Abstract
We identified a recessive, brassinolide-insensitive mutant caused by a deletion allele (bri1-201) of the brassinosteroid (BR) receptor BRI1. The bri1-201 mutant displayed altered expression levels of genes differentially regulated by gibberellin (GA). RNA-blot analysis revealed that BR and GA antagonistically regulate the accumulation of mRNAs of the GA-responsive GASA1 gene, as well as the GA-repressible GA5 gene. Expression studies with cycloheximide indicated that the antagonistic effects of GA and BR on GA5 require de novo protein synthesis. Reporter transgene analyses and RNA-blot analysis showed that BR and GA modulate GA5 expression, at least in part, at the transcriptional level, and that the signals are independent and subtractive.
Brassinosteroids (BR) and gibberellins (GA) are plant growth regulators controlling cell and plant size, and mutations impairing their biosynthesis or sensitivity result in dwarfism. Early physiological work on wild-type (WT) tissues of different plants showed that GA and BR additively enhance growth, indicating that the two hormones act independently at the cellular level (Gregory and Mandava, 1982). However, recent molecular work indicates that cross-talk may occur between BR- and GA-signaling pathways. For example, mRNA of the GA-responsive γ-TIP gene accumulates ectopically in BR-deficient and BR-signaling mutants, suggesting that BR and GA antagonistically regulate γ-TIP expression (Kauschmann et al., 1996). γ-TIP encodes a tonoplast-intrinsic aquaporin or water channel, and its antagonistic regulation by BR and GA may reflect differences in the mechanisms by which the two hormones modulate cell growth and size by regulating turgor pressure or solute flow. In contrast, mRNA levels of the MERI-5 gene (Medford et al., 1991) are regulated positively by either BR or GA treatment (Kauschmann et al., 1996). MERI-5 probably encodes a xyloglucan-endohydrolase involved in cell wall loosening, thereby modulating cell expansion and growth. If so, MERI-5 expression may be required for growth processes mediated by both GA and BR.
Feedback control of the expression of BR and GA biosynthetic genes regulates endogenous levels of the growth hormones. For example, BR negatively controls transcription of the CPD steroidogenic cytochrome P450 (Mathur et al., 1998). In a similar manner, GA negatively regulates the expression of the GA4 3β-hydroxylase (Chiang et al., 1995) and the GA5 GA 20-oxidase-1 (Phillips et al., 1995; Xu et al., 1995), which encode enzymes converting inactive GA precursors into active GAs. If cross-talk occurs between GA and BR signaling, biosynthetic enzymes such as CPD, GA4, and GA5 may be potential regulatory targets. Such cross-talk could occur via shared signaling components, interactions between components specific for each pathway, or via control of the transcription or stability of common targets by distinct factors.
