 1723-1730. [DOI] [PubMed] [Google Scholar]
- 5.Uno Y., Furihara, T., Abe, H., Yoshida, R., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2000) Proc. Natl. Acad. Sci. USA 97 11632-11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockinger E., Gilmour, S. J. & Thomashow, M. F. (1997) Proc. Natl. Acad. Sci. USA 94 1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1998) Plant Cell 10 1391-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaglo-Ottosen K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O. & Thomashow, M. F. (1998) Science 280 104-106. [DOI] [PubMed] [Google Scholar]
- 9.Xiong L., Gong, Z., Rock, C. D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D. & Zhu, J.-K. (2001) Dev. Cell 1 771-781. [DOI] [PubMed] [Google Scholar]
- 10.Hugouvieux V., Kwak, J. M. & Schroeder, J. I. (2001) Cell 106 477-487. [DOI] [PubMed] [Google Scholar]
- 11.Xiong L., Ishitani, M., Lee, H. & Zhu, J.-K. (2001) Plant Cell 13 2063-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong L., Lee, B.-H., Ishitani, M., Lee, H., Zhang, C. & Zhu, J.-K. (2001) Genes Dev. 15 1971-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B. & Zhu, J.-K. (2001) Genes Dev. 15 912-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J.-K. (2001) Curr. Opin. Plant Biol. 4 401-406. [DOI] [PubMed] [Google Scholar]
- 15.Hirose Y. & Manley, J. L. (2000) Genes Dev. 14 1415-1429. [PubMed] [Google Scholar]
- 16.Conaway J. W., Shilatifard, A., Dvir, A. & Conaway, R. C. (2000) Trends Biochem. Sci. 25 375-380. [DOI] [PubMed] [Google Scholar]
- 17.Dahmus M. E. (1996) J. Biol. Chem. 271 19009-19012. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich M. A., Prenger, J. P. & Guilfoyle, T. J. (1990) Plant Mol. Biol. 15 207-223. [DOI] [PubMed] [Google Scholar]
- 19.Cho E.-J., Kobor, M. S., Kim, M., Greenblatt, J. & Buratowski, S. (2001) Genes Dev. 15 3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder S. C., Schwer, B., Schuman, S. & Bentley, D. (2000) Genes Dev. 14 2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archambault J., Chambers, R., Kobor, M. S., Ho, Y., Bolotin, D., Andrews, B., Kane, C. M. & Greenblatt, J. (1997) Proc. Natl. Acad. Sci. USA 94 14300-14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobor M. S., Archambault, J., Lester, W., Holstege, F. C. P., Gileadi, O., Jansma, D. B., Jennings, E. G., Kouyoumdjian, F., Davidson, A. R., Young, R. A., et al. (1999) Mol. Cell 4 55-62. [DOI] [PubMed] [Google Scholar]
- 23.Cho H., Kim, T.-K., Mancebo, H., Lane, W. S., Flores, O. & Reinberg, D. (1999) Genes Dev. 13 1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengal E., Flores, O., Krauskopf, A., Reinberg, D. & Aloni, Y. (1991) Mol. Cell. Biol. 11 1195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton Z. F., Kileen, M., Sopta, M., Ortolan, L. G. & Greenblatt, J. (1988) Mol. Cell. Biol. 8 1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores O., Lu, H., Killeen, M., Greenblatt, J., Burton, Z. F. & Reinburg, D. (1991) Proc. Natl. Acad. Sci. USA 88 9999-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killeen M., Coulombe, B. & Greenblatt, J. (1992) J. Biol. Chem. 267 9463-9466. [PubMed] [Google Scholar]
- 28.Price D. H., Sluder, A. E. & Greenleaf, A. (1989) Mol. Cell. Biol. 9 1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall N. F., Dahmus, G. K. & Dahmus, M. E. (1998) J. Biol. Chem. 273 31726-31730. [DOI] [PubMed] [Google Scholar]
- 30.Dubois M. F. & Bensaude, O. (1998) Cell Stress Chaperones 3 147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishitani M., Xiong, L., Stevenson, B. & Zhu, J.-K. (1997) Plant Cell 9 1935-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel D. & Ahn, J.-H. (2000) Plant Physiol. 122 1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y.-G., Mitsukawa, N., Oosumi, T. & Whittier, R. (1995) Plant J. 8 457-463. [DOI] [PubMed] [Google Scholar]
- 34.Cushman J. C. (1995) Methods Cell Biol. 50 113-128. [DOI] [PubMed] [Google Scholar]
- 35.Becker D. (1990) Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koiwa H., D'Urzo, M. P., Zhu-Salzman, K., Ibeas, J. I., Shade, R. E., Murdock, L. L., Bressan, R. A. & Hasegawa, P. M. (2000) Anal. Biochem. 282 153-155. [DOI] [PubMed] [Google Scholar]
- 37.Trayhurn P., Thomas, M. E., Duncan, J. S., Black, D., Beattie, J. H. & Rayner, D. V. (1995) Biochem. Soc. Trans. 23 494S. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L., Lee, H., Ishitani, M., Tanaka, Y., Stevenson, B., Kiowa, H., Bressan, R. A., Hasegawa, P. M. & Zhu, J.-K. (2002) Proc. Natl. Acad. Sci. USA 99 10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thaller M. C., Schippa, S. & Rossolini, G. M. (1998) Protein Sci. 7 1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston D. S., Brown, N. H., Gall, J. G. & Jantsch, M. (1992) Proc. Natl. Acad. Sci. USA 89 10979-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu C. & Fedoroff, N. (2000) Plant Cell 12 2351-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gatignol A., Buckler-White, A., Berkhout, B. & Jeang, K.-T. (1991) Science 251 1597-1600. [DOI] [PubMed] [Google Scholar]
- 43.Bork P., Hofmann, K., Bucher, P., Neuwald, A. F., Altschul, S. F. & Koonin, E. V. (1997) FASEB J. 11 68-76. [PubMed] [Google Scholar]
- 44.Archambault J., Pan, G., Dahmus, G. K., Cartier, M., Marshall, N., Zhang, S., Dahmus, M. E. & Greenblatt, J. (1998) J. Biol. Chem. 273 27593-27601. [DOI] [PubMed] [Google Scholar]
- 45.Isel C. & Karn, J. (1999) J. Mol. Biol. 290 929-941. [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., Hasegawa, J. & Handa, H. (1999) Cell 97 41-51. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi Y., Filipovska, J., Yano, K., Furuya, A., Inukai, N., Narita, T., Wada, T., Sugimoto, S., Konarska, M. M. & Handa, H. (2001) Science 293 124-127. [DOI] [PubMed] [Google Scholar]
- 48.Ramos A., Grunert, S., Adams, J., Micklem, D., Proctor, M., Freund, S., Bycroft, M., St. Johnston, D. & Varani, G. (2000) EMBO J. 19 997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwer B. & Shuman, S. (1996) Gene Exp. 5 331-344. [PMC free article] [PubMed] [Google Scholar]
- 50.McCourt P. (2001) Mol. Cell 8 1157-1158. [DOI] [PubMed] [Google Scholar]
- 51.Kim D.-k., Yamaguchi, Y., Wada, T. & Handa, H. (2001) Mol. Cells 11 267-274. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.