Abstract
Eukaryotic ribosomes are made of two components, four ribosomal RNAs, and approximately 80 ribosomal proteins (r-proteins). The exact number of r-proteins and r-protein genes in higher plants is not known. The strong conservation in eukaryotic r-protein primary sequence allowed us to use the well-characterized rat (Rattus norvegicus) r-protein set to identify orthologues on the five haploid chromosomes of Arabidopsis. By use of the numerous expressed sequence tag (EST) accessions and the complete genomic sequence of this species, we identified 249 genes (including some pseudogenes) corresponding to 80 (32 small subunit and 48 large subunit) cytoplasmic r-protein types. None of the r-protein genes are single copy and most are encoded by three or four expressed genes, indicative of the internal duplication of the Arabidopsis genome. The r-proteins are distributed throughout the genome. Inspection of genes in the vicinity of r-protein gene family members confirms extensive duplications of large chromosome fragments and sheds light on the evolutionary history of the Arabidopsis genome. Examination of large duplicated regions indicated that a significant fraction of the r-protein genes have been either lost from one of the duplicated fragments or inserted after the initial duplication event. Only 52 r-protein genes lack a matching EST accession, and 19 of these contain incomplete open reading frames, confirming that most genes are expressed. Assessment of cognate EST numbers suggests that r-protein gene family members are differentially expressed.
The eukaryotic ribosome is a complex structure composed of four rRNAs and about 80 ribosomal proteins (r-proteins). It represents an essential piece of the cell machinery, responsible for protein synthesis, and as such plays a major role in controlling cell growth, division, and development. For example, previous studies have shown that genetic defects in ribosomal components, such as reduction of the levels of individual r-proteins, can cause deleterious effects on the development and physiology of an organism. In Drosophila melanogaster, mutations in r-proteins genes cause the haplo-insufficient Minute phenotype with reduced growth and cell division rates, characterized by a reduced body size and short, thin bristles (Lambertsson, 1998). In contrast, a conditional deletion in the gene encoding r-protein S6 in adult mice (Mus musculus) affects cell cycle progression but not cell growth (Volarevic et al., 2000). In humans, a quantitative reduction in synthesis of the X-linked form of r-protein S4 is observed in individuals with Turner syndrome (monosomic for X) and may contribute to this complex phenotype, which includes short stature and infertility (Zinn and Ross, 1998). In plants, mutations in r-protein genes affect embryo viability or plant development (Van Lijsebettens et al., 1994; Tsugeki et al., 1996; Revenkova et al., 1999; Ito et al., 2000). In addition, a positive correlation was reported between the level of r-protein gene transcript accumulation and cell division in suspension culture cells (Joanin et al., 1993; Garo et al., 1994) or tissues such as auxin-treated hypocotyls, apical meristems, young leaves, and lateral roots (Gantt and Key, 1985; Williams and Sussex, 1995).
Numerous analyses on prokaryotic ribosomes and r-proteins have provided significantly to our knowledge of ribosome structure and composition. Three-dimensional structures of the 30S and 50S ribosomal subunits of thermophilic eubacteria (30S, Thermus thermophilus; 50S, Haloarcula marismortui) have recently been described at 5.5- and 2.5-Å resolution, respectively, from crystallographic data (Ban et al., 1999, 2000; Clemons et al., 1999). In Escherichia coli, 55 r-proteins have been identified and their amino acid sequences determined (Wittmann, 1982; Wittmann-Liebold et al., 1990). The ordered assembly process of eubacterial ribosomes is also reasonably well known (Nomura et al., 1984; Culver et al., 1999). It is generally accepted that ribosomes of an archaebacterial ancestor gave rise to the cytosolic ribosomes of eukaryotes (Matheson et al., 1990; Wittmann-Liebold et al., 1990; Wool et al., 1995). By contrast, the r-proteins of plastids and mitochondria show strong evolutionary similarity to those of eubacteria and include organelle-specific proteins (Graack and Wittmann-Liebold, 1998; Koc et al., 2000; Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000). In eukaryotes, the protein composition of rat (Rattus norvegicus) ribosomes was determined by direct protein sequencing followed by gene cloning and a presumed complete set of 79 proteins was compiled (Wool et al., 1995). In addition, genes corresponding to 78 Saccharomyces cerevisiae r-proteins were identified through genome sequencing efforts (Goffeau et al., 1996; Planta and Mager, 1998). Eukaryotic r-proteins can be classified based on homology to r-proteins of archae- and eubacteria (Wool et al., 1995). The 80S rat ribosome contains 33 proteins for which orthologues can be found in eubacteria, archaebacteria, and eukaryotes (Group I); 35 proteins with orthologues in archaebacteria and other eukaryotes (Group II); and 21 proteins that appear to be unique to eukaryotes (Group III). The striking evolutionary conservation of r-proteins is not surprising given the constraints of rRNA-protein interactions, coordinated ribosome assembly, and ribosome function. In fact, phylogenetic relationships between animal, fungi, and plant kingdoms have been inferred from comparison of orthologous r-proteins (Veuthey and Bittar, 1998).
The expression and distribution of r-protein genes of both prokaryotes and eukaryotes has also been examined. In eubacteria, most of the r-protein genes are clustered in a few operons, which allows for coordinated regulation (Nomura et al., 1984). Kenmochi et al. (1998b) recently mapped 75 human r-protein genes and showed that they are distributed over all chromosomes, with a bias toward chromosome 19. Synthesis of r-proteins in eukaryotes undoubtedly requires coordination of now unlinked genes. It is striking that the regulation of r-protein gene expression appears to occur at the transcriptional level in yeast (Saccharomyces cerevisiae; Planta and Mager, 1998) and predominantly at the translational level in animals (Meyuhas, 2000; Meyuhas and Hornstein, 2000).
In contrast to the information available on r-proteins and r-protein genes in prokaryotes and a few eukaryotic models (rats and yeast), limited information is available on r-proteins and the number, distribution, and expression of r-protein genes in plants. Gantt and Key (1983) resolved 40 and 51 proteins of the small (40S) and large (60S) subunits of the cytosolic ribosomes of soybean (Glycine max) by two-dimensional gel electrophoresis. In addition, plant genes encoding 77 orthologues to rat cytosolic r-proteins were identified (Bailey-Serres, 1998), including an r-protein (P3) that is apparently limited to plants (Szick et al., 1998). Information describing the genomic distribution of r-protein genes in plants is limited to the mapping of 57 loci for r-protein genes in rice (Oryza sativa; Wu et al., 1995). However, because this study relied on RFLPs, many loci may have been missed due to lack of polymorphism and cross hybridization between members of gene families. Reconstruction of full-length Arabidopsis r-protein cDNAs from redundant overlapping expressed sequence tags (ESTs) demonstrated that the occurrence of small gene families with several transcribed genes seems to be the rule rather than an exception (Cooke et al., 1997).
Several studies on plant r-protein genes have revealed the presence of multigene families in which members show both overlapping and differential patterns of mRNA accumulation (Larkin et al., 1989; Van Lijsebettens et al., 1994; Williams and Sussex, 1995; Dresselhaus et al., 1999; Revenkova et al., 1999). Evidence that r-protein gene expression may be controlled at a posttranscriptional level was observed for L13 in rapeseed (Brassica napus) and Arabidopsis (Saez-Vasquez et al., 2000), P2 in anoxic roots of maize (Zea mays) seedlings (Fennoy and Bailey-Serres, 1998), as well as S4, S6, L3, and L16 following imbibition in embryos of maize (Beltran-Pena et al., 1995). From these analyses, it appears that r-protein expression in plants may be regulated at the transcriptional and posttranscriptional levels.
The international Arabidopsis Genome Initiative (AGI; Bevan et al., 1997; Lin et al., 1999; Mayer et al., 1999; AGI, 2000) has led to the to the accumulation of an enormous quantity of genomic sequence data, in addition to more than 112,500 ESTs (Höfte et al., 1993; Newman et al., 1994; Cooke et al., 1996; Asamizu et al., 2000). The essentially complete genome sequence is publicly accessible through The Arabidopsis Information Resource (TAIR) database (http://www.Arabidopsis.org/). This situation provided a unique opportunity for analyzing r-protein gene number, chromosomal location, and expression. Here, we report the identification and map positions of 249 r-protein genes of Arabidopsis. Location of the genes was initially determined by physical mapping using ESTs and subsequently confirmed from the genomic sequence data, in some cases of genomic regions that were not completely annotated. Analysis of r-protein gene distribution initially allowed us to discover duplications of several very large DNA sequences, which shed light on Arabidopsis genome evolution (Blanc et al., 2000). Comparison of the distribution of these gene families in the Arabidopsis genome and in other organisms and its implications on the understanding of multigene family organization and genome evolution are discussed. The systematic identification of ESTs representing different gene family members as well as reverse transcriptase (RT)-PCR on RNA obtained from different tissues and PCR on a cDNA library (Newman et al., 1994) revealed that levels of r-protein pseudogenes are very low and indicated that many of genes family members are differentially expressed. Variation in r-protein gene family member sequences and expression patterns raises the possibility of ribosome heterogeneity at subcellular and intracellular levels.
RESULTS
Identification of 249 Cytoplasmic r-Protein Genes in Arabidopsis
To identify r-protein genes in the Arabidopsis genome, we chose rat as the eukaryotic model because its r-protein genes have been extensively studied and corresponding genes in plants had been identified (Bailey-Serres, 1998). We collected all 79 rat r-protein sequences from the Swiss-PROT library (Bairoch and Apweiler, 2000) and carried out TBLASTN (Altschul et al., 1997) searches on Arabidopsis EST and cDNA sequences in GenBank (Release 65.0, November 2000). Most of the 79 rat protein genes had several orthologues in Arabidopsis based on high probability BLAST scores (data not shown). An estimate of the number of expressed genes in each family was determined by constructing contigs from ESTs. The accuracy of EST contig construction was tested as described by Cooke et al. (1997) and redundancy within families was eliminated by careful comparison of the contigs to one another and to genomic sequences. In this manner, we identified 200 r-protein genes. In addition, TBLASTN alignment against Arabidopsis genomic sequence data released through the AGI allowed us to identify a total of 249 r-protein genes, including 101 encoding 32 putative small-subunit proteins and 148 encoding 48 putative large-subunit proteins (Table I). Genes identified from ESTs and genomic sequences were compared and a nonredundant set of r-proteins was collated. A perfect match to a genomic sequence was found for all 200 EST contigs. Therefore, this approach revealed an additional 49 genomic sequences that were not identified by EST contigs, including those that appear to contain an incomplete ORF. This analysis also resulted in discovery of 36 r-protein genes that were not detected by automated annotation or in which the annotation was incorrect (Table I, indicated with an asterisk after the gene name). Because no orphaned EST contigs were identified, it seems unlikely that additional r-protein genes will be identified in the centromeric regions that have not been fully sequenced.
Table I.