During the course of a phenotypic screen of gamma-mutagenized plants, we isolated a dwarf mutant similar to GA or BR biosynthetic/signaling mutants. Here, we demonstrate that this mutant phenotype is caused by a novel deletion allele (bri1-201) of the BRI1 BR receptor (Li and Chory, 1997; He et al., 2000). RNA-blot analysis with RNAs from WT, BR-insensitive bri1-201, and BR-deficient cpd mutants showed that BR and GA antagonistically regulate GASA1 and GA5. Expression studies with the protein synthesis inhibitor cycloheximide (CHX) indicated that GA5 regulation by BR or GA requires de novo protein synthesis. Transgene reporter analysis and RNA-blot analysis also indicated that GA5 regulation by BR and GA occurs, at least in part, at the transcriptional level.
RESULTS
The bri1-201 Mutant and Allele
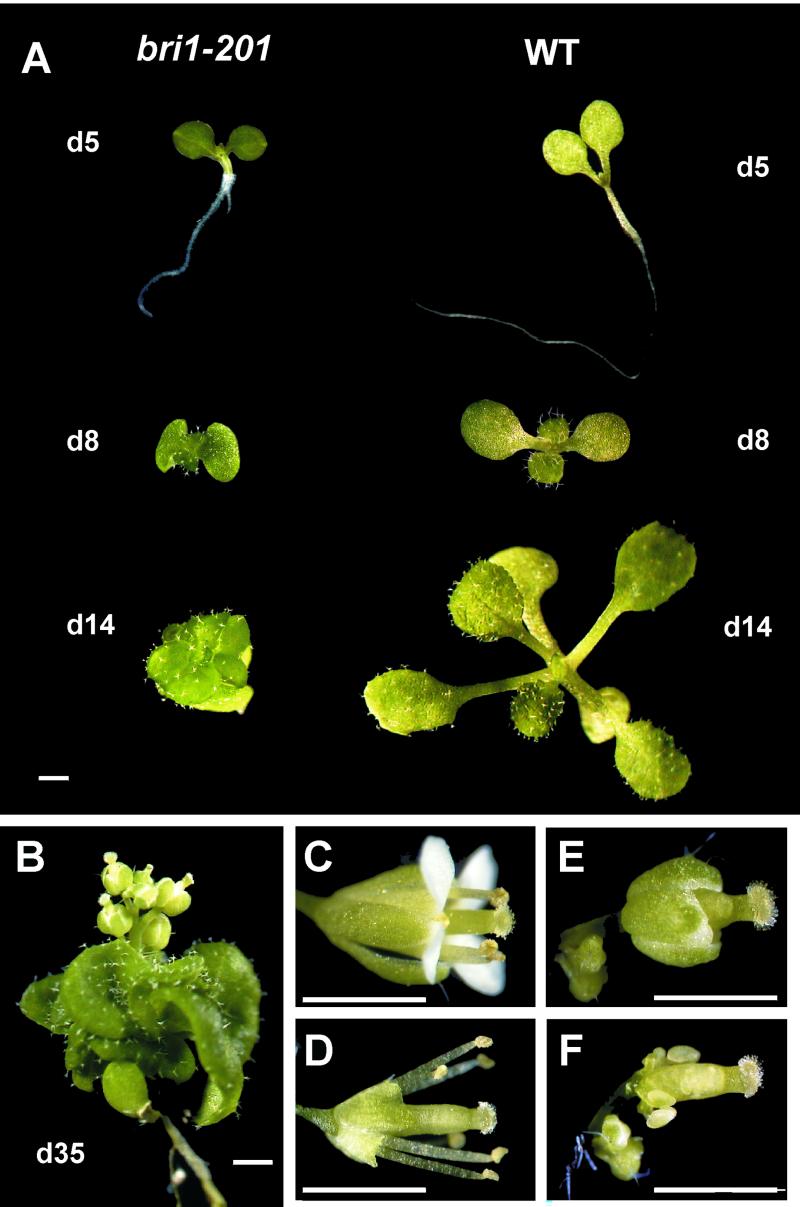
A screen of 100,000 progeny from gamma-mutagenized plants for visible mutant phenotypes identified the dwarf shown in Figure 1. This mutant exhibited reduced size at early stages of development, reduced apical dominance, extreme dwarfism at flowering, and delayed flowering and leaf senescence (Fig. 1, A and B). The mutant was apparently male sterile (Fig. 1, E and F) because homozygous seed was not produced by selfing, although pollination of the dwarf with WT pollen produced viable seed. F2 progeny from such crosses segregated in a 3:1 ratio (210 mutants out of 949 plants, χ2 = 0.04), indicating that the mutant phenotype was caused by a single recessive allele. The phenotype of the mutant suggested that it might be the result of a lesion in the biosynthesis or sensitivity to growth hormones such as BR or GA.
Figure 1.
Phenotype of the bri1-201 mutant under long-day conditions (16 h light/8 h dark). A, Comparative development of bri1-201 and WT. B, bri1-201 35 d after germination. C through F, WT (C and D) and bri1-201 flowers (E and F). In D and F, sepals and petals were removed to show reproductive organs. Bars represent 1 mm.
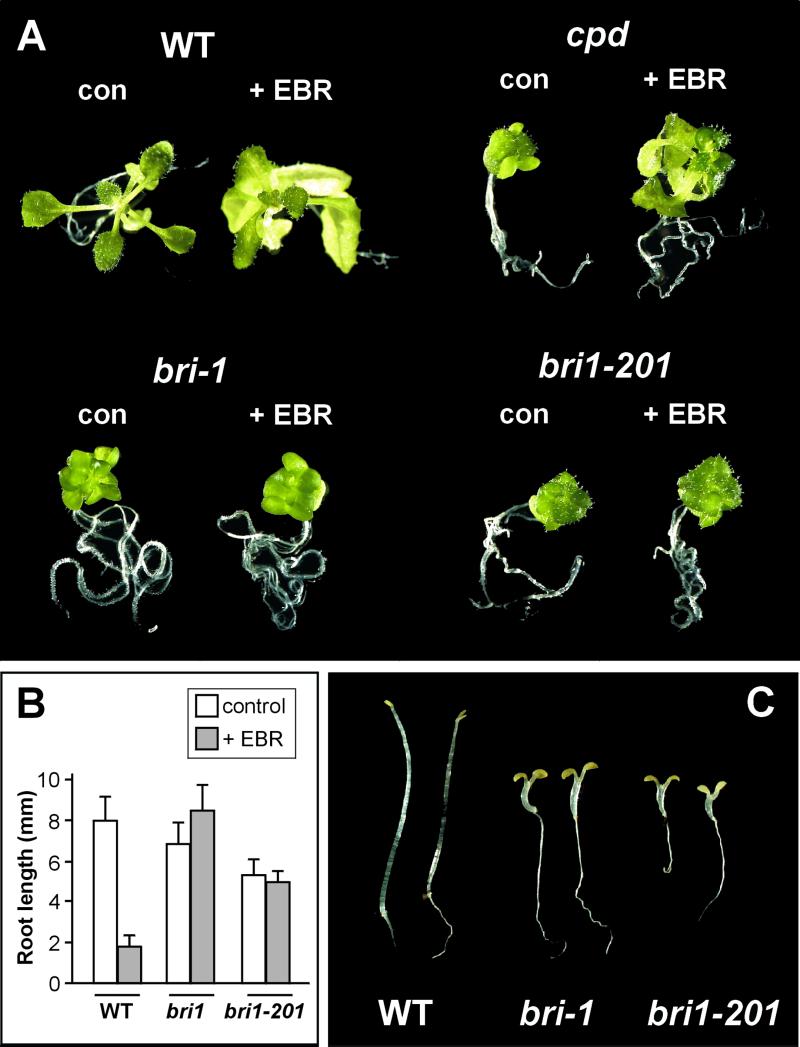
We used two approaches to elucidate the cause of the mutant phenotype: physiological and molecular studies of BR and GA responses and physical mapping. To investigate sensitivity to BR, the dwarf, as well as WT, BR-deficient cpd (Szekeres et al., 1996), and BR-insensitive bri1-1 (Clouse et al., 1996) controls were grown on Murashige and Skoog medium for 2 weeks and then transferred to plates supplemented with 1 μm 24-epibrassinolide (EBR). WT and cpd plantlets exhibited BR-sensitive growth, whereas the dwarf and bri1-1 failed to normalize leaf development (Fig. 2A). When grown on EBR concentrations inhibiting WT root elongation, both the dwarf and bri1-1 seedlings also maintained equivalent root growth (Fig. 2B). Moreover, dark-grown dwarf and bri1-1 seedlings were shorter than WT and both exhibited skotomorphogenesis, a characteristic of BR mutants (Fig. 2C). Taken together, these results indicate that the dwarf is BR insensitive.
