Abstract
The sensing of Gram-negative Extracellular Vesicles (EVs) by the innate immune system has been extensively studied in the past decade. In contrast, recognition of Gram-positive EVs by innate immune cells remains poorly understood. Comparative genome-wide transcriptional analysis in human monocytes uncovered that S. pyogenes EVs induce proinflammatory signatures that are markedly distinct from those of their parental cells. Among the 209 genes exclusively upregulated by EVs, caspase-5 prompted us to study inflammasome signaling pathways in depth. We show that lipoteichoic acid (LTA), a structural component of Gram-positive bacterial membranes present on EVs from S. pyogenes and other Gram-positive species, is sensed by TLR2 which triggers the alternative inflammasome composed of NLRP3 and the inflammatory caspases-4/-5 to mount an IL-1β response without inducing cell death. For S. pyogenes, we identify TLR8 as a sensor to mediate caspase-4/-5-dependent IL-1β secretion. Notably, inflammasome activation by intact bacteria is independent of the global virulence regulator CovS in monocytes. Overall, our study highlights a new role for TLR2 and caspase-4/-5 in the recognition of Gram-positive EVs in human monocytes.
Keywords: Streptococcus pyogenes, Monocytes, Extracellular Vesicles, Caspases, Inflammasome
Subject terms: Immunology; Membranes & Trafficking; Microbiology, Virology & Host Pathogen Interaction
Synopsis

The recognition of Gram-positive EVs by innate immune cells remains poorly understood. This study identifies alternative inflammasome activation by S. pyogenes and Gram-positive EVs in human monocytes and highlights a new role for TLR2 and caspase-4/-5 in this process.
Lipoteichoic acid (LTA) as a constituent of Gram-positive EVs activates the alternative inflammasome via TLR2.
IL-1β secretion is regulated by MYD88, RIPK1, CASP8, and CASP4/5.
EVs promote monocyte survival by inhibiting caspase-3 activation.
The recognition of Gram-positive EVs by innate immune cells remains poorly understood. This study identifies alternative inflammasome activation by S. pyogenes and Gram-positive EVs in human monocytes and highlights a new role for TLR2 and caspase-4/-5 in this process.

Introduction
Streptococcus pyogenes (S. pyogenes) is a strict human pathogen that causes superficial infections (e.g., impetigo) as well as life-threatening invasive disease (e.g., Streptococcal toxic shock syndrome), with a global burden of at least 500,000 deaths annually (Carapetis et al, 2005). S. pyogenes expresses a wide array of virulence components that facilitate its survival within the host by hijacking the activity of immune cells (Graham et al, 2002; Tsatsaronis et al, 2014; Hynes and Sloan, 2016). Factors associated with bacterial virulence can be classified according to their location: membrane-bound, membrane-anchored, or cytosolic. Interestingly, several virulence mediators present in S. pyogenes extracellular vesicles (Spy EVs) do not possess a secretion signal peptide and are therefore not secreted through the canonical bacterial secretory pathway (Sec) (Lei et al, 2000; Resch et al, 2016). Since no alternative secretion mechanism has been identified in S. pyogenes, it is likely that EVs contribute to the release of components that cannot be secreted by the Sec pathway (Resch et al, 2016).
EVs are nanoparticles consisting of a lipid bilayer shell encapsulating proteins and nucleic acids derived from their parental cells. The release of EVs is conserved across all domains of life to coordinate intra- and interspecies communication via cellular components (Deatheragea et al, 2012; Yoon et al, 2014; Woith et al, 2019). In Gram-negative bacteria, the composition, biogenesis, and functions of outer membrane vesicles (OMVs) in bacterial physiology have been extensively characterized (Schwechheimer and Kuehn, 2015; Orench-Rivera and Kuehn, 2016). Numerous reports have addressed the intricate communication process that occurs between the host and OMVs of pathogens or microbiota species (Kaparakis-Liaskos and Ferrero, 2015; Ñahui Palomino et al, 2021). Similarly, over the last few decades, numerous studies have demonstrated that Gram-positive model organisms and pathogens also release EVs (Lee et al, 2009; Rivera et al, 2010; Brown et al, 2015). However, only a few of these studies have addressed the impact of Gram-positive EVs on the host (Gurung et al, 2011; Olaya-Abril et al, 2014; Wang et al, 2020).
Previously, our laboratory and others have shown that Spy EVs encapsulate virulence factors, highlighting their potential role in modulating the host-pathogen interface (Biagini et al, 2015; Resch et al, 2016; Uhlmann et al, 2016; Murase et al, 2021). Several groups have reported that enrichment and exclusion of bacterial components in EVs, a process termed cargo selectivity, commonly occurs (Haurat et al, 2011; Elhenawy et al, 2014; Veith et al, 2014). For example, Spy EVs are enriched in the Streptococcal inhibitor of the complement (Sic) and Streptolysin O (SLO) compared to their parental cells (Resch et al, 2016), which are components that have been associated with pathogenicity and virulence (Harder et al, 2009; Pence et al, 2010; Nasser et al, 2014). Due to their small size and resistance to degradation, bacterial EVs are able to diffuse and travel long distances in the human body, reaching locations that their parental cells might not access (Tulkens et al, 2020). In addition, microbial EVs can be internalized and/or recognized through pathogen recognition receptors (PRRs), which may differ from the PRRs participating in the recognition of their parental cells (Vanaja et al, 2016).