Identification of Arabidopsis orthologues of rat small (40S) and large (60S) ribosomal subunit proteins
| r-Protein
|
Evolutionary Group | Genomic
|
MATDB AGI Gene Name | EST
|
Chromosome No. | Mbp | Nearest Marker
|
Deduced Polypeptide
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein name | Gene name | GenBank accession no. | Clone position | GenBank accession no. | Frequency | Marker name | Map position | % ID rat | kD | Amino acids | pI | ||||
| Sa | RPSaA | I | AC016529 | T10D10.16 | At1g72370 | T14000 | 22 | 1 | 27.0 | nga111 | 115.5 | 54.1 | 32.3 | 298 | 4.9 |
| RPSaB | AC011437 | F7O18.26 | At3g04770 | U66223 | 1 | 3 | 1.25 | GAPC | 8.4 | 56.9 | 30.7 | 280 | 4.9 | ||
| S2 | RPS2A | I | AC082643 | F9K23.9 | At1g58380 | AV550768 | 3 | 1 | 21.3 | SGCSNP301 | 85.9 | 75.3 | 30.7 | 284 | 11.0 |
| RPS2B | AC027036 | T4M14.3 | N.A. | N.F. | 0 | 1 | 21.4 | ARR3 | 87 | 76.3 | 30.8 | 284 | 11.0 | ||
| RPS2C | AC002339 | T11A7.6 | At2g41840 | B10274 | 5 | 2 | 17.7 | COR15 | 76.8 | 74.5 | 30.9 | 285 | 11.1 | ||
| RPS2D | AL133248 | T8H10.90 | At3g57490 | F14347 | 2 | 3 | 21.8 | SNP7 | 77 | 74.9 | 30.1 | 276 | 11.0 | ||
| S3 | RPS3A | I | AC007071 | T9H9.13 | At2g31610 | AV553035 | 7 | 2 | 13.7 | nga361 | 63 | 81.5 | 27.5 | 250 | 10.4 |
| RPS3B | AL132960 | F5K20.170 | At3g53870 | T04067 | 17 | 3 | 20.4 | AFC1 | 73.9 | 81.9 | 27.3 | 249 | 10.4 | ||
| RPS3C | AB015477 | MOK9.14 | At5g35530 | AV550513 | 8 | 5 | 13.8 | PHYC | 71.1 | 76.3 | 27.5 | 248 | 10.4 | ||
| S3a | RPS3aA | II | AC009465 | T9J14.21 | At3g04840 | AV545036 | 13 | 3 | 1.4 | GAPC | 8.4 | 67.6 | 29.9 | 262 | 10.6 |
| RPS3aB | AL023094 | T4L20.250 | At4g34670 | AJ001342 | 2 | 4 | 15.8 | g3088 | 83.3 | 69.6 | 29.8 | 262 | 10.5 | ||
| S4 | RPS4A* | II | AC002329 | F5J6.12 | At2g17360 | F20029 | 7 | 2 | 7.8 | m216 | 33.1 | 69.7 | 30.1 | 263 | 11.0 |
| RPS4B | AL163652 | T28J14.30 | At5g07090 | AV554668 | 16 | 5 | 2.2 | SGCSNP21 | 18 | 69.8 | 29.9 | 262 | 10.9 | ||
| RPS4C* | AB017068 | MJG14.8 | N.A. | N.F. | 0 | 5 | 14.8 | gln1-1 | 77.3 | iORF | – | – | – | ||
| RPS4D* | AB025632 | MQJ2.2 | At5g58420 | AV551244 | 14 | 5 | 23.7 | mi184 | 113.7 | 69.8 | 29.8 | 262 | 11.0 | ||
| S5 | RPS5A | I | AC005896 | F3G5.6 | At2g37270 | AI099638 | 8 | 2 | 15.9 | Ve018 | 69.7 | 78.0 | 23.0 | 207 | 10.5 |
| RPS5B | AC016795 | F26K24.23 | At3g11940 | BE038477 | 20 | 3 | 3.7 | SGCSNP245 | 14.8 | 76.7 | 22.9 | 207 | 10.4 | ||
| S6 | RPS6A | II | AL031004 | F28M20.110 | At4g31700 | AV550020 | 6 | 4 | 14.5 | g8300 | 81.2 | 67.6 | 28.4 | 250 | 11.4 |
| RPS6B | AL353995 | F12B17.290 | At5g10360 | AV37347 | 18 | 5 | 3.4 | ve033 | 25.42 | 67.6 | 28.1 | 249 | 11.5 | ||
| S7 | RPS7A | III | AC073555 | F11I4.1 | N.A. | AV549012 | 4 | 1 | 17.6 | mi441 | 72.9 | 55.4 | 21.9 | 191 | 10.6 |
| RPS7B | AC021640 | F16B3.19 | At3g02560 | Z47625 | 2 | 3 | 0.5 | MI74B | 5.8 | 53.8 | 22.2 | 191 | 10.6 | ||
| RPS7C | AL391148 | T21H19.50 | At5g16130 | AV544115 | 5 | 5 | 5.3 | nga106 | 33.3 | 54.3 | 22.1 | 190 | 10.6 | ||
| S8 | RPS8A* | II | AF296825 | F5O24 | At5g20290 | BE037738 | 10 | 5 | 6.9 | mi433 | 42.2 | 62.4 | 24.1 | 213 | 11.2 |
| RPS8B | AB016890 | MNC17.15 | At5g59240 | AI999676 | E | 5 | 24.0 | mi184 | 113.7 | 64.5 | 23.8 | 210 | 11.3 | ||
| S9 | RPS9A | I | AL161533 | F16J13.230 | At4g12160 | N.F. | 0 | 4 | 6.4 | g4108 | 43.5 | iORF | – | – | – |
| RPS9B | AL353993 | F8M21.90 | At5g15200 | AV54959 | 30 | 5 | 5.0 | SNP13 | 30.3 | 74.7 | 23.0 | 198 | 10.9 | ||
| RPS9C | AB010077 | MYH19.1 | At5g39850 | AV010077 | 1 | 5 | 16.1 | SNP150 | 83.2 | 74.2 | 23.2 | 197 | 11.1 | ||
| S10 | RPS10A | III | AL049480 | F14M19.20 | At4g25740 | AI997138 | 6 | 4 | 12.4 | RPS2 | 75.6 | 58.9 | 19.4 | 177 | 10.5 |
| RPS10B | AB005233 | MBK23.4 | At5g41520 | AV536209 | 11 | 5 | 16.7 | g4028 | 86.2 | 52.9 | 19.7 | 180 | 10.6 | ||
| RPS10C | AB025606 | F6N7.14 | At5g52650 | AI999527 | 4 | 5 | 21.4 | SGCSNP242 | 107.1 | 53.5 | 19.8 | 181 | 10.4 | ||
| S11 | RPS11A | I | AL132967 | T2J13.230 | At3g48930 | Z26185 | 5 | 3 | 18.6 | SGCSNP352 | 68.3 | 61.0 | 18.0 | 160 | 11.3 |
| RPS11B | AL022198 | F6I18.290 | At4g30800 | N.F. | E | 4 | 14.3 | PRHA | 78.9 | 64.7 | 17.9 | 159 | 11.5 | ||
| RPS11C | AB005244 | MRO11.22 | At5g23740 | AV561164 | 2 | 5 | 8.2 | CDPK9 | 44.5 | 61.4 | 17.7 | 159 | 11.4 | ||
| S12 | RPS12A | III | AC010924 | T24D18.3 | At1g15930 | T14030 | 9 | 1 | 5.5 | srp54a | 18.9 | 52.6 | 15.4 | 144 | 5.3 |
| RPS12B | AC011713 | F23A5.10 | At1g80750 | N.F. | 0 | 1 | 30.1 | mi157 | 124.3 | iORF | – | – | – | ||
| RPS12C | AC006223 | F22D22.19 | At2g32060 | AI999579 | 6 | 2 | 13.9 | ASP1 | 62.7 | 52.6 | 15.3 | 144 | 5.8 | ||
| S13 | RPS13A | I | AL162295 | T4C21.180 | At3g60770 | Z17784 | 10 | 3 | 22.9 | snp74 | 84.6 | 77.3 | 17.0 | 150 | 11.2 |
| RPS13B(A) | AF069299 | F6N15.7 | At4g00100 | Z29915 | 6 | 4 | 0.5 | NOR4 | 0.0 | 78.0 | 16.9 | 150 | 11.2 | ||
| S14 | RPS14A | I | AC007135 | F9C22.9 | At2g36160 | AV552523 | 1 | 2 | 5.5 | ve016 | 67.6 | 85.3 | 16.3 | 150 | 11.3 |
| RPS14B | AC008153 | F24K9.19 | At3g11510 | R89968 | 3 | 3 | 3.6 | SNP245 | 14.7 | 85.3 | 16.3 | 150 | 11.3 | ||
| RPS14C | AL050300 | F2206.40 | At3g52580 | AV55346 | 3 | 3 | 19.9 | mi456 | 72.7 | 85.3 | 16.2 | 150 | 11.3 | ||
| S15 | RPS15A | I | AC000104 | F19P19.29 | At1g04270 | AV544758 | 17 | 1 | 1.1 | SGCSNP151 | 3.3 | 75.4 | 17.1 | 152 | 11.1 |
| RPS15B | AL391712 | T5E8.290 | At5g09490 | N.F. | 0 | 5 | 2.9 | ve033 | 25.4 | 71.7 | 17.1 | 152 | 10.9 | ||
| RPS15C | AL391712 | T5E8.300 | At5g09500 | N.F. | 0 | 5 | 2.9 | ve033 | 25.4 | 73.9 | 16.7 | 150 | 11.3 | ||
| RPS15D | AL391712 | T5E8.310 | At5g09510 | AV549585 | 6 | 5 | 2.9 | ve033 | 25.4 | 75.4 | 17.1 | 152 | 11.1 | ||
| RPS15E | AB016875 | K9D7.14 | At5g43640 | N.F. | 0 | 5 | 17.6 | mi194 | 90.5 | 74.7 | 16.8 | 149 | 11.3 | ||
| RPS15F | AB008265 | MCD12.3 | At5g63070 | N.F. | 0 | 5 | 25.3 | mi211A | 119 | iORF | – | – | – | ||
| S15a | RPS15aA | I | AC007583 | F24B9.12 | At1g07770 | AV538172 | 9 | 1 | 2.5 | ve004 | 7.76 | 77.7 | 14.8 | 130 | 10.7 |
| RPS15aB | AC005169 | F6F22.25 | At2g19720 | Z26126 | 1 | 2 | 8.9 | MI148 | 36.1 | 47.6 | 14.7 | 129 | 10.6 | ||
| RPS15aC | AC004218 | F12L6.25 | At2g39590 | N.F. | NE | 2 | 16.8 | M429 | 73.1 | 73.1 | 15.3 | 130 | 10.0 | ||
| RPS15aD | AL355775 | F12M12.10 | At3g46040 | AW004284 | 3 | 3 | 17.5 | M249 | 61.3 | 77.7 | 14.8 | 130 | 10.8 | ||
| RPS15aE | AL161575 | F27B13 | At4g29430 | N.F. | 0 | 4 | 13.8 | prha | 78.9 | 48.0 | 14.9 | 129 | 10.7 | ||
| RPS15aF | AB015475 | MMN10.8 | At5g59850 | AV554198 | 7 | 5 | 24.2 | SNP2 | 115.9 | 77.7 | 14.8 | 130 | 10.7 | ||
| S16 | RPS16A | I | AC006586 | F7B19.13 | At2g09990 | AV536848 | 2 | 2 | 4.1 | mi421 | 19.1 | 73.3 | 16.6 | 146 | 11.0 |
| RPS16B | AC016829 | T6K12.15 | At3g04230 | Z17479 | 2 | 3 | 1.1 | GAPC | 8.4 | 73.3 | 16.6 | 146 | 11.0 | ||
| RPS16C* | AC051626 | F20L16 | At5g18380 | AV534112 | 1 | 5 | 6.1 | GDH1 | 33.29 | 74.0 | 16.6 | 146 | 11.0 | ||
| S17 | RPS17A | II | AC006951 | T1O3.20 | At2g04390 | AV550538 | 3 | 2 | 1.6 | Igs1 | 13.2 | 61.1 | 16.0 | 141 | 10.8 |
| RPS17B* | AC007018 | F5G3.12 | At2g05220 | AV534112 | 7 | 2 | 1.9 | m497A | 13.3 | 61.9 | 16.0 | 140 | 10.8 | ||
| RPS17C | AC011560 | F13M14.10 | At3g10610 | AV534760 | 4 | 3 | 3.3 | SNP11 | 14.7 | 60.2 | 16.0 | 140 | 10.8 | ||
| RPS17D | AB008271 | MUK11.12 | At5g04800 | AV553023 | 8 | 5 | 1.4 | nga225 | 14.3 | 61.1 | 16.0 | 141 | 10.8 | ||
| S18 | RPS18A (A) | I | AC003979 | T22J18.5 | At1g22780 | AV552655 | 12 | 1 | 8.1 | m235 | 34.0 | 74.3 | 17.5 | 152 | 11.3 |
| RPS18B (B) | AC015446 | F12G12.15 | At1g34030 | BE037678 | 10 | 1 | 12.5 | AIG1 | 55.62 | 74.3 | 17.5 | 152 | 11.3 | ||
| RPS18C (C) | AL049482 | F17A8.150 | At4g09800 | AV530846 | 4 | 4 | 5.4 | DET1 | 31.4 | 74.3 | 17.5 | 152 | 11.3 | ||
| S19 | RPS19A | II | AC011664 | F1C9.13 | At3g02080 | AV536148 | 7 | 3 | 0.3 | mi74b | 5.8 | 56.9 | 15.8 | 143 | 10.9 |
| RPS19B | AL391143 | T20K14.130 | At5g15520 | AV559770 | 1 | 5 | 5.1 | nga106 | 33.2 | 56.5 | 15.8 | 143 | 11.0 | ||