Figure 2.
bri1-201 exhibits insensitivity to EBR and constitutive skotomorphogenesis. A, Two-week-old WT and bri1-201 were transferred to plates without (con) or with (+EBR) 0.5 μm EBR and grown for 5 d. B, Root length of WT, bri1-1 and bri1-201 seedlings grown 5 d without (control) or with (+EBR) 0.2 μm EBR. Each value represents the mean of 50 independent measurements. C, Skotomorphogenesis of WT and mutant seedlings. Seedlings were germinated and grown for 5 d in darkness on Murashige and Skoog medium.
The dwarf locus was mapped by scoring F2 mutant progeny, from a cross between plants heterozygous for the dwarfing allele (Columbia-0 [Col-0] ecotype) to WT Landsburg erecta, for segregation of using simple sequence-length polymorphism markers spanning the 10 Arabidopsis chromosome arms (Bell and Ecker, 1994). The mutation thus was localized to the lower arm of chromosome IV, south of marker nga1107 (three recombinants out of 60 mutant plants examined). Because BRI1 maps in this region (Li and Chory, 1997), this suggested that the dwarf might be caused by an allele of bri1. To test this, the BRI1 genes from three dwarf plants were PCR amplified and sequenced. An 8-bp deletion was found in all three mutants 22 nucleotides downstream of the start ATG, which produced a frame shift in the open reading frame resulting in a stop codon after 44 amino acid residues. Therefore, we designated this allele and the mutant bri1-201. RNA-blot analysis showed that WT and bri1-201 plantlets accumulated equivalent levels of mRNA hybridizing to a BRI1 3′-untranslated region probe (data not shown). This indicates that transcription and stability of BRI1 mRNA are not affected by BR levels or BR-signaling intermediates in WT Arabidopsis plants. Because bri1-201 lacks any BRI1 protein and is slightly more dwarfed than bri1 (Fig. 2, B and C), bri1-201 constitutes a null allele similar to the deletion mutant bri1-4, which exhibits a frame shift in the BRI1 open reading frame at 140 amino acid residues (Noguchi et al., 1999).
Expression of GA-Regulated Genes in bri1-201
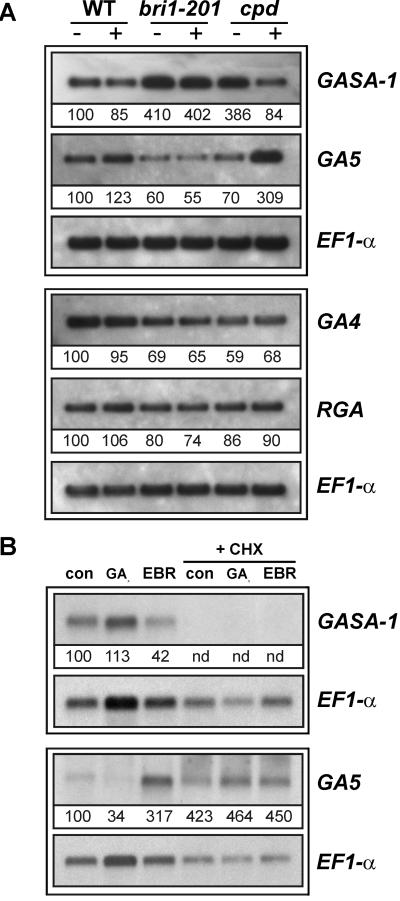
Our interest in plant responses to GAs (Raventos et al., 2000; Meier et al., 2001) initially prompted us to examine whether the mutant was sensitive to the application of exogenous GA prior to our identification of the bri1-201 allele. Growth assays with seedlings and rosette-stage plants showed that aerosol treatment with 10−5 m GA3 did not restore WT growth to the mutant, suggesting that it might be insensitive to GA. We attempted to confirm this by examining the expression of several GA-regulated genes in the mutant by RNA-blot analysis. These included the GA-responsive GASA1 gene (Herzog et al., 1995), the GA feedback-regulated, biosynthetic genes GA5 and GA4 (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995), and the RGA and GAI genes encoding GA-signaling components (Peng et al., 1997; Silverstone et al., 1998). This revealed that differential gene expression by GA was not affected in the mutant because accumulation of GASA1/GAI/RGA was enhanced by GA3, whereas accumulation of GA5/GA4 mRNAs was still negatively feedback regulated by GA3 (Fig. 3). This indicated that the mutant was sensitive to GA because it was not qualitatively compromised in responses to GA at the molecular level. Nonetheless, we noted that the control mRNA levels of GASA1 and GA5 were significantly different between the mutant and WT. For example, GA5 mRNA was three times less abundant in bri1-201 than in WT, whereas GASA1 mRNA was more than 7-fold higher in the mutant. Moreover, the levels of GA4, RGA and GAI mRNAs were slightly lower in bri1-201 than in WT.
Figure 3.
RNA-blot analysis of GA5, GA4, GASA1, GAI, and RGA mRNA accumulation in bri1-201 and WT upon GA3 treatment. Poly(A+) RNA (1 μg lane−1) from 16-d-old plants grown on Murashige and Skoog medium without or with 50 μm GA3 added for the last 48 h. Ribonucleic 32P-CTP antisense probes were synthesized using T7 RNA polymerase from partial cDNA 3′ sequences cloned in the pGEM-Teasy vector. GA5, GA4, and GASA1 hybridizations were performed on the same filter, whereas GAI and RGA hybridizations were performed on independent filters. Radioactive signals were quantified on all membranes and standardized (WT con = 100) by comparison to signals obtained after subsequent blotting with the EF1-α probe.
These results would be consistent with a lesion affecting the amplitude of GA-dependent responses controlling the expression of both GA inducible and repressible genes such as GASA1 and GA5. However, our subsequent finding that the mutant is BR insensitive indicated that BR affects the expression of these genes antagonistically to GA. This would explain why GA5 mRNA levels are lower, whereas GASA1 levels are higher in the bri1-201 mutant than in WT. In an alternate manner, the alteration in GASA1 and GA5 mRNA accumulation levels observed in bri1-201 could be due to pleiotropic effects of the mutation.
Effect of BR and GA on GA5 and GASA1 Steady-State mRNA Levels
To investigate whether altered GA-related gene expression in bri1-201 is correlated to deficiencies in BR signaling, EBR treatments were performed on WT, bri1-201, and the BR-deficient cpd mutant (Szekeres et al., 1996). mRNA levels were again monitored by EF1-α-normalized RNA-blot analysis. This showed that EBR treatment increased GA5 mRNA slightly in WT and more strongly in the BR-deficient cpd mutant, but did not affect GA5 mRNA levels in bri1-201 (Fig. 4A). In contrast, accumulation of the GA responsive GASA1 mRNA was reduced by EBR slightly in WT and very strongly in cpd. These results indicate that BR and GA antagonistically affect GA5 and GASA1 mRNA levels.
Figure 4.
GASA1 and GA5 mRNA accumulation is antagonistically controlled by GA and BR. Two-week-old seedlings grown on Murashige and Skoog were transferred to 50 mL liquid one-half-strength Murashige and Skoog in flasks for 1 d prior to treatment. RNA-blot analyses were performed and normalized with EF1-α as described in Figure 3. A, WT, bri1-201 and cpd seedlings were treated with 1 μm EBR for 48 h. B, cpd seedlings were treated with GA or BR in presence (CHX) or not of translational inhibitors. CHX (50 μm) and chloramphenicol (50 μm) were added to the medium 2 h before GA or BR treatments (50 μm GA3 or 1 μm EBR for 16 h).
Although GA4 and RGA mRNA levels were somewhat lower in bri1-201 and cpd than in WT, EBR treatment failed to restore their mRNA levels in the cpd mutant, in contrast to GA5 (Fig. 4A). This result indicates that BR effects on GA4 expression are less pronounced than BR control of GA5.
To assess whether de novo protein synthesis affects the accumulation of steady-state mRNA levels of GASA1 and GA5, GA and BR treatments were performed in the presence of translational inhibitors, and mRNA levels monitored by EF1-α-normalized RNA-blot analysis. As shown in Figure 4B (top, lanes 1 and 2 versus lanes 4 and 5), CHX blocked both basal levels of GASA1 mRNA accumulation in control tissue, as well as the enhancement of GASA1 mRNA accumulation by GA treatment. This suggests that GA-responsive GASA1 mRNA accumulation requires the de novo synthesis of an activator or a factor that stabilizes GASA1 mRNA. However, because CHX abolished and BR treatment decreased GASA1 mRNA accumulation, these experiments do not indicate whether BR repression of GASA1 mRNA accumulation requires de novo protein synthesis.
In contrast to GASA1, RNA-blot analysis showed that CHX alone induced GA5 mRNA accumulation to above-control levels, and that CHX blocked the repression of GA5 accumulation by GA (Fig. 4B; bottom, lane 2 versus lane 5). This is consistent with a model in which a labile repressor is required for GA negative feedback regulation of GA5 (Meier et al., 2001). However, BR induction apparently was blocked by CHX because GA5 mRNA levels were similar following treatment with CHX alone, or CHX together with GA or BR (Fig. 4B; bottom, lanes 4–6). These results do not distinguish between whether BR enhancement of GA5 mRNA accumulation is effected via a de novo-synthesized activator or an mRNA-stabilizing factor, or via derepression of GA negative feedback regulation.