| RPS19C | AB006696 | MAF19.17 | At5g61170 | AI996699 | 1 | 5 | 24.7 | LFY3 | 116.8 | 59.0 | 15.7 | 143 | 11.0 | ||
| S20 | RPS20A | I | AL353992 | F14D17.100 | At3g45030 | AV353992 | 3 | 3 | 16.9 | TOPP5 | 59.2 | 74.1 | 13.1 | 117 | 10.5 |
| RPS20B* | AL096860 | T21L8.120 | At3g47370 | AV532791 | 3 | 3 | 17.9 | ASN1 | 61.4 | 74.1 | 13.7 | 117 | 10.5 | ||
| RPS20C | AB019235 | MMI9.13 | At5g62300 | AV533085 | 3 | 5 | 25.1 | LFY3 | 116.8 | 74.1 | 13.1 | 117 | 10.5 | ||
| S21 | RPS21A* | III | AB024028 | K1G2 | At3g27450 | N.F. | 0 | 3 | 10.1 | mi287 | 43.6 | iORF | – | – | – |
| RPS21B | AL132960 | F5K20.190 | At3g53890 | AI997498 | 2 | 3 | 20.4 | Ve042 | 76.2 | 46.3 | 9.1 | 82 | 8.1 | ||
| RPS21C* | AC069556 | T1G16 | At5g27700 | AV536952 | 6 | 5 | 9.9 | SO262 | 65.2 | 43.8 | 9.0 | 81 | 8.4 | ||
| S23 | RPS23A* | I | AC016661 | F11F8.27 | At3g09680 | N.F. | 0 | 1 | 2.9 | mi357 | 16.2 | 76.1 | 15.8 | 142 | 11.1 |
| RPS23B | AL162973 | F9G14.270 | At5g02960 | AV553972 | 19 | 5 | 0.6 | SNP241 | 3.7 | 78.9 | 16.2 | 146 | 11.1 | ||
| S24 | RPS24A | II | AC009465 | T9J14.13 | At3g04920 | BE038406 | 4 | 3 | 1.4 | GAPC | 8.4 | 67.5 | 15.4 | 133 | 11.0 |
| RPS24B* | AC007627 | F15F15 | At5g28060 | BE037704 | 4 | 5 | 10.1 | SO262 | 65.2 | 65.1 | 15.4 | 133 | 11.3 | ||
| S25 | RPS25A | III | AC007047 | F16F14.14 | At2g16360 | N.F. | NE | 2 | 7.4 | mi398 | 29.2 | iORF | – | – | – |
| RPS25B | AC007119 | F2G1.15 | At2g21580 | BE038441 | 19 | 2 | 9.5 | mi238 | 39.9 | 59.4 | 12.1 | 108 | 11.5 | ||
| RPS25C | AP002066 | T4A2.5 | At3g30740 | N.F. | 0 | 3 | 12.4 | atpox | 52.4 | iORF | – | – | – | ||
| RPS25D | AL023094 | T4L20.250 | At4g34670 | N.F. | 0 | 4 | 15.7 | SNP232 | 83.4 | iORF | – | – | – | ||
| RPS25E | AL050351 | T22F8.100 | At4g39200 | AV533470 | 29 | 4 | 17.5 | AP2 | 95.9 | 58.5 | 12.1 | 108 | 11.5 | ||
| S26 | RPS26B | III | AC002336 | T2P4.14 | At2g40510 | BE038315 | 11 | 2 | 17.2 | g4514 | 73.7 | 67.3 | 14.8 | 133 | 11.7 |
| RPS26A | AC002336 | T2P4.6 | At2g40590 | Z26184 | 1 | 2 | 17.2 | g4514 | 73.7 | 67.3 | 14.8 | 133 | 11.7 | ||
| RPS26C | AL163763 | F18O21.300 | At3g56340 | AI998355 | 11 | 3 | 21.3 | SNP189 | 77.2 | 70.9 | 14.6 | 130 | 11.7 | ||
| S27 | RPS27A (C) | II | AC004665 | F4I18.31 | At2g45710 | AA712867 | 4 | 2 | 19.1 | ve019 | 82.1 | 75.3 | 9.5 | 84 | 9.1 |
| RPS27B (A) | AL137898 | T20K12.10 | At3g61110 | AL137898 | 6 | 3 | 23.2 | SNP221 | 85.8 | 77.9 | 9.5 | 85 | 8.7 | ||
| RPS27C (ϕ)* | AL137898 | T20K12 | N.A. | N.F. | 0 | 3 | 23.2 | SNP221 | 85.8 | iORF | – | – | – | ||
| RPS27D (B) | AB024025 | K16F13.1 | At5g47930 | AV531451 | 12 | 5 | 19.5 | SGCSNP147 | 99.5 | 79.2 | 9.5 | 84 | 9.1 | ||
| S27a | RPS27aA | II | AC007945 | F28C11.5 | At1g23410 | Z25557 | 2 | 1 | 8.4 | m235 | 34 | 81.4 | 17.7 | 156 | 10.6 |
| RPS27aB | AC004411 | F14M4.6 | At2g47100 | AV548497 | 3 | 2 | 19.6 | Athb7 | 84.5 | 84.9 | 17.8 | 157 | 10.6 | ||
| RPS27aC | AL138651 | T17J13.210 | At3g62250 | AA728493 | 6 | 3 | 23.5 | mi424 | 82.8 | 84.2 | 17.8 | 157 | 10.6 | ||
| S28 | RPS28A | II | AC010927 | T22K18.8 | At3g10090 | N.F. | NE | 3 | 3.1 | mi357 | 16.2 | 78.3 | 7.4 | 64 | 11.5 |
| RPS28B | AB005235 | MED24.15 | At5g03850 | AV530936 | 2 | 5 | 1.9 | SGCSNP396 | 9.28 | 78.3 | 7.4 | 64 | 11.5 | ||
| RPS28C | AB008266 | MHJ24.12 | At5g64140 | Z17569 | 2 | 5 | 25.7 | ve032 | 123.3 | 80.0 | 7.3 | 64 | 11.7 | ||
| S29 | RPS29A | I | AL163975 | T15B3.120 | At3g43980 | T22180 | 3 | 3 | 16.2 | TOPP5 | 59.2 | 72.2 | 6.4 | 56 | 10.8 |
| RPS29B | AL163975 | T15B3.150 | At3g44010 | Z47604 | 3 | 3 | 16.2 | TOPP5 | 59.2 | 72.2 | 6.4 | 56 | 10.8 | ||
| RPS29C* | AL161584 | F17I5 | N.A. | AI996253 | 5 | 4 | 15.5 | pCITd104 | 83.3 | 70.4 | 6.1 | 54 | 10.9 | ||
| RPS29D* | AL161584 | F17I5 | N.A. | N.F. | 0 | 4 | 15.5 | pCITd104 | 83.3 | iORF | – | – | – | ||
| S30 | RPS30A* | II | AC005169 | F6F22.22 | At2g19750 | AV532814 | 4 | 2 | 8.9 | mi148 | 36.1 | 75.9 | 6.9 | 62 | 12.8 |
| RPS30B | AL161575 | F19B15 | At4g29390 | N.F. | 0 | 4 | 13.5 | mi232 | 76.7 | 76.3 | 6.9 | 62 | 12.8 | ||
| RPS30C | AB013392 | M1K19.12 | At5g56670 | AI100293 | 2 | 5 | 23.0 | mi69 | 114.3 | 75.9 | 6.9 | 62 | 12.8 | ||
| P0 | RPP0A | I | AF002109 | T28M21.17 | At2g40010 | N.F. | 0 | 2 | 16.9 | SGCSNP214 | 74.7 | 51.6 | 33.7 | 317 | 5.0 |
| RPP0B | AC011436 | F3L24.7 | At3g09200 | T21000 | 40 | 3 | 2.8 | mi467 | 15.6 | 53.8 | 34.1 | 320 | 4.8 | ||
| RPP0C | AC073395 | F11B9.17 | At3g11250 | AV561267 | 6 | 3 | 3.5 | SGCSNP11 | 14.7 | 55.7 | 34.4 | 323 | 4.9 | ||
| P1 | RPP1A | I | AC007323 | T25K16.9 | At1g01100 | AV536016 | 4 | 1 | 12.1 | Ve001 | 2.9 | 58.2 | 11.2 | 112 | 4.1 |
| RPP1B | AL161472 | T18A10.9 | At4g00810 | AV522332 | 3 | 4 | 0.3 | mi122 | 5 | 56.1 | 11.0 | 110 | 4.0 | ||
| RPP1C | AB016886 | MCA23.2 | At5g47700 | AV530633 | 3 | 5 | 19.4 | SGCSNP147 | 99.4 | 57.7 | 11.2 | 113 | 4.1 | ||
| P2 | RPP2A | I | AC005824 | F15K20.18 | At2g27720 | AV532448 | 17 | 2 | 12.0 | nga1126 | 50.6 | 50.4 | 11.4 | 115 | 4.4 |
| RPP2B | AC005824 | F15K20.19 | At2g27710 | AV535852 | 13 | 2 | 12.0 | nga1126 | 50.6 | 50.4 | 11.4 | 115 | 4.4 | ||
| RPP2C | AP002059 | T20D4.1 | At3g28500 | F19923 | 2 | 3 | 10.7 | AIG2 | 50.59 | 39.1 | 11.7 | 115 | 4.4 | ||
| RPP2D | AL353818 | F14L2.140 | At3g44590 | AV534715 | 2 | 3 | 16.6 | m249 | 61.3 | 57.7 | 11.0 | 111 | 4.2 | ||
| RPP2E | AB022222 | MUD12.2 | At5g40040 | Z17443 | 2 | 5 | 16.1 | SGCSNP150 | 83.2 | 53.2 | 11.8 | 114 | 4.3 | ||
| P3 | RPP3A | I | AL049480 | F14M19 | At4g25890 | AV556500 | 1 | 4 | 12.3 | RSP2 | 75.6 | Planta | 11.8 | 119 | 4.2 |
| RPP3B | AB019233 | MJB24.10 | At5g57290 | AV535058 | 7 | 5 | 23.3 | m558a | 113.8 | Planta | 11.9 | 120 | 4.3 | ||
| L3 | RPL3A (1) | I | AC005687 | F1I21.L3 | At1g43170 | AV562764 | 21 | 1 | 15.8 | SGCSNP163 | 63 | 66.2 | 44.6 | 389 | 11.0 |
| RPL3B(2) | AC005850 | T25B24.7 | At1g61580 | AV557676 | 1 | 1 | 22.3 | mi230 | 86.5 | 67.4 | 44.5 | 390 | 11.1 | ||
| RPL3C | AB016888 | MDH9.14 | N.A. | N.F. | E | 5 | 17.0 | DFR | 89.5 | iORF | – | – | – | ||
| L4 | RPL4A | II | AC016661 | F11F8.22 | At3g09630 | AV551524 | 8 | 1 | 2.9 | APX1B | 16.2 | 58.1 | 44.7 | 406 | 11.1 |
| RPL4B | AC079605 | T32G9.26 | At1g35200 | N.F. | 0 | 1 | 12.9 | mi342 | 58.7 | iORF | – | – | – | ||
| RPL4C | AC007266 | F27A10.4 | N.A. | N.F. | 0 | 2 | 10.7 | SNP203 | 44.4 | iORF | – | – | – | ||
| RPL4D | AL162973 | F9G14.180 | At5g02870 | AV541474 | 6 | 5 | 0.6 | SNP241 | 3.7 | 56.7 | 44.7 | 407 | 11.1 | ||
| L5 | RPL5A | I | AB025639 | MWL2.17 | At3g25520 | AV5645486 | 5 | 3 | 9.2 | m433 | 38 | 57.0 | 34.2 | 300 | 10.1 |
| RPL5B | AB016876 | MKM21.5 | At5g39740 | AV525399 | 9 | 5 | 16.0 | SGCSNP150 | 83.2 | 57.3 | 34.4 | 301 | 10.0 | ||
| RPL5C | AB010699 | MSN9.3 | At5g40130 | N.F. | 0 | 5 | 16.1 | SGCSNP164 | 83.7 | iORF | – | – | – | ||
| L6 | RPL6A | III | AC026238 | F25I16.12 | At1g18540 | AV566810 | 22 | 1 | 6.4 | mi348 | 23.6 | 52.6 | 26.2 | 233 | 10.9 |
| RPL6B | AC016662 | F2P9.7 | At1g74060 | H36726 | 6 | 1 | 27.5 | bw54 | 116 | 54.4 | 26.0 | 233 | 11.2 | ||
| RPL6C | AC016662 | F2P9.8 | At1g74050 | AV442576 | 11 | 1 | 27.5 | bw54 | 116 | 54.8 | 26.1 | 233 | 11.2 | ||
| L7 | RPL7A | I | AC011713 | F23A5.10 | At1g80750 | AV561722 | 1 | 1 | 30.1 | SGCSNP355 | 131.1 | 37.7 | 28.3 | 247 | 10.5 |
| RPL7B | AC006200 | F10A8.13 | At2g01250 | AV550374 | 6 | 2 | 0.2 | rga | 1.7 | 60.8 | 28.1 | 242 | 10.7 | ||
| RPL7C | AC004005 | F6E13.25 | At2g44120 | AI100283 | 3 | 2 | 18.4 | m336 | 79 | 60.3 | 28.5 | 242 | 10.7 | ||
| RPL7D | AP002038 | K20M4.2 | At3g13580 | N.F. | 0 | 3 | 4.4 | nga162 | 20.5 | 61.7 | 28.4 | 240 | 10.8 | ||
| L7a | RPL7aA | II | AC002535 | T30B22.8 | At2g47610 | T76559 | 7 | 2 | 19.7 | mi79a | 87.5 | 57.9 | 29.1 | 257 | 10.9 |
| RPL7aB | AL162651 | F26K9.300 | At3g62870 | AV536728 | 16 | 3 | 23.7 | SNP264 | 89.3 | 57.1 | 29.0 | 256 | 11.0 | ||
| L8 | RPL8A | I | AC006201 | T27K22.11 | At2g18020 | T44362 | 8 | 2 | 8.1 | m216 | 33.1 | 71.4 | 27.9 | 258 | 11.6 |
| RPL8B | AL132980 | F24M12.230 | At3g51190 | N.F. | 0 | 3 | 19.5 | MUR 1 | 72.7 | 71.0 | 28.0 | 260 | 11.3 | ||