BR Regulation of the GA5 Promoter
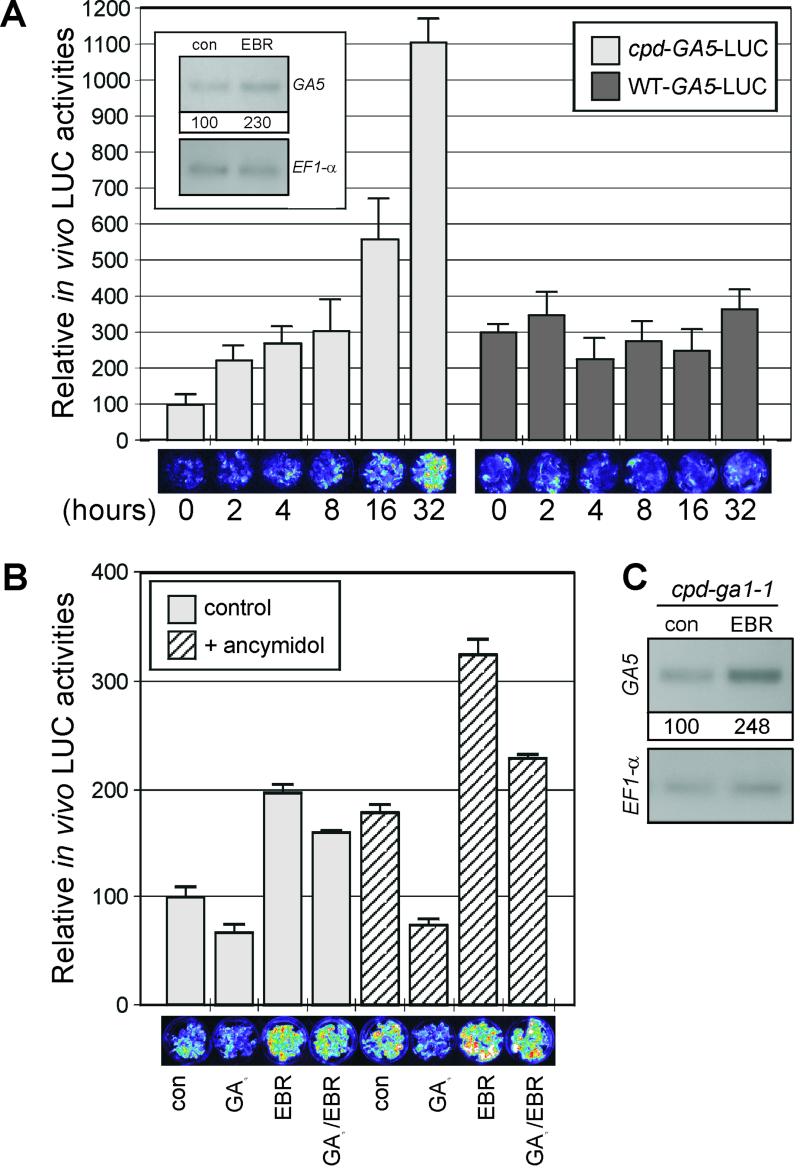
We previously have generated transgenic plants expressing the firefly luciferase (LUC) gene under the control of 0.4 kb of the GA5 promoter, and demonstrated that this promoter fragment contains cis sequences required for transcriptional regulation of GA5 by GA (Meier et al., 2001). This GA5-LUC reporter was introduced into the BR-deficient cpd mutant by crossing. F3 seedlings, homozygous for the GA5-LUC reporter and exhibiting either WT or cpd phenotypes, were treated with EBR and their in vivo LUC expression measured with a CCD camera. EBR treatment of WT plants carrying GA5-LUC did not significantly affect LUC reporter activity, although the control level of LUC activity was 3-fold higher in these WT plants than in the cpd plants expressing the reporter (Fig. 5A). This result is consistent with the low GA5 transcript accumulation level observed in bri1-201, and suggests that endogenous BR levels in the WT contribute to basal GA5 expression levels. In contrast, expression of the GA5-LUC reporter in cpd plants increased after 2 h of BR treatment and reached up to 11-fold induction after 32-h treatment (Fig. 5A). This indicates that the GA5 gene is, at least in part, transcriptionally regulated by BR, and in an opposite manner to GA. Accumulation of GA5 transcript in cpd seedlings was monitored by RNA-blot analysis after a short-term treayment (2 h) with EBR and confirmed the rapid regulation of GA5 transcript levels by BR observed in the reporter assays (Fig. 5A, inset).
Figure 5.
Effects of BR and GA on GA5-LUC expression. A, LUC imaging of the GA5-LUC reporter in 16-d-old WT-GA5-LUC and cpd-GA5-LUC seedlings treated with 1 μm EBR for various times. Insert in A, GA5 transcript accumulation monitored by RNA-blot analysis from 16-d-old cpd seedlings treated with 1 μm EBR for 2 h. Hybridization conditions and normalization with EF1-α were performed as described in Figure 3. B, One-week-old cpd-GA5-LUC seedlings grown on Murashige and Skoog plates were transferred to liquid one-half-strength Murashige and Skoog in presence or absence of ancymidol (1 mg L−1) and grown for 7 more d. BR (0.1 μm EBR) and GA (50 μm GA3) treatments were performed for 16 h by adding the hormones to the medium. C, GA5 transcript accumulation in 16-d-old cpd-ga1-1 double mutant seedlings treated with 1 μm EBR for 16 h. Hybridization conditions and normalization with EF1-α were performed as described in Figure 3. In A and B, bioluminescent LUC images (displayed in pseudocolors) acquired by CCD camera are shown below the quantification (average gray values) of LUC images.
If GA and BR antagonistically regulate GA5 transcription via independent pathways, the two signals may be subtractive. To investigate this possibility, cpd-GA5-LUC seedlings were subjected to GA3 and EBR treatments in the presence or absence of the GA biosynthesis inhibitor ancymidol (Rademacher, 1989). As expected, GA3 treatment down-regulated GA5-LUC activity, whereas EBR treament up-regulated reporter activity (Fig. 5B, bar 1 versus bars 2 and 3). In the presence of both hormones, LUC activity was reduced to the same extent in WT and WT treated with GA3 (Fig. 5B, bars 1 and 2 versus bars 3 and 4). As we have observed previously (Meier et al., 2001), LUC reporter activity was also up-regulated by ancymidol treatment (bar 1 versus bar 5). This indicates that the basal levels of active GAs in seedlings contribute to negative feedback of the GA5 gene. It is interesting that reporter activity was increased to the same extent upon EBR treatment of cpd seedlings grown in the presence or absence of ancymidol (Fig. 5B, bars 1 and 3 versus bars 5 and 7). Moreover, in the presence of ancymidol, GA3 reduced reporter activity to the same extent in plants treated with (Fig. 5B, bar 7 versus bar 8) or without (bar 5 versus bar 6) EBR.
To assess the specificity of the ancymidol treatment on the inhibition of GA biosynthesis, and to rule out the possibility of side effects on other hormone biosynthetic pathways (i.e. BR), we generated cpd-ga1-1 double mutants in which both active GA and BR levels are dramatically reduced. Accumulation of GA5 mRNA upon EBR treatment in the cpd-ga1-1 double mutant was investigated by means of RNA-blot analysis. As shown in Figure 5C, EBR treatment induced GA5 mRNA accumulation in cpd-ga1-1 seedlings, indicating that BR-induced GA5 expression is GA independent. This result is consistent with the additivity of the EBR and ancymidol treatments on the GA5-LUC reporter activation observed in Figure 5B. Taken together, these results show that GA and BR modulate GA5 transcription in a subtractive manner.
DISCUSSION
Dwarfism is a commonly identified mutant phenotype and can result from lesions in phytohormone synthesis or sensitivity (Li and Chory, 1997; Peng et al., 1997; Johnson and Ecker, 1998), or in potentially pleiotropic stress signaling (Bowling et al., 1994). Therefore, we undertook two approaches to determine the lesion causing dwarfism in a mutant identified in a phenotypic screen of gamma-mutagenized seeds. First, map-based cloning and genomic sequencing identified the mutation as a novel deletion allele (bri1-201) of the BRI1 gene encoding the BRI1 BR receptor (Li and Chory, 1997; Friedrichsen et al., 2000; He et al., 2000). Second, physiological analyses showed that bri1-201 is insensitive to BR, as expected. In addition, and prior to our identification of the bri1-201 allele, our interest in GA action (Raventos et al., 2000; Meier et al., 2001) prompted us to examine GA responses at the molecular level in bri1-201. RNA-blot analysis revealed that mRNA levels of GA-inducible GASA1 (Herzog et al., 1995) were higher, whereas mRNA levels of GA-repressible GA5 (Phillips et al., 1995; Xu et al., 1995) were lower in bri1-201 compared with WT. These results indicated that although bri1-201 is qualitatively sensitive to GA, the expression of specific genes is antagonistically affected by BR and GA. This was confirmed by transgenic experiments showing that expression of a fusion between the GA5 promoter and the LUC reporter was antagonistically regulated by GA and BR in cpd seedlings. In addition, endogenous GA5 up-regulation was observed in cpd-ga1-1 double mutants upon EBR treatment, confirming that BR and GA effects on GA5 expression are independent and subtractive.
These results indicate that cross-talk exists between these two important growth hormones, and that GA and BR modulate the expression of GA5, at least in part, at the transcriptional level. A result of this cross-talk may be that BR potentiates GA action by positively affecting GA5, a key GA biosynthetic enzyme whose activity regulates active GA levels (Phillips et al., 1995; Xu et al., 1995). This is consistent with phenotypic studies of the BR-deficient dwf4 mutant showing that a fully active BR pathway is required for cell elongation as a response to GA (Azpiroz et al., 1998). Using reporter transgenes, we have observed that GA4 is expressed in the hypocotyl of young seedlings (data not shown), which is also the case for GA1 (Silverstone et al., 1997) and for GA5 (Meier et al., 2001). This suggests that active GAs are produced in the hypocotyl. These data are consistent with the results of (Ephritikhine et al., 1999) showing that in the BR-deficient sax1 mutant, GA-insensitive cell elongation is restricted to the hypocotyl and is reversible by BR application. Moreover, Goetz et al. (2000) have recently shown that the Lin6 gene, encoding an extracellular invertase responsible for phloem unloading, was specifically induced in the hypocotyl in response to BR treatment.
The antagonistic effects of BR and GA on GA5 transcription may be explained via interaction(s) between upstream signaling components, or transcription factor(s) binding to common or distinct GA5 promoter elements. Our data do not distinguish between these possibilities, although reporter analysis and RNA-blot analysis, performed either in the presence of ancymidol, which depletes endogenous GA levels, or in a double mutant impaired in GA and BR synthesis, show that the two hormones affect GA5 transcription in a subtractive manner. More detailed promoter analysis is required to determine whether cis elements responsive to GA or BR are separable. To this end, we are introducing deletion derivatives of GA5-LUC into the cpd mutant.