| RPL8C | AL022141 | F23E13.20 | At4g36130 | H37035 | 8 | 4 | 16.3 | fah1 | 86.3 | 70.6 | 27.9 | 258 | 11.5 | ||
| L9 | RPL9A | I | AC027035 | T16O9.23 | N.A. | AV533409 | 19 | 1 | 12.1 | mi2532 | 51.8 | 57.1 | 22.0 | 194 | 10.2 |
| RPL9B | AC021045 | T9L6.2 | At1g33120 | AV549042 | 15 | 1 | 12.0 | mi2532 | 51.8 | 57.1 | 22.0 | 194 | 10.2 | ||
| RPL9C | AC021045 | T9L6.5 | At1g33140 | AV549555 | 28 | 1 | 12.0 | mi2532 | 51.8 | 57.1 | 22.0 | 194 | 10.2 | ||
| RPL9D | AL049524 | F7L13.30 | At4g10450 | AV541541 | 6 | 4 | 5.6 | SGCSNP24 | 30.88 | 58.3 | 22.0 | 194 | 10.3 | ||
| L10 | RPL10A | II | AC012188 | F14L17.9 | At1g14320 | AV553316 | 25 | 1 | 4.9 | SGCSNP303 | 14.6 | 69.9 | 24.9 | 220 | 11.3 |
| RPL10B | AC005508 | T2P11.10 | At1g26910 | N.F. | E | 1 | 9.4 | mi192 | 41.1 | 69.4 | 24.9 | 221 | 11.2 | ||
| RPL10C* | AC079285 | T12I7.3 | At1g66580 | AI998557 | 3 | 1 | 24.4 | mi185 | 102.1 | 69.8 | 24.1 | 214 | 11.3 | ||
| L10a | RPL10aA* | I | AC006932 | T27G7.6 | At1g08360 | AV554254 | 14 | 1 | 2.6 | SGCSNP308 | 0.89 | 63.0 | 24.3 | 215 | 10.7 |
| RPL10aB* | AC005824 | F15K20.37 | At2g27530 | AV551399 | 11 | 2 | 12.1 | ngal126 | 50.65 | 64.2 | 24.3 | 215 | 10.7 | ||
| RPL10aC | AB007651 | MWD9.24 | At5g22440 | AV553355 | 16 | 5 | 7.5 | mi433 | 42 | 62.8 | 24.5 | 217 | 10.6 | ||
| L11 | RPL11A(A) | I | AC006931 | F7D19.26 | At2g42740 | N.F. | 0 | 2 | 18.0 | COR15 | 76.8 | 72.0 | 20.9 | 182 | 10.7 |
| RPL11B | AL353032 | T20N10.50 | At3g58700 | Z29916 | 3 | 3 | 22.2 | SNP7 | 77.1 | 70.1 | 20.9 | 182 | 10.8 | ||
| RPL11C(B) | AL035526 | F28A21.140 | At4g18730 | AA712813 | 18 | 4 | 9.4 | AG | 63 | 70.3 | 21.1 | 184 | 10.8 | ||
| RPL11D | AB012245 | MRA19.21 | At5g45775 | AV532938 | 6 | 5 | 18.5 | mi61 | 98.1 | 70.1 | 20.9 | 182 | 10.8 | ||
| L12 | RPL12A | I | AC006260 | T2N18.5 | At2g37190 | AV540047 | 5 | 2 | 15.8 | ve018 | 69 | 70.6 | 18.0 | 166 | 9.6 |
| RPL12B | AL132966 | F4P12.130 | At3g53430 | BE038784 | 8 | 3 | 20.2 | AFC1 | 73.9 | 69.3 | 18.0 | 166 | 9.6 | ||
| RPL12C | AB005246 | MUP24.9 | At5g60670 | AV530701 | 7 | 5 | 24.4 | SGCSNP2 | 115.9 | 69.9 | 17.8 | 166 | 9.6 | ||
| L13 | RPL13A | III | AL096856 | T24C20.10 | At3g48130 | N.F. | 0 | 3 | 18.3 | m409 | 64 | iORF | – | – | – |
| RPL13B | AL132967 | T2J13.150 | At3g49010 | AV553216 | 10 | 3 | 18.6 | SGCSNP291 | 68.2 | 55.2 | 23.8 | 206 | 11.7 | ||
| RPL13C | AL132967 | T2J13.200 | At3g48960 | Z34694 | 18 | 3 | 18.6 | SGCSNP291 | 68.2 | 51.2 | 23.5 | 206 | 11.3 | ||
| RPL13D | AB005244 | MRO11.6 | At5g23900 | AI100098 | 11 | 5 | 8.1 | CDKP9 | 44.5 | 57.1 | 23.5 | 206 | 11.7 | ||
| L13a | RPL13aA | I | AC012395 | T1B9.24 | At3g07110 | AV541696 | 14 | 3 | 2.3 | SGCSNP115 | 3.32 | 60.5 | 23.5 | 206 | 11.2 |
| RPL13aB | AB028609 | K7P8.12 | At3g24830 | AA042521 | 4 | 3 | 9.1 | g4711 | 38 | 60.5 | 23.5 | 206 | 11.1 | ||
| RPL13aC | AL049751 | F17N18.60 | At4g13170 | AI999348 | 1 | 4 | 6.8 | mi465 | 45 | 61.1 | 23.6 | 206 | 11.2 | ||
| RPL13aD | AB012242 | K24G6.9 | At5g48760 | AV542288 | 6 | 5 | 19.9 | M331 | 102.6 | 60.5 | 23.6 | 206 | 11.2 | ||
| L14 | RPL14A | III | AC007109 | T13C7.4 | At2g20450 | N.F. | 0 | 2 | 9.1 | SNP71 | 35.8 | 46.9 | 15.5 | 134 | 10.9 |
| RPL14B | AL161566 | T24A18.40 | At4g27090 | BE038422 | 8 | 4 | 12.8 | mi123 | 75.6 | 44.6 | 15.5 | 134 | 10.8 | ||
| L15 | RPL15A | III | Z97341 | FCA6 | At4g16720 | BE039553 | 6 | 4 | ∼8.0 | SGCSNP272 | 56 | 70.4 | 24.2 | 204 | 12.0 |
| RPL15B* | Z97343 | FCA8 | At4g17390 | AV549804 | 7 | 4 | ∼8.0 | mi112 | 58.1 | 70.0 | 24.2 | 204 | 12.0 | ||
| L17 | RPL17A | I | AC004557 | F17L21.19 | At1g27400 | AI996162 | 8 | 1 | 9.6 | ve008 | 47.7 | 66.7 | 19.3 | 172 | 11.0 |
| RPL17B | AC004393 | T1F15.11 | At1g67430 | BE03992 | 12 | 1 | 25.0 | mi185 | 102.2 | 67.3 | 19.9 | 175 | 10.9 | ||
| L18 | RPL18A | II | AC002535 | T30B22.13 | At2g47570 | N.F. | E | 2 | 19.7 | mi79a | 87 | 62.5 | 20.8 | 187 | 11.3 |
| RPL18B | AC011620 | F18C1.14 | At3g05590 | AV550190 | 6 | 3 | 1.7 | mi355 | 13.9 | 64.9 | 20.9 | 187 | 11.7 | ||
| RPL18C* | AC007399 | F14I23 | At5827850 | AV552450 | 8 | 5 | 10.0 | SO262 | 65.2 | 63.8 | 20.9 | 187 | 11.7 | ||
| L18a | RPL18aA* | II | AC022455 | T1P2.8 | At1g29970 | N.F. | 0 | 1 | 10.5 | m215 | 41.6 | 53.3 | 21.4 | 178 | 11.2 |
| RPL18aB | AC004077 | T31E10.18 | At2g34480 | AV549659 | 12 | 2 | 14.8 | ve016 | 67.6 | 53.3 | 21.3 | 178 | 11.3 | ||
| RPL18aC | AB023038 | MIE1.10 | At3g14600 | AV542705 | 3 | 3 | 4.8 | SNP20 | 20 | 51.5 | 21.3 | 178 | 11.1 | ||
| L19 | RPL19A | II | AC009525 | F22D16.23 | N.A. | AV536229 | 25 | 1 | 0.7 | GST1 | 3.9 | 68.9 | 24.6 | 214 | 12.0 |
| RPL19B | AB022217 | MGL6.25 | At3g16780 | AV549838 | 3 | 3 | 5.7 | m228 | 23 | 69.5 | 24.3 | 209 | 11.9 | ||
| RPL19C | AF075597 | T2H3.3 | At4g02230 | T04719 | 5 | 4 | 1.0 | ve023 | 11.9 | 71.2 | 23.3 | 200 | 12.0 | ||
| L21 | RPL21A | II | AC003970 | F14J9.25 | At1g09590 | AV552764 | 5 | 1 | 3.0 | phyA | 11.3 | 48.7 | 18.7 | 164 | 11.3 |
| RPL21B* | AC003970 | F14J9 | N.A. | N.F. | 0 | 1 | 3.0 | phyA | 11.3 | iORF | – | – | – | ||
| RPL21C | AC000132 | F21M12.8 | At1g09690 | AV537606 | 7 | 1 | 3.1 | ve005 | 11.4 | 48.7 | 18.7 | 164 | 11.3 | ||
| RPL21D | AC007654 | T19E23.15 | N.A. | BE527706 | 2 | 1 | 11.2 | UFO | 47 | iORF | – | – | – | ||
| RPL21E | AC079733 | T8L23.13 | At1g57660 | R65045 | 7 | 1 | 20.9 | nga280 | 83.8 | 48.7 | 18.7 | 164 | 11.3 | ||
| RPL21F | AL132977 | T10K17.30 | At3g57820 | AA585876 | 1 | 3 | 21.9 | SNP7 | 77.1 | iORF | – | – | – | ||
| L22 | RPL22A | III | AC009525 | F22D16.17 | At1g02830 | N.F. | NE | 1 | 0.6 | GST1 | 3.9 | 58.4 | 14.5 | 127 | 10.6 |
| RPL22B | AC011620 | F18C1.17 | At3g05560 | T88520 | 1 | 3 | 1.7 | mi355 | 13.9 | 68.7 | 14.0 | 124 | 10.4 | ||
| RPL22C* | AC069556 | T1G16 | At5g27770 | Z33746 | 1 | 5 | 9.8 | SO262 | 65.2 | 64.1 | 14.0 | 124 | 10.1 | ||
| L23 | RPL23A* | I | AC000104 | F19P19.5 | At1g04480 | AV557949 | 9 | 1 | 1.1 | SGCSNP151 | 3.3 | 84.4 | 14.5 | 136 | 11.1 |
| RPL23B | AC002332 | F4P9.14 | At2g33370 | Z33670 | 10 | 2 | 14.4 | ve015 | 63.9 | 84.9 | 15.0 | 140 | 11.2 | ||
| RPL23C | AC022287 | T27C4.4 | At3g04400 | BE037765 | 4 | 3 | 1.1 | GAPC | 8.4 | 84.9 | 15.0 | 140 | 11.2 | ||
| L23a | RPL23aA(2) | I | AC004218 | F12L6.12 | At2g39460 | BE039409 | 5 | 2 | 16.7 | SGCSNP37 | 72.4 | 74.8 | 17.4 | 154 | 11.0 |
| RPL23aB(3) | AL132954 | T26I12.160 | At3g55280 | AV544539 | 7 | 3 | 20.9 | ve022 | 76.8 | 74.1 | 17.9 | 158 | 11.0 | ||
| L24 | RPL24A* | II | AC006282 | F13K3.2 | At2g36620 | AV551827 | 4 | 2 | 15.6 | ve017 | 64.1 | 47.0 | 18.4 | 160 | 11.5 |
| RPL24B | AL132969 | F8J2.190 | At3g53020 | Z26463 | 4 | 3 | 20.1 | SGCSNP188 | 74.4 | 48.0 | 18.6 | 163 | 11.5 | ||
| L26 | RPL26A | I | AL132965 | T16K5.260 | At3g49910 | AW004134 | 6 | 3 | 18.9 | SGCSNP398 | 72.2 | 73.4 | 16.9 | 146 | 11.5 |
| RPL26B | AB013390 | K9I9.7 | At5g67510 | Z26419 | 1 | 5 | 27.0 | m555 | 132.6 | 76.7 | 16.8 | 146 | 11.8 | ||
| L27 | RPL27A | III | AC006223 | F22D22.3 | At2g32220 | AI995587 | 2 | 2 | 13.8 | SGCSNP26 | 63.27 | 57.8 | 15.5 | 135 | 11.0 |
| RPL27B | AP001306 | MKA23.13 | At3g22230 | AV550432 | 7 | 3 | 7.8 | PAP606 | 30 | 56.3 | 15.6 | 135 | 11.1 | ||
| RPL27C | AL161540 | FCA2 | At4g15000 | T76226 | 5 | 4 | ∼7.5 | mi198 | 49.6 | 55.6 | 15.6 | 135 | 11.1 | ||
| L27a | RPL27aA | I | AC012187 | F13K23.22 | At1g12960 | N.F. | 0 | 1 | 4.3 | ve006 | 16.1 | 61.5 | 16.5 | 144 | 11.0 |
| RPL27aB | AC005292 | F26F24.13 | At1g23290 | Z26208 | 8 | 1 | 8.3 | m235 | 34 | 67.6 | 16.3 | 146 | 11.3 | ||
| RPL27aC | AC010796 | F24J13.17 | N.A. | AV537006 | 6 | 1 | 26.3 | mi462 | 110.7 | 67.6 | 16.5 | 146 | 11.4 | ||
| L28 | RPL28A | III | AC005169 | F6F22.24 | At2g19730 | BE038429 | 4 | 2 | 8.8 | mi148 | 36.1 | 34.9 | 15.9 | 143 | 11.4 |
| RPL28B* | AP000600 | MAG2 | N.A. | N.F. | 0 | 3 | 4.7 | nga162 | 20.5 | iORF | – | – | – | ||
| RPL28C | AL161574 | F19B15 | At4g29410 | AV545939 | 8 | 4 | 13.5 | mi232 | 76.7 | 35.7 | 15.9 | 143 | 11.6 | ||
| L29 | RPL29A | III | AC023912 | F3E22.16 | At3g06700 | T46465 | 3 | 3 | 2.1 | SGCSNP115 | 3.32 | 69.2 | 7.0 | 61 | 12.0 |