Our current CHX experiments do not determine whether GASA1 is a primary GA or BR response gene because the presence of translational inhibitors abolished its transcript levels. This does, however, suggest that labile factor(s) are required either for GASA1 transcription or RNA stability. The effect of BR on GASA1 expression appears similar to BR repression of transcription of the CPD steroid hydroxylase, whose expression also requires de novo protein synthesis (Mathur et al., 1998). We recently showed that GA induction of GASA1 occurs at the transcriptional level using a GASA1 promoter-LUC fusion (Raventos et al., 2000). However, it is unlikely that GASA1 regulation by GA and BR are mediated through a common cis element because GASA1 mRNA accumulation was repressed by BR, whereas BR failed to affect GASA1-LUC reporter activity in a transgenic cpd mutant line (data not shown). GASA1 regulation by BR therefore may occur via a silencer located elsewhere in the GASA1 gene, or at the posttranscriptional level. In a similar manner, posttranscriptional regulation of the BRU1 gene by BR has been reported (Zurek and Clouse, 1994).
In contrast to GASA1, GA5 mRNA levels were increased by CHX alone. In addition, mRNA levels seemed unaffected by GA or BR in the presence of CHX. This indicates that de novo protein synthesis is required for GA repression and BR induction of GA5 transcription or mRNA accumulation. Regardless of whether GA5 is a primary or late GA reponse gene, this is consistent with a simple model in which a labile repressor is required for GA negative feedback regulation of GA5 (Meier et al., 2001).
Both GA5-LUC activity and endogenous GA5 transcripts levels increased upon BR treatment, indicating that GA5 is transcriptionally regulated by this hormone. However, GA5 mRNA levels are only moderately increased by BR in the cpd mutant upon long-term EBR treatment, whereas GA5-LUC activity was induced up to 11-fold after 32 h (Fig. 5A). This suggests that GA5 mRNA levels are regulated both at the transcriptional and posttranscriptional levels, whereas the GA5-LUC transgene lacks the corresponding posttranscriptional control. We have observed a similar result with new alleles of the late-flowering fpa mutant (Koornneef et al., 1991) in which the GA5-LUC reporter is highly expressed, whereas GA5 transcripts levels are only slightly higher than in WT (Meier et al., 2001). In an alternate manner, it is possible that the GA5-LUC reporter used here lacks cis elements that negatively regulate GA5 transcriptional induction by BR. In addition, BR regulation of both GA5 and GASA1 was clearly visible in cpd, whereas it was only moderate in WT (Fig. 4A). Moreover, the GA5-LUC reporter lacked apparent BR regulation in WT seedlings, whereas a strong BR up-regulation was detected in cpd (Fig. 5A). In a similar manner, BR induction of the xyloglucan endotransglycosylase homologs MERI-5 and TCH4 was only visible in BR-deficient mutants (Kauschmann et al., 1996). Two complementary explanations may account for these differences. First, hormonal desensitization pathways necessary for homeostatic growth control may be less active in hormone deficient mutants than in WT. Second, tissue-specific hormonal effects may be masked in WT if both hormonal and other signals contribute to basal levels of target gene expression.
In conclusion, results presented here confirm and extend earlier reports of interactions between the important plant growth regulators GA and BR. Although certain of these interactions appear to be antagonistic, BR induction of GA5 indicates that BR potentiates GA activity, at least in certain tissues. Elucidation of both transcriptional and posttranscriptional mechanisms will be required to understand how BR and GA regulate common targets involved in the control of cell elongation and plant growth.
MATERIALS AND METHODS
Plant Material and Hormone Treatments
The T4 Arabidopsis ecotype Col-0 line carrying 400 bp of the GA5 promoter driving the expression of the FF-LUC reporter (GA5-LUC) is described elsewhere (Meier et al., 2001). This line is homozygous for a single locus carrying the kanamycin resistance marker and exhibits sufficient LUC activity for bioluminescence imaging. The GA5-LUC reporter was introduced by crossing into the T-DNA-tagged cpd mutant (a gift from Csaba. Koncz, Max-Planck Institut fur Zuchtungsforschung, Koln, Germany), followed by segregation analysis for kanamycin resistance and cpd dwarfism in the F3 population. Double mutants impaired in both GA and BR synthesis were generated by fertilizing the GA-treated ga1-1 mutant (Koornneef and van der Veen, 1980) with cpd pollen. F1 generation seedlings were selected for hygromycin resistance, which cosegregates with the cpd mutation. F2 generation seedlings homozygous for ga1-1 (i.e. required exogenous GA for flowering) and heterozygous for cpd (i.e. hygomycin resistant, not cpd phenotype) were amplified to produce the F3 population, from which double mutants (25%) were visually selected. Joanne Chory (The Salk Institute for Biological Studies, La Jolla, CA) provided bri1-1 seeds. Seeds were surface sterilized and germinated on Murashige and Skoog medium supplemented with 0.7% (w/v) agar and 1% (w/v) Suc. Unless specified, hormone treatments were performed on 2-week-old seedlings, which were transferred to liquid Murashige and Skoog medium 24 h prior to treatment. Exogenous application of GA (50 μm GA3, Sigma-Aldrich, Vallenbæk, Strand, Denmark) and BR (0.1–1 μm 24-EBR, Sigma) was performed by adding the hormones to the medium for various times as indicated.
Generation of Arabidopsis Mutants
Approximately 25,000 bulked seeds were γ-irradiatiated (60 kRad) from a cobalt60 source (RISØ Industrial Irradiation, Risø, Denmark). M1 seeds were bulked in pools of 1,250 seeds each and propagated in a long-day greenhouse. The bri1-201 mutant was visually selected from M2 progeny.