| RPL29B | AC023912 | F3E22.18 | At3g06680 | N.F. | 0 | 3 | 2.1 | SGCSNP115 | 3.32 | 69.2 | 7.0 | 61 | 12.0 | ||
| L30 | RPL30A | II | AC025781 | F15C21.6 | At1g36240 | N.F. | 0 | 1 | 13.7 | SGCSNP279 | 61.13 | 72.5 | 12.3 | 112 | 10.6 |
| RPL30B* | AC009243 | F28K19.15 | At1g77940 | AV532452 | 13 | 1 | 29.0 | ve011 | 119.4 | 69.7 | 12.4 | 112 | 10.1 | ||
| RPL30C | AB026654 | MVE11.10 | At3g18740 | N.F. | 0 | 3 | 6.4 | ve039 | 24.6 | 69.7 | 12.3 | 112 | 10.5 | ||
| L31 | RPL31A | II | AC005169 | F6F22.23 | At2g19740 | AA712836 | 4 | 2 | 8.8 | mi148 | 36.1 | 58.8 | 13.7 | 119 | 10.7 |
| RPL31B | AL049171 | T25K17.40 | At4g26230 | BE526625 | 2 | 4 | 12.4 | RPS2 | 75.6 | 59.3 | 13.8 | 119 | 10.6 | ||
| RPL31C | AB013392 | MIK19.16 | At5g56710 | AF162852 | 4 | 5 | 23.0 | mi69 | 114.2 | 57.5 | 13.8 | 119 | 10.7 | ||
| L32 | RPL32A | II | AL110123 | F15J5.70 | At4g18100 | Z17739 | 3 | 4 | 9.2 | mi32 | 60.9 | 66.9 | 15.5 | 133 | 11.6 |
| RPL32B | AB019223 | K11I1.2 | At5g46430 | AA042212 | 3 | 5 | 18.9 | SGCSNP219 | 96.8 | 64.6 | 14.5 | 133 | 11.5 | ||
| L34 | RPL34A | III | AC005508 | T2P11.7 | At1g26880 | F20073 | 3 | 1 | 9.3 | mi92 | 41.1 | 52.2 | 13.7 | 120 | 12.2 |
| RPL34B | AC013289 | T6C23.18 | At1g69620 | AI013289 | 9 | 1 | 26.0 | mi462 | 110.7 | 51.3 | 13.7 | 119 | 12.2 | ||
| RPL34C | AP000386 | MLD15.7 | At43g28900 | N.F. | 0 | 3 | 10.9 | AIG2 | 50.5 | 49.6 | 13.6 | 120 | 12.0 | ||
| L35 | RPL35A | I | AC016661 | F11F8.7 | At3g09500 | N.F. | 0 | 1 | 2.9 | APX1B | 16.2 | 64.8 | 14.3 | 123 | 11.6 |
| RPL35B | AC004218 | F12L6.5 | At2g39390 | BE038438 | 6 | 2 | 16.7 | SGCSNP37 | 72.4 | 64.8 | 14.3 | 123 | 11.6 | ||
| RPL35C | AL132954 | T26I12.50 | At3g55170 | BE038964 | 2 | 3 | 20.9 | SGCSNP134 | 75.75 | 63.9 | 14.2 | 123 | 11.6 | ||
| RPL35D | AL162971 | T22P11.200 | At5g02610 | AV549599 | 6 | 5 | 0.5 | SNP241 | 3.7 | 64.8 | 14.3 | 123 | 11.6 | ||
| L35a | RPL35aA | II | AC067971 | F10K1.22 | At1g06980 | N.F. | 0 | 1 | 2.1 | GT45 1 | 8.52 | 55.9 | 12.9 | 112 | 11.5 |
| RPL35aB | AC008046 | F5A13.4 | At1g41880 | AV535617 | 6 | 1 | 15.2 | mi133 | 61 | 55.9 | 12.8 | 111 | 11.5 | ||
| RPL35aC | AC020579 | F1O17.6 | At1g74270 | N.F. | 0 | 1 | 27.6 | SGCSNP380 | 117.2 | 56.9 | 12.9 | 112 | 11.5 | ||
| RPL35aD | AL161667 | F1I16.160 | At3g55750 | AI994336 | 6 | 3 | 21.1 | SGCSNP189 | 77.2 | 55.9 | 12.8 | 111 | 11.5 | ||
| L36 | RPL36A | III | AC004684 | F13M22.10 | At2g37600 | AI999791 | 1 | 2 | 16.0 | ve018 | 69.7 | 58.3 | 12.7 | 113 | 12.3 |
| RPL36B | AL132960 | F5K20.40 | At3g53740 | AV533586 | 14 | 3 | 20.3 | ve042 | 76.29 | 60.0 | 12.7 | 112 | 12.3 | ||
| RPL36C | AL162971 | T22P11.40 | At5g02450 | T04630 | 2 | 5 | 0.5 | SGCSNP241 | 3.7 | 59.6 | 12.2 | 108 | 12.1 | ||
| L36a | colRPL36aA | II | AB015474 | MLM24.12 | At3g23390 | AV541635 | 10 | 3 | 8.3 | mi386 | 36.3 | 76.8 | 12.1 | 105 | 11.1 |
| RPL36aB | Z97336 | FCA1 | At4g14320 | BE528949 | 6 | 4 | 7.2 | ve024 | 51.9 | 76.8 | 12.1 | 105 | 11.1 | ||
| L37 | RPL37A* | II | AC007591 | F9L1 | At1g15250 | F20017 | 3 | 1 | 5.2 | SRP54A | 18.9 | 66.7 | 10.6 | 93 | 12.4 |
| RPL37B | AC037424 | F19K6.12 | At1g52300 | AV524548 | 17 | 1 | 19.1 | PAP240 | 81.1 | 67.4 | 10.8 | 95 | 12.4 | ||
| RPL37C* | AB012247 | MSL1 | At3g16080 | AI998492 | 5 | 3 | 5.4 | m228 | 23.4 | 63.8 | 10.7 | 95 | 12.4 | ||
| L37a | RPL37aA | III | AC004667 | T4C20.10 | N.A. | N.F. | E | 2 | 15.1 | m323 | 67.9 | iORF | – | – | – |
| RPL37aB | AC009991 | F9F8.23 | At3g10950 | N.F. | 0 | 3 | 3.5 | MNSOD | 14.7 | 69.3 | 10.4 | 92 | 11.1 | ||
| RPL37aC* | AL163852 | F27H5 | N.A. | BE577732 | 13 | 3 | 22.7 | SGCSNP74 | 84.6 | 70.9 | 10.0 | 89 | 11.0 | ||
| L38 | RPL38A | III | AC002335 | T1O24.20 | At2g43460 | N96748 | 5 | 2 | 18.2 | COR15 | 76.8 | 78.3 | 8.1 | 69 | 10.7 |
| RPL38B | AL138659 | T16L24.90 | At3g59540 | N.F. | 0 | 3 | 22.4 | SNP74 | 84.6 | 78.3 | 8.1 | 69 | 10.7 | ||
| L39 | RPL39A* | II | AC007070 | T22F11.20 | At2g25210 | Z17538 | 3 | 2 | 11.0 | g6842 | 46.7 | 72.5 | 6.4 | 51 | 12.8 |
| RPL39B* | AC009755 | F14P3.16 | At3g02190 | N.F. | NE | 3 | 0.4 | mi74b | 5.8 | 74.5 | 6.4 | 51 | 12.8 | ||
| RPL39C* | AL021636 | F10N7 | At4g31981 | AV536940 | 3 | 4 | 14.7 | g8300 | 81.2 | 72.9 | 6.3 | 50 | 12.8 | ||
| L40 | RPL40A | III | AC006921 | F9C22.10 | At2g36170 | AV533842 | 4 | 2 | 14.4 | SGCSNP333 | 67.97 | 92.2 | 14.7 | 128 | 10.7 |
| RPL40B | AL050300 | F22O6.30 | At3g52590 | Z35369 | 15 | 3 | 19.9 | mi456 | 72.7 | 92.2 | 14.7 | 128 | 10.7 | ||
| L41 | RPL41A | III | AC009894 | T6H22.15 | N.A. | AI998257 | 2 | 1 | 3.4 | mi3030 | 83.7 | 96.0 | 3.4 | 25 | N.D. |
| RPL41B* | AC002986 | YUP8H12R | N.A. | N.F. | 0 | 1 | 29.5 | SNP253 | 120.4 | iORF | – | – | – | ||
| RPL41C* | AC018721 | T7M7 | N.A. | N.F. | 0 | 2 | 17.0 | SNP241 | 74.4 | 96.0 | 3.4 | 25 | N.D. | ||
| RPL41D | AC074395 | T8G24.5 | At3g08520 | N.F. | 0 | 3 | 2.5 | SNP192 | 11 | 96.0 | 3.4 | 25 | N.D. | ||
| RPL41E | AC009991 | F9F8.7 | At3g11120 | N.F. | 0 | 3 | 3.4 | SNP11 | 14.7 | 96.0 | 3.4 | 25 | N.D. | ||
| RPL41F* | AC024128 | MGH6 | N.A. | T41975 | 3 | 3 | 4.1 | nga162 | 20.5 | 96.0 | 3.4 | 25 | N.D. | ||
| RPL41G | AL163832 | F27K19.200 | At3g56020 | AI998878 | 1 | 3 | 21.2 | SNP189 | 77.2 | 96.0 | 3.4 | 25 | N.D. | ||
Ribosomal protein type corresponding to rat nomenclature. Asterisk, Gene and gene family member designation and genes that were not annotated or incorrectly annotated. NA, Standard AGI gene name and genes that have not been annotated. Classification based on evolutionary group (Group I, represented in eubacteria, archaebacteria, and eukaryotes; Group II, represented in archaebacteria and eukaryotes; and Group III, represented in eukaryotes only). GenBank accession no. corresponding to genomic sequence. BAC clone and position of annotated gene corresponding to genomic sequence. NF, Representative EST or cDNA GenBank accession no. and genes with no corresponding EST are indicated none found. No. of ESTs present in GenBank release 65.0, 0 if no EST is identified, NE if no expression is detected by RT-PCR, and E if expression is detected by RT-PCR. Chromosomal location, AGI map position (Mbp), nearest genetic marker as determined from the AGI map and AGI map position of nearest genetic marker (cM). Percent identity to the rat orthologue determined by the BESTFIT algorithm. iORF, Incomplete open reading frame. Predicted molecular mass (kDa), no. of amino acids of deduced ORF (A.A.), and predicted pI.
Arabidopsis Cytoplasmic r-Proteins Are Encoded by Small Gene Families
We identified multiple Arabidopsis r-protein genes for all 79 r-protein types of rat. We propose a unifying r-protein gene nomenclature in which Arabidopsis r-protein gene names contain the prefix RP (r-protein) and the suffix S or L referring to r-protein type (small or large) modeling that found for the mammalian nomenclature. For example, RPL3 encodes r-protein L3. The one exception to this rule is the conventional nomenclature for the acidic ribosomal phosphoproteins, known as the P proteins (here, RPP2 encodes P2). For each distinct gene family member a letter is provided (i.e. RPL3A and RPL3B are distinct genes that encode L3). This alphabetic designation of gene family members is ordered by chromosomal location. In addition, previously published gene designations are included in Table I in parentheses. The number of genes within an r-protein gene family varies between two and seven (L41), with most families containing three or four genes (Table I and Fig. 1). In 21 instances, the genomic sequences lacked a complete ORF (for example, the deduced ORF encoded a truncated protein due to a premature translational stop codon, a frameshift in the ORF, or an internal deletion) and these were designated an incomplete ORF; in most of these cases (19), there was no cognate EST identified for these presumed pseudogenes. The copy number of r-protein genes is apparently random. There was no bias based on ribosomal subunit or r-protein group classification (see Table I).
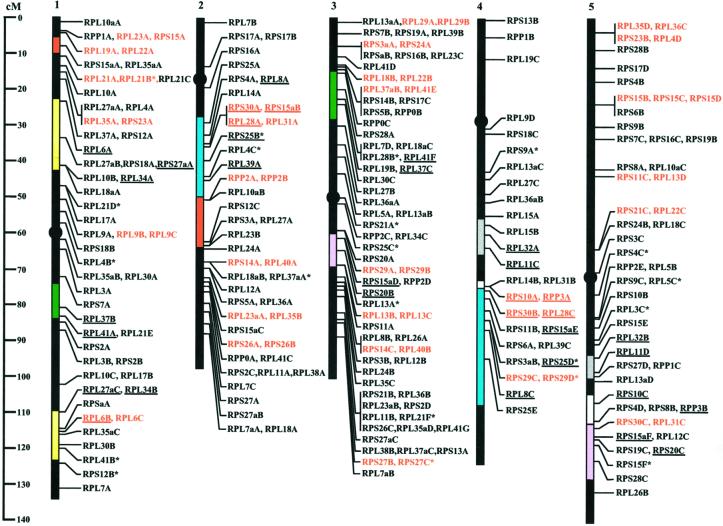
Figure 1.
Genomic location of Arabidopsis r-protein genes. The 249 Arabidopsis r-protein genes are mapped by distance (centiMorgans) to nearest genetic marker from the distal short arm on the genetic map of each chromosome (Lister et al., 1993). Centromeres are shown as black circles. Genes listed linearly are tandemly arranged on the same chromosome and those located on the same BAC clone are depicted in red. An asterisk indicates genes with an incomplete ORF. Duplicated regions corresponding to numbers 1, 2, 3, 4, 5, 6, and 7 from Table III are indicated in yellow, red, blue, green, pink, gray, and white, respectively. Genes conserved between duplicated regions are underlined. (Continued from p. 400)
Arabidopsis r-Protein Genes Are Not Distributed Randomly
Database mining allowed us to identify bacterial artificial chromosome (BAC) or phage artificial chromosome (P1) clones carrying one or several genes for r-proteins (Table I). In addition, existing knowledge of the location of these clones allowed us to identify the positions of the r-protein genes on the AGI map (http://www.Arabidopsis.org). A composite map of the 249 r-protein genes, integrating genomic sequence information and nearest genetic marker data available through AGI, was constructed (Fig. 1). Chromosome map positions are given in centiMorgans from the top of the chromosome, and the nearest genetic marker to each r-protein gene is indicated in Table I. Mapping results are also summarized in Table II. We observed differences in the number of genes per chromosome as the number of r-protein genes located on chromosomes 1, 2, 3, 4, and 5 are 54, 45, 71, 29, and 50, respectively. The distribution of the r-protein genes is visible on the gene map (Fig. 1; r-protein gene density is 538 Kb per r-protein gene for chromosome 1, 436 Kb per r-protein gene for chromosome 2, 326 Kb per r-protein gene for chromosome 3, 605 Kb per r-protein gene for chromosome 4, and 519 Kb per r-protein gene for chromosome 5. This situation appears to contrast with the even distribution of all protein coding sequences observed for the five chromosomes (AGI, 2000); however, statistical analysis (g test, P value = 0.4522) indicated that these differences are not significant. If the r-protein genes were randomly distributed, approximately one gene per 500 kb would be expected; however, in 29 instances, two to four r-protein genes were found on a single BAC (Table II). In eight instances, genes that encode different r-proteins are within 5 kb. In several additional cases, r-protein genes have been duplicated and found on the same BAC, and in one instance the genes are triplicated within the same BAC (S15 on chromosome 5). In addition, there are several examples where only one r-protein gene is found within a BAC; nevertheless, the density of r-protein genes within that region may still be rather high (Fig. 1). These data indicate that localized duplication of these genes has occurred infrequently.
Table II.
Arabidopsis BAC clones containing more than one r-protein gene
| Chromosome No. | BAC Clone | Genes | Intergene Distance |
|---|---|---|---|
| Kb | |||
| 1 | F19P19 | RPL23A,RPS15A | 73.2 |
| F22D16 | RPL22A,RPL19A | 15.7 | |
| F14J9 | RPL21A,RPL21B | 44.3 | |
| F11F8 | RPL35A,RPS23A | 49.3 | |
| T9L6 | RPL9B,RPL9C | 11.1 | |
| T2P11 | RPL34A,RPL10B | 5.0 | |
| F2P9 | RPL6B,RPL6C | 1.2 | |
| 2 | F6F22 | RPS15aB,RPL28A | 0.3 |
| RPL28A,RPL31A | 0.3 | ||
| RPL31A,RPS30A | 0.6 | ||
| F15K20 | RPP2B,RPP2A | 0.7 | |
| F9C22 | RPS14A,RPL40A | 1.0 | |
| F12L6 | RPL35B,RPL23aA | 23.2 | |
| T2P4 | RPS26A,RPS26B | 15.5 | |
| 3 | F3E22 | RPL29A,RPL29B | 2.6 |
| T9J14 | RPS3aA,RPS24A | 29.4 | |
| F18C1 | RPL18B,RPL22B | 6.2 | |
| F9F8 | RPL37aB,RPL41E | 56.5 | |
| T15B3 | RPS29A,RPS29B | 20.9 | |
| T2J13 | RPL13B,RPL13C | 15.5 | |
| F22O6 | RPS14C,RPL40B | 0.9 | |
| T20K12 | RPS27B,RPS27C | 0.6 | |
| 4 | F14M19 | RPS10A,RPP3A | 50.8 |
| F19B15 | RPS30B,RPL28C | 2.5 | |
| F17I5 | RPS29C,RPS29D | 14.5 | |
| 5 | T22P11 | RPL35D,RPL36C | 53.2 |
| F9G14 | RPS23B,RPL4D | 3.2 | |
| T5E8 | RPS15B,RPS15C | 0.8 | |
| RPS15C,RPS15D | 1.6 | ||
| MRO11 | RPS11C,RPL13D | 54.8 | |
| T1G16 | RPS21C,RPL22C | 28.1 | |
| MIK19 | RPS30C,RPL31C | 8.0 |
In the analysis of the distribution of r-protein genes, we observed that RPL28A and RPS30A are on chromosome 2 and RPL28C and RPS30B are on chromosome 4. This observation led us to compare adjacent genes in these two BACs (Table III, Fig. 1, genes conserved between duplications are underlined; about one-half of the 249 r-protein genes are in currently identified duplicated regions; in Fig. 1, large duplicated regions are shown). However, the percentage of genes encoding the same type of r-protein found in conserved positions in both copies of the duplicated regions is 25% to 30% with a range between 0% to 66% (Table III). This observation is consistent with another study that found only 28% of genes in duplicated regions are actually present in duplicate copies (Vision et al., 2000). The most extreme situation is illustrated by two duplicated segments on chromosomes 1 (6.1–10.8 cM) and 2 (50.6–63.9 cM), which contain two and seven r-protein genes, respectively, of which none are paralogous (Table III, Duplicated Region 2; Fig. 1, red colored regions). In summary, analysis of the distribution of the r-protein genes in the Arabidopsis genome showed no evident clustering of these genes. However, r-protein gene density in some regions of the Arabidopsis genome is much higher than that expected for a uniform distribution.
Table III.
Large duplicated regions of the Arabidopsis genome-containing r-protein genes
| Duplicate No. | Duplicated Regions
|
No. of Genes within Duplicated Regions | No. of Genes Conserved between Duplicated Regions | % Genes Conserved between Duplicated Regions | ||
|---|---|---|---|---|---|---|
| Chromosome | Border BAC clones | Position | ||||
| cM | ||||||
| 1 | 1 | F20D23-T7N9 | 23.6–41.1 | 6 | 3 | 50 |
| 1 | T6C23-F18B13 | 110.7–123.8 | 8 | – | 38 | |
| 2 | 1 | F19P19-F22O13 | 6.1–10.8 | 2 | 0 | 0 |
| 2 | T22O13-F4P9 | 50.6–63.9 | 7 | – | 0 | |
| 3 | 2 | F16F14-T19L18 | 30.9–50.6 | 10 | 6 | 60 |
| 4 | T13J8-T5J17 | 76.8–108.5 | 12 | – | 50 | |
| 4 | 1 | F27J15-T6H22 | 73.5–83.8 | 3 | 2 | 66 |
| 3 | MBK21-MOE17 | 16.2–28.1 | 8 | – | 25 | |
| 5 | 3 | T6H20-F24M12 | 60.5–68.2 | 7 | 2 | 29 |
| 5 | K19M22-K1L20 | 113.7–128 | 8 | – | 25 | |
| 6 | 4 | FCA8-T13K14 | 57.6–65.4 | 3 | 2 | 66 |
| 5 | K23L20-MNJ7 | 94.1–99.4 | 4 | – | 50 | |
| 7 | 4 | F22K18-T27E11 | 72.4–76.8 | 4 | 3 | 50 |
| 5 | K2I5-MJB24 | 105.4–113.7 | 4 | – | 50 | |
Expression of Arabidopsis r-Protein Genes Appears to Be Differentially Regulated
The occurrence of r-protein gene families raises the question of whether the genes are differentially regulated. The frequency of ESTs available in GenBank (database of expressed sequence tags) has been proposed as a useful tool for preliminary analysis of gene expression (Adams et al., 1995). Despite the limited number of Arabidopsis ESTs (112,500; release 022301, February 2001) available in GenBank, we used this approach to obtain a first assessment of r-protein gene expression. All gene families have at least one EST for at least one gene, but the frequency of ESTs for individual genes varies greatly between different gene family members and gene families. Many r-protein genes (approximately 20%) apparently are very highly expressed, as indicated by the EST number in Table I (10–40 ESTs). The frequency of ESTs observed per gene was variable among genes from the same family. For example, in the P0 gene family, the three genes encode complete ORFs but were represented by 40, 6, and 0 ESTs. On the other hand, in many cases a representative EST was observed for each member of a given family. Cognate ESTs were not found for 52 of the r-protein genes (approximately 20%). Of these, 19 lack a complete ORF and hence are most likely pseudogenes. Genes with a complete deduced ORF may lack a representative EST due to low levels of mRNA accumulation solely in specific cell types or at a specific developmental stage. To examine this possibility, PCR and RT-PCR (with gene specific primers) using a cDNA library or RNA prepared from 3-week-old plants was performed on a subset of r-protein genes lacking a corresponding EST. A PCR (or RT-PCR) product was observed for many (72%) of these genes (data not shown), suggesting that they may be transcribed at some stage in development. Consistent with analyses from other groups, we observed differential levels of expression of individual gene family members.
Global analysis of the expression of the 54, 45, 71, 29, and 50 r-protein genes located on chromosomes 1, 2, 3, 4, and 5, respectively, showed that the percentage of these r-protein genes for which an EST is available is 74.1%, 80%, 77.4%, 79.3%, and 84%, respectively. The average numbers of ESTs per mapped r-protein gene per chromosome are 7.8, 5.3, 5.4, 5.3, and 6.1 (chromosomes 1, 2, 3, 4, and 5, respectively). These results suggest a positive bias in favor of chromosome 1 and 5: The r-protein genes on the two chromosomes, in average, seemed to be more abundantly expressed. However, statistical analysis using a non-parametric ANOVA (Kruskal-Wallis test, performed because the data failed to meet the assumption of normality [data not shown] for a standard ANOVA) indicates that there is no significant difference (P value = 0.6087) in the expression of the r-protein genes, among the five chromosomes, based on EST frequency (SAS Institute Inc., 1989).
Biochemical Characteristics of Deduced Arabidopsis r-Proteins
The deduced amino acid sequence for each of the 80 types of r-proteins was determined. In addition, for each r-protein, the predicted molecular mass and pI was calculated, and the percent identity to the rat ortholgue was determined. The deduced Arabidopsis r-proteins range in size from 44.7 (L4) to 3.4 (L31) kD. Of the deduced proteins, Sa, P0, P1, P2, P3, and S12 were acidic (pI 4.0–5.8) and the remainder were basic, ranging in pI from 8.1 (S27) to 12.8 (S30 and L39). The positive charge of the majority of r-proteins is consistent with their interaction with rRNA. The identity between Arabidopsis and rat orthologues averaged 66% and ranged from 96% for L41% to 35% for L28. It is interesting that an L28 orthologue was not identified in the genomic sequence of S. cerevisiae (Planta and Mager, 1998), indicating that it is a rather divergent r-protein. A final observation was that the identity between rat and individual Arabidopsis orthologues (deduced proteins from the same gene family) were usually within 0% to 5.0% of one another, indicating that members of individual r-protein families are highly conserved. However, there were a few exceptions where the identities within an r-protein family varied 14.1%, 24.0%, and 30.1%, corresponding to the r-proteins P2, L7, and S15a, respectively. These distinctions in proteins encoded by these classes could result in ribosomal heterogeneity or may reflect the evolution of proteins with extra-ribosomal function.