Reporter Assays
LUC bioluminescence imaging was performed as previously described (Raventos et al., 2000; Meier et al., 2001). In brief, bioluminescence was measured after spraying transgenic plants expressing either the GA5-LUC or the GA4-LUC reporters uniformly with 5 mm luciferin (JBL/Promega, Madison, WI) in 0.01% (w/v) Triton X-100. Plates were imaged three times for 5 min, and a bright-field reference image was taken thereafter. The first image was discarded due to chlorophyll phosphorescence, and the two remaining LUC images analyzed with the image-1/Metamorph system version 3.0 (Universal Imaging Corp., Downingtown, PA).
RNA Analysis
Total RNA was extracted from plant tissue using the RNAgent kit (Promega) and Poly-A RNAs isolated using the PolyA-tract kit IV (Promega). Poly-A RNA samples (1 μg) were size fractionated on 1.2% (w/v) agarose/formaldehyde gel and blotted onto Hybond N+ membranes (Amersham, Pharmacia Biotech, Horsholm, Denmark). Transcript levels of genes involved in GA and BR biosynthesis (GA5, GA4, and CPD), genes encoding GA-signaling pathway components (RGA and GAI), as well as GA-regulated genes (GASA-1) were investigated. Ribonucleic 32P-CTP antisense probes were synthesized using T7 RNA polymerase (Ribokit, Promega) from partial cDNA 3′ sequences cloned in the pGEM-Teasy vector (Promega) using the following primer combinations: GA5, aaggcctttgtggtcaatatcggc and gagatgctaaaaggtgttattgcc; GA4, ggtccgaaggtttcaccatcac and gagctttgttgaagtgagttgc; RGA, tggttcgtccggtttagcgccg and cagttcggtttaggtcttggtcc; GAI, cgggtctgctgggtttgcgg and tagtttggcttcggtcggaaatc; and GASA-1, ctctccttggagaatcatggct and acactcacaacgacaacgtacg. Hybridization and washing conditions were performed as recommended by the manufacturer. As a control, radioactive signals were quantified on all membranes by comparison to signals obtained after subsequent blotting with an elongation factor alpha probe (EF1-α, Axelos et al., 1989), whose transcript levels appeared unaffected by either GA3 or EBR treatment (not shown).
Mapping of the Dwarf Mutant
Plants heterozygous for the bri1-201 allele conferring dwarfism (Col-0 ecotype) were crossed to the Landsburg erecta ecotype. Mapping of the mutation was performed on F2 progeny exhibiting the dwarf phenotype using simple sequence-length polymorphism markers (Bell and Ecker, 1994). Sequencing of the BRI gene from the bri1-201 mutant was performed with an ABI prism sequencer (Perkin Elmer, Nærum, Denmark).
ACKNOWLEDGMENTS
We are grateful to Joanne Chory and Csaba Koncz for supplying bri1-1 and cpd seeds, Suksawad Vongvisuttikun for excellent technical assistance, and Henrik Næsted for stimulating discussions.
Footnotes
This research was funded by the European Union (grant nos. CT96–0062 and CT96–0621 to J.M.) and by a Danish research grant (no. 93524444A98000040 to R.F.). T.B. was funded by a postdoctoral Marie Curie Research training grant.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010173.
LITERATURE CITED
- Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1 alpha: molecular cloning, characterization and expression. Mol Gen Genet. 1989;219:106–112. doi: 10.1007/BF00261164. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. ; erratum Chiang HH, Hwang I, Goodman HM (1997) Plant Cell 9: 979–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Roitsch T. Tissue-specific induction of the mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role for steroid hormones in assimilate partitioning. Plant J. 2000;22:515–522. doi: 10.1046/j.1365-313x.2000.00766.x. [DOI] [PubMed] [Google Scholar]
- Gregory LE, Mandava N. The activity and interaction of brassinolide and gibberellic acid in mung bean (Phaseolus aureus) epicotyls. Physiol Plant. 1982;54:239–243. [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- Herzog M, Dorne AM, Grellet F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol. 1995;27:743–752. doi: 10.1007/BF00020227. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Medford JI, Elmer JS, Klee HJ. Molecular cloning and characterization of genes expressed in shoot apical meristems. Plant Cell. 1991;3:359–370. doi: 10.1105/tpc.3.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C, Bouquin T, Nielsen ME, Raventos D, Mattsson O, Rocher A, Schomburg F, Amasino R, Mundy J. Gibberellin response mutants identified by luciferease imaging. Plant J. 2001;25:509–519. doi: 10.1046/j.1365-313x.2001.00980.x. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher M. Gibberellins: metabolic pathways and inhibitors of biosynthesis. In: Sandman B, editor. Target Sites of Herbicide Action. Boca Raton, FL: CRC Press, Inc.; 1989. pp. 127–145. [Google Scholar]
- Raventos D, Meier C, Jensen AB, Mattsson O, Mundy J. Fusion genetic analysis of gibberellin signaling mutants. Plant J. 2000;22:427–438. doi: 10.1046/j.1365-313x.2000.00759.x. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Chang C, Krol E, Sun TP. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997;12:9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulatorrepressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L) epicotyls. Plant Physiol. 1994;104:161–170. doi: 10.1104/pp.104.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]