DISCUSSION
Arabidopsis Ribosomes Contain at Least 80 r-Protein Types, Encoded by 249 Genes
Previous work from our two groups identified 106 Arabidopsis r-protein genes by contig construction from EST sequences coding for 50 orthologues of yeast r-proteins (Cooke et al., 1996) and 77 Arabidopsis orthologues of rat r-proteins (Bailey-Serres, 1998). This report extends the parallel analyses of our two groups on the set of Arabidopsis r-proteins that can be defined by homology to the 79 known eukaryotic r-proteins. All rat r-protein genes have an orthologue in Arabidopsis; however, plants possess an additional r-protein, P3, that appears to be limited to the plant kingdom (Szick et al., 1998). A total of 80 r-protein types encoded by 249 genes were classified, positioned on the AGI map, and the nearest genetic marker identified. Based on this study, Arabidopsis has at least 32 small ribosomal subunit proteins encoded by 101 genes and 48 large ribosomal subunit proteins encoded by 148 genes. Due to the extensive segmental duplication of the Arabidopsis genome, all r-protein genes have between two and several paralogues. Our study included analysis of genomic sequences and ESTs encoding r-proteins. Because all ESTs were assigned to specific genomic sequences, it is unlikely that additional genes that encode rat r-protein orthologues will be identified in the unsequenced centromeric and rDNA regions. Based on this analysis of Arabidopsis r-protein genes, the protein composition of plant ribosomes is very similar to that of other eukaryotes. Our study provides an entry to several important issues such as systematic annotation of r-protein genes; normalization of nomenclature; evolutionary studies of gene structure; analysis of gene expression at the transcriptional, posttranscriptional, and translational levels; examination of r-protein transport to the nucleolus; and ribosome biogenesis.
Analysis of Arabidopsis r-Protein Gene Distribution Provides Insight into r-Protein Gene Evolution
In humans, r-protein genes are found on all chromosomes but with a bias toward chromosome 19 (Kenmochi et al., 1998b). In prokaryotic genomes, r-protein gene clustering is found in the form of operons in which expression of several genes is coordinately regulated under a single promoter (Nomura et al., 1984). No obvious similar clustering has been reported in eukaryotic genomes and recent results (Kenmochi et al., 1998a) showed only one example of local clustering in the human genome, three genes encoding L13A, S11, and L18 being located within 0.6 cM. It is noteworthy that in the Arabidopsis genome, r-protein gene density is much higher in several regions than would be expected from a uniform distribution. For example, the chromosome 2 BAC clone F6F22 contains four different r-protein gene types within 1.2 kb (Table II). Whether this grouping corresponds to a fossil functional clustering remains to be established by the analysis of different plant genomes.
Analysis of r-protein gene organization has served as a starting point for new insights on genome organization and dynamics in Arabidopsis. It has become obvious that the Arabidopsis genome is a mosaic of duplicated regions (AGI, 2000; Blanc et al., 2000; Paterson et al., 2000; Vision et al., 2000). These data have extended observations made by comparison of chromosomes 2 and 4 (Lin et al., 1999; Mayer et al., 1999). These duplications are either the result of reciprocal translocations between Arabidopsis chromosomes or of an ancient polyploidisation event. It can be reasonably assumed that large duplications constitute one of the main factors of gene duplication in Arabidopsis and have certainly contributed to the increase in r-protein gene number because one-half of the 249 mapped genes are located in duplicated regions. However, closer examination of r-protein genes in duplicated regions shows that considerable rearrangements involving r-protein genes have taken place following duplication of chromosomal segments. Genes encoding the same r-protein are found in conserved positions in both duplicated segments for only approximately 25% of the genes. This observation indicates that although many r-protein genes occur in large duplicated segments, the story is much more complex. It appears that one copy frequently was lost for many of the pairs following duplication of a large chromosomal region, or r-protein genes have been inserted following duplication events. However, the relatively low number of intron-less genes having an intron-containing paralogue argues against the latter mechanism (Martinez et al., 1989).
Because r-proteins form a complex macromolecule in which coordinated regulation of protein levels as well as steric constraints are essential, it is possible that negative selection has led to the elimination of duplicated copies of certain genes. However, the Group I class of r-proteins are found to occur within eubacteria, archaebacteria, and eukaryotes (Wool et al., 1995), yet do not show any bias toward lower copy number than Group II and III r-proteins. Our analysis has shown in addition that tandem duplication, which is another mechanism to increase gene copy number, does not appear to have been important in the expansion of r-protein gene families. Because Arabidopsis is a model genome that will be used to investigate the genomes of many cultivated crops, and because r-protein genes have been conserved throughout evolution, this work should serve as a basis to analyze the distribution and expression of r-protein genes in crop plant species.
The Majority of Arabidopsis r-Protein Genes Appear to Be Expressed
An important question raised by the occurrence of multigene families is the regulation and level of expression of each member in the family. Assessing r-protein gene expression by the presence of an EST showed that at least 77% of r-protein genes (not including the 21 genes with incomplete ORFs) are expressed at a level detectable by an EST. Most or all copies of genes in the individual families have been tagged. The r-protein genes for which no EST is yet available could correspond either to genes that are rarely transcribed or to pseudogenes. As shown in Table I, several r-protein genes for which an EST was not identified have truncated ORFs or deletions within their ORFs. Analysis of expression, PCR, or RT-PCR indicated that many of these genes are in fact expressed (Table I, EST column, represented with an E or NE). Only 7% of r-protein genes were not expressed in the tissues tested. The infrequent nature (7%) of potential r-protein pseudogenes is in agreement with previous data of Lin et al. (1999), who reported that only 10% of all the genes identified or predicted on chromosome 2 correspond to pseudogenes. Our observation that the majority of r-protein genes are expressed in plants is notably different from the situation reported in mammals, in which multiple pseudogenes and only one functional, intron-containing gene was observed for most r-proteins (Wiedemann and Perry, 1984; Wagner and Perry, 1985; Baker and Board, 1992).
The large number of expressed genes in multigene families in plants is probably due to the fact that plants have evolved by polyploidy (Dornelas et al., 1998), followed by specialization of the function or expression patterns of gene family members, thus allowing increased plasticity in response to non-optimal growth conditions. The high degree of sequence identity between different r-proteins suggests specialization by different temporal or spatial expression patterns to increase protein synthesis at certain developmental times. To date, all detailed analyses of Arabidopsis r-protein genes have illustrated distinctions in regulation of expression of gene family members. For example, high levels of expression of one Arabidopsis L11 gene (RPL11C, previously called RPL16B) was observed in shoot and primary root meristems and lateral root primordia in response to auxin treatment, whereas expression of another L11 gene (RPL11A, previously called RPL16A) showed more cell type-specific gene expression (Williams and Sussex, 1995). Mutations in Arabidopsis S13 and S18 genes were shown to cause a pointed first leaf (pfl) phenotype, remarkably indicating that mutations that alter the expression of r-protein genes may confer a similar phenotype (Van Lijsebettens et al., 1994; Ito et al., 2000). In pfl1, a T-DNA insertion into the S18A (RPS18A) gene results in complete abrogation of gene expression (Van Lijsebettens et al., 1994). Although S18 is encoded by three genes that appear to have overlapping expression, synthesis in mitotically active tissues seems to be required for normal leaf development. In pfl2, caused by a Ds insertion into the S13A (RPS13B) gene, a significantly reduced number and increased size of subepidermal palisade cells of the first leaf was observed (Ito et al., 2000). Consistent with the apparent effects on cell division, a conditional deletion of r-protein S6 gene in mice does not impair the growth of liver cells following partial hepatectomy but does block the progression through the cell cycle (Volarevic et al., 2000). In this example, existing levels of ribosomes are sufficient for cell growth. In contrast, r-protein gene mutations in Drosophila melanogaster are known to cause the haploinsufficient Minute phenotype that shows slower rates of cell growth and division (Lambertsson, 1998). Further studies using DNA microarray studies, r-protein gene promoter fusions to a reporter gene, and r-protein gene mutants will be necessary to assess the regulation and role of individual r-protein genes. These studies hopefully will shed light on the role of r-proteins and ribosome biogenesis on regulation of cell growth and proliferation in plants.
Our results show varying numbers of r-protein genes in different families, although it is clear that control mechanisms must exist to ensure the presence of stoichiometric levels of each protein in the ribosomes. This could be achieved by higher expression levels of members of smaller gene families. However, expression levels of different members deduced from the number of cognate ESTs show no clear inverse relationship between the level of expression and the number of genes. Therefore, it is likely that r-protein synthesis is also controlled at a posttranscriptional step. It has been determined that vertebrate r-protein levels are regulated at the translational level, possibly by sequences around a polypyrimidine tract present at the 5′ end of the mRNA, through the regulation of r-protein S6 phosphorylation (Fumagalli and Thomas, 2000; Meyuhas, 2000; Meyuhas and Hornstein, 2000). In plants, posttranscriptional regulation of rapeseed L13 r-protein (Sáez-Vásquez et al., 2000), maize P2a (Fennoy et al., 1998), and maize S6 (Sanchez de Jimenez et al., 1999) expression was reported. Preliminary surveys suggest that a number of plant r-protein mRNAs possess 5′-polypyrimidine tracts (A. Williams and J. Bailey-Serres, unpublished data). In addition, studies with a cell-free wheat germ translation system confirmed that translation of an mRNA with a 5′-polypyrimidine tract was regulated by levels of a titratable repressor protein (Shama and Meyuhas, 1996). Furthermore, the phosphorylation of r-protein S6 is regulated in plants (Turck et al., 1998; A. Williams and J. Bailey-Serres, unpublished data). These observations indicate that the role of translational regulation in r-protein synthesis needs to be rigorously examined.
The existence of differentially regulated multigene families encoding r-proteins raises the additional possibility of ribosomal heterogeneity and its possible functional significance. Here, we observed that the frequency of ESTs for different r-protein gene family members is variable (Table I). Szick-Miranda and Bailey-Serres (2001) recently demonstrated developmentally and environmentally regulated heterogeneity of the composition of the P2-type of r-protein in ribosomes of maize. This, along with our results, raises the intriguing possibility that microheterogeneity in the protein composition of ribosomes may occur at the tissue or cellular level. Such heterogeneity might be used for fine tuning of the efficiency of the translational machinery during development or under specific growth conditions.
In conclusion, this work reports a number of original findings: (a) 249 r-protein genes encoding 79 rat orthologues, and one plant-specific r-protein (P3), were identified and mapped in Arabidopsis; (b) the analysis revealed that r-protein genes are distributed over all Arabidopsis chromosomes; (c) the examination of frequency of ESTs for the different r-proteins gene family members and RT-PCR analysis of a several r-protein genes families demonstrated differential patterns of gene expression with no clear relationship between expression levels and gene number; (d) the expression analysis utilizing the number of ESTs suggest that there is no significant bias in the expression of the r-protein genes among the five chromosomes; and (e) large duplications of chromosomal segments have contributed to the increase in gene copy number but is insufficient to account for all copies because it seems that many duplicated genes have been eliminated during evolution. The identification of the r-protein genes and the determination of their primary structure and organization constitutes a first step to determine their biological role, mechanisms controlling their expression, and modeling of ribosome structure and function in plants.
MATERIALS AND METHODS
Identification and Mapping of ESTs Corresponding to r-Protein Genes
The 79 rat (Rattus norvegicus) r-protein sequences were obtained from Swiss-PROT (Bairoch and Apweiler, 2000) and the corresponding Arabidopsis ESTs were identified by TBLASTN alignment (Altschul et al., 1997) against all Arabidopsis sequences available in the database of expressed sequence tags and GenBank (http://www.ncbi.nlm.nih.gov). Sequences whose putative translation product showed significant similarity to the rat sequence were collected using the Query server at NCBI (http://www.ncbi.nlm.nih.gov/GenBank/GenBankEmail.html), imported into Sequencher (Gene Codes Corp. Ann Arbor, MI), trimmed at the 3′ end to remove ambiguous sequences, and contigs were constructed with 90% identity in 30-nucleotide steps. Assembled contigs were manually adjusted to identify members of the same gene family as described by Cooke et al. (1997). ESTs were also compared with genomic sequences to confirm identity. From this analysis, the minimal number of genes expressed in each r-protein gene family was determined. The sequence of each identified contig is available on request.
At the beginning of this work, the easiest strategy to map available EST contigs was by PCR on yeast artificial chromosome (YAC) DNA pools using gene-specific primers (Camilleri et al., 1998). Because most of the YACs in the library have been progressively anchored with respect to the genetic map (Lister and Dean, 1993), positioning of an EST on a YAC immediately gave an approximate map position.
Identification of r-Protein Genes and Mapping by Genomic Sequencing
Arabidopsis r-protein genes were identified in the genomic sequence using the same approach as for ESTs using TBLASTN of rat r-proteins against Arabidopsis genomic sequences. Despite the fact that gene annotation lagged behind sequencing, it became easiest to retrieve r-protein genes from the genomic sequence. Careful attention was paid to identify gene exons based on perfect match to ESTs (so that the same gene was not counted twice). Genes encoding plastidic or mitochondrial r-proteins were frequently identified by similarity to known chloroplast or mitochondiral proteins. These genes usually possessed targeting sequences and had higher identity to Escherichia coli r-protein genes than those of rat, and were excluded. Identification of a gene by genomic sequence mining allowed for positioning the gene on the AGI map. The percent identity to rat r-protein genes was determined by BESTFIT algorithm available through GCG (University of Wisconsin Genetics Computer Group, Madison, WI). The predicted molecular mass and pI of deduced r-proteins was determined by use of PEPTIDESORT (University of Wisconsin Genetics Computer Group). Genes that were not annotated or were annotated incorrectly were translated using MBS Translator (available at http://mbshortcuts.com/translator/) and intron/exon boundaries were determined by visual inspection of translated sequences comparing genes within a given family that were correctly annotated.
Expression Analysis of r-Protein Genes
Expression levels were estimated based on the number of ESTs in contigs, constructed as described by Cooke et al. (1997), corresponding to individual r-protein genes. Expression analysis of r-protein genes lacking a corresponding EST was examined using PCR or RT-PCR, with gene-specific primers. PCR analysis was performed on an Arabidopsis cDNA library (Newman et al., 1994). RT-PCR was performed on RNA extracted from 3-week-old Arabidopsis ecotype Col 0 plants. Total RNA extraction was performed as previously described (Raynal et al., 1999). Amplification products were resolved on agarose gels and visualized by staining with ethidium bromide. Specific primers for Arabidopsis r-protein genes were designed using regions presenting a sequence polymorphism. Primer sequences are available on request.
ACKNOWLEDGMENTS
We thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the gift of ESTs, and Mike Bryant (Department of Biology, University of California, Riverside) for his expert assistance with the statistical analysis. We gratefully acknowledge all our colleagues from the AGI consortium for their immediate release of sequence data. Without this policy, such work would not have been possible.
Footnotes
This work was supported by the European Union EudicotMap program (contract no. BIO 4CT 97–2170); by the Centre National de la Recherche Scientifique and the French Ministry of National Education, Research, and Technology (grants to M.D.); and by the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program (grant no. 00–35301–9108 to J.B.-S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010265.
LITERATURE CITED
- Adams MD, Kerlavage RD, Fleischmann RA, Fuldner CJ, Bult NH, Lee EF, Kirkness KG, Weinstock JD, Gocayne O, White Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–17. [PubMed] [Google Scholar]
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J. Cytoplasmic ribosomes of higher plants. In: Bailey-Serres J, Gallie D, editors. A Look Beyond Transcriptionii: Mechanisms Determining mRNA Stability and Translation in Plants. Rockville, MD: American Society of Plant Physiologists; 1998. pp. 125–144. [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RT, Board PG. The human ubiquitin/52-residue ribosomal protein fusion gene subfamily (UbA52) is composed primarily of processed pseudogenes. Genomics. 1992;14:520–522. doi: 10.1016/s0888-7543(05)80258-7. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Capel M, Moore PB, Steitz TA. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–846. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Beltran-Pena E, Ortiz-Lopez A, Sanchez de Jimenez E. Synthesis of ribosomal proteins from stored mRNAs early in seed germination. Plant Mol Biol. 1995;28:327–336. doi: 10.1007/BF00020251. [DOI] [PubMed] [Google Scholar]
- Bevan M, Ecker J, Theologis S, Federspiel N, Davis R, McCombie D, Martienssen R, Chen E, Waterston B, Wilson R. Objective: the complete sequence of a plant genome. Plant Cell. 1997;9:476–478. doi: 10.1105/tpc.9.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsis thaliana genome. Plant Cell. 2000;12:1095–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri CJ, Lafleuriel C, Macadre F, Varoquaux Y, Parmentier G, Picard M, Caboche M, Bouchez D. A YAC contig map of Arabidopsis thaliana chromosome 3. Plant J. 1998;14:633–642. doi: 10.1046/j.1365-313x.1998.00159.x. [DOI] [PubMed] [Google Scholar]
- Clemons WM, May JL, Jr, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- Cooke R, Raynal M, Laudié M, Delseny M. Identification of members of gene families in Arabidopsis thaliana by contig constructions from partial cDNA sequences: 106 genes encoding 50 cytoplasmic ribosomal proteins. Plant J. 1997;11:1127–1140. doi: 10.1046/j.1365-313x.1997.11051127.x. [DOI] [PubMed] [Google Scholar]
- Cooke R, Raynal M, Laudie M, Grellet F, Delseny M, Morris PC, Guerrier D, Giraudat J, Quigley F, Clabault G. Further progress towards a catalogue of all Arabidopsis genes: analysis of a set of 5000 non-redundant ESTs. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- Culver GM, Cate JH, Yusupova GZ, Yusupov MM, Noller HF. Identification of an RNA-protein bridge spanning the ribosomal subunit interface. Science. 1999;285:2133–2136. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Lejeune B, Dron M, Kreis M. The Arabidopsis SHAGGY-related protein kinase (ASK gene family: structure organization and evolution) Gene. 1998;212:249–257. doi: 10.1016/s0378-1119(98)00147-4. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Heuer S, Sauter M, Lorz H, Kranz E. Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet. 1999;261:416–427. doi: 10.1007/s004380050983. [DOI] [PubMed] [Google Scholar]
- Fennoy SL, Nong T, Bailey-Serres J. Transcriptional and post-transcriptional processes regulate gene expression in oxygen-deprived roots of maize. Plant J. 1998;15:727–735. doi: 10.1046/j.1365-313X.1998.00249.x. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G. S6 phosphorylation and signal transduction. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. pp. 695–717. [Google Scholar]
- Gantt JS, Key JL. Auxin-induced changes in the level of translatable ribosomal protein messenger ribonucleic acids in soybean hypocotyl. Biochemistry. 1983;22:4131–4139. [Google Scholar]
- Gantt JS, Key JL. Coordinate expression of ribosomal protein mRNAs following auxin treatment of soybean hypocotyls. J Biol Chem. 1985;260:6175–81. [PubMed] [Google Scholar]
- Garo J, Kim SR, Chung YY, Lee JM, An G. Developmental and environmental regulation of two ribosomal protein genes in tobacco. Plant Mol Biol. 1994;25:761–770. doi: 10.1007/BF00028872. [DOI] [PubMed] [Google Scholar]
- Graack HR, Wittmann-Liebold B. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem J. 1998;329:433–448. doi: 10.1042/bj3290433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Höfte H, Desprez T, Amselem J, Chiapello H, Caboche M, Moisan A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier D. An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–64. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- Joanin P, Gigot C, Phillips G. cDNA nucleotide sequence and expression of a maize cytoplasmic ribosomal protein S13 gene. Plant Mol Biol. 1993;21:701–704. doi: 10.1007/BF00014553. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Ashworth LK, Lennon G, Higa S, Tanaka T. High-resolution mapping of ribosomal protein genes to human chromosome 19. Genome Res. 1998b;5:229–233. doi: 10.1093/dnares/5.4.229. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC. A map of 75 human ribosomal protein genes. Genome Res. 1998a;8:509–523. doi: 10.1101/gr.8.5.509. [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL. A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J Biol Chem. 2000;275:32585–32591. doi: 10.1074/jbc.M003596200. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Hunsperger JP, Culley D, Rubenstein I, Siflow CD. The organization and expression of maize ribosomal protein gene family. Genes Dev. 1989;3:500–509. doi: 10.1101/gad.3.4.500. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean D. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Martinez P, Martin W, Cerff R. Structure, evolution and anaerobic regulation of a nuclear gene encoding cytosolic glyceraldehyde 3-phosphate dehydrogenase from maize. J Mol Biol. 1989;208:551–565. doi: 10.1016/0022-2836(89)90147-2. [DOI] [PubMed] [Google Scholar]
- Matheson AT, Auer J, Ramíerez C, Böck A. Structure and evolution of archaebacterial ribosomal proteins. In: Hill WE, Dahlberg A, Garrett RE, Moore PB, Schlessinger D, Warner, editors. , The Ribosome, Structure, Function and Evolution. Washington, DC: American Society of Microbiologists; 1990. pp. 617–633. [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian KD, Terryn N. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Hornstein E. Translational control of TOP mRNAs. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Press, eds; 2000. pp. 671–693. [Google Scholar]
- Newman T, Debruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Banghman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming R. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–40. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal M, Guilleminot J, Gueguen C, Cooke R, Delseny M, Gruber V. Structure, organization and expression of two closely related novel Lea (late-embryogenesis-abundant) genes in Arabidopsis thaliana. Plant Mol Biol. 1999;40:153–165. doi: 10.1023/a:1026403215270. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 1999;18:490–499. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J, Gallois P, Delseny M. Accumulation and nuclear targeting of BnC24 a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci. 2000;156:35–46. doi: 10.1016/s0168-9452(00)00229-6. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS User's Guide: Statistics, Version 6. Ed 4. Vol. 2. Cary, NC: SAS Institute Inc.; 1989. [Google Scholar]
- Sanchez de Jimenez E, Aguilar R, Dinkova T. S6 ribosomal protein phosphorylation and translation of stored mRNA in maize. Biochemie. 1999;79:187–194. doi: 10.1016/s0300-9084(97)83505-5. [DOI] [PubMed] [Google Scholar]
- Shama S, Meyuhas O. The translational cis-regulatory element of mammalian ribosomal protein mRNAs is recognized by the plant translational apparatus. Eur J Biochem. 1996;236:383–388. doi: 10.1111/j.1432-1033.1996.00383.x. [DOI] [PubMed] [Google Scholar]
- Szick K, Springer M, Bailey-Serres J. Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes. Proc Natl Acad Sci USA. 1998;95:2378–2383. doi: 10.1073/pnas.95.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szick-Miranda K, Bailey-Serres J. Regulated heterogeneity in 12-kDa P-protein phosphorylation and composition of ribosomes in maize (Zea mays L.) J Biol Chem. 2001;276:10921–10928. doi: 10.1074/jbc.M011002200. [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F. A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70(s6k) function in vivo. Mol Cell Biol. 1998;18:2038–2044. doi: 10.1128/mcb.18.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuthey AL, Bittar G. Phylogenetic relationships of fungi Plantae and Animalia inferred from homologous comparison of ribosomal proteins. J Mol Evol. 1998;47:81–92. doi: 10.1007/pl00006365. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Wagner MM, Perry RP. Characterization of multigene family encoding the mouse S16 ribosomal protein: strategy for distinguishing an expressed gene from its processed pseudogene countreparts by an analysis of total genomic. DNA Mol Cell Biol. 1985;5:3560–3576. doi: 10.1128/mcb.5.12.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann LM, Perry RP. Characterization of expressed gene and several processed pseudogenes for the mouse ribosomal protein gene family. Mol Cell Biol. 1984;4:2518–2528. doi: 10.1128/mcb.4.11.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Sussex LM. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- Wittmann HG. Components of bacterial ribosomes. Ann Rev Biochem. 1982;51:155–183. doi: 10.1146/annurev.bi.51.070182.001103. [DOI] [PubMed] [Google Scholar]
- Wittmann-Leibold B, Kopke AKE, Arndt E, Kromer W, Hatakeyama T, Wittmann H-G. Sequence comparison and evolution of ribosomal proteins and their genes. In: Hill WE, Dahlberg A, Garrett RE, Moore PB, Schlessinger D, Warner JR, editors. The Ribosome, Structure, Function and Evolution. Washington, DC: American Society of Microbiologists; 1990. pp. 598–616. [Google Scholar]
- Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- Wu JW, Matsui E, Yamamoto K, Nagamura Y, Kurata N, Sasaki T, Minobe Y. Genomic organization of 57 ribosomal protein genes in rice (Oryza sativa) through RFLP mapping. Genome. 1995;38:1189–1200. doi: 10.1139/g95-157. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR. The plastid ribosomal proteins: identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast) J Biol Chem. 2000;275:28466–28482. doi: 10.1074/jbc.M005012200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR. The plastid ribosomal proteins: identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast) J Biol Chem. 2000;275:28455–28465. doi: 10.1074/jbc.M004350200. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Ross JL. Turner syndrome and haploinsufficiency. Curr Opin Genet Dev. 1998;8:322–327. doi: 10.1016/s0959-437x(98)80089-0. [DOI] [PubMed] [Google Scholar]