The delivery of Gram-negative bacterial components including lipopolysaccharide (LPS) to the cytosol of host cells via OMVs has been shown to promote canonical as well as non-canonical inflammasome assembly, leading to caspase-1-dependent cleavage and subsequent release of the proinflammatory cytokines IL-1α, IL-1β, and IL-18 as well as the induction of pyroptotic cell death (Cecil et al, 2017; Finethy et al, 2017; Yang et al, 2020). Activation of this large multiprotein complex requires existing or de-novo synthetized inflammasome protein components and their signal-induced oligomerization: PRR-engagement on the cell surface stimulates NFκB-dependent gene expression of inactive precursor molecules for the cytokines (priming). Subsequently, the presence of bacterial products within the cytosol is sensed by various NOD-like receptors (NLRs) triggering oligomerization and recruitment of caspase-1 (activation). However, unlike macrophages, LPS has been shown to trigger an alternative one-step inflammasome activation pathway in human monocytes, which is dependent on TLR4 signaling and does not require LPS internalization or priming (Netea et al, 2009; Gaidt et al, 2016; Gritsenko et al, 2020). In addition to TLR4, a more recent study implicated multiple TLRs to activate the alternative inflammasome in human monocytes leading to IL-1β secretion, indicating that pathogen-associated molecular patterns (PAMPs) other than LPS can be recognized by this pathway (Unterberger et al, 2023). However, to date, no publication reports activation of the alternative inflammasome in response to live, Gram-positive bacteria or their secreted EVs.
In the present study, we have comparatively examined the response of primary human monocytes to Spy EVs and their parental cells. Interestingly, although both stimuli induce a significant overlap in the monocyte’s response, we identified specific signatures triggered by either stimulus. We provide evidence, that Spy EVs are sensed by TLR2 on monocytes which triggers the alternative inflammasome comprised of NLRP3 and the inflammatory caspases-4 and -5, leading to the release of IL-1β without inducing cell death. We identify lipoteichoic acid (LTA), a structural component of Gram-positive bacterial membranes, present on EVs but also in EV-depleted culture supernatant, as the ligand for TLR2. Accordingly, EVs from other Gram-positive species stimulate caspase-4/-5-dependent IL-1β secretion, underlining the conservation of this mechanism. In contrast, inflammasome activation in response to infection with S. pyogenes requires TLR8 and is characterized by ASC speck formation and GSDMD-dependent pyroptotic cell death, indicating that distinct innate immune pathways are involved in IL-1β production in response to S. pyogenes or its EVs.
Results
Transcriptome analysis reveals a unique set of genes that are upregulated upon encounter with Spy EVs
Our laboratory has previously characterized the content of Spy EVs derived from the hypervirulent clinical isolate ISS3348 by proteomic, RNA sequencing, and lipidomic analysis (Resch et al, 2016). Spy EVs and their parental cells show an asymmetrical distribution in their composition (e.g., of virulence factors) (Resch et al, 2016), supporting the idea that EVs might trigger distinct responses than S. pyogenes cells. To characterize and compare the response of innate immune cells when encountering Spy EVs or intact bacteria, we performed RNA sequencing on human primary monocytes (Dataset EV1) and B-cell Leukemia C/EBPα Estrogen Receptor clone 1 (BLaER1)-derived monocytes (Dataset EV2) after incubation with Spy EVs or infection with S. pyogenes. Spy EV purification and physiochemical characterization is summarized in Fig. EV1A,B. EV-quantification on the basis of size and concentration was initially performed using Nanoparticle tracking analysis (NTA) (Fig. EV1C) and confirmed by Tunable Resistive Pulse Sensing (TRPS), both methodologies proven to be more sensitive as compared to protein-content based quantification. To rule out any potential contamination of the Spy EVs with LPS, vesicle preparations were tested using the Pro-Q™ Emerald 300 lipopolysaccharide gel stain (Fig. EV1D) and mass spectrometry (Appendix Fig. S1). We did not detect any LPS in our EV preparations. Transmission electron microscopy (TEM) was used to visualize the EVs (Fig. EV1E) as previously described (Resch et al, 2016). While there are no published reports showing quantitative data on the number of EVs released by a bacterium during an in vivo infection, we found that the ratio between S. pyogenes and its EVs in THB culture is ~20 (17.7) EVs per bacterial cell. To assess a suitable dose for Spy EVs, we challenged human monocytes with increasing particle-concentrations of EVs and measured IL-1β and IL-6 in cell culture supernatants at 18 h post-stimulation (Fig. EV1F). We observed an EV-particle-dose-dependent release of both cytokines, however, secretion of IL-1β was considerably lower than that of IL-6, which explains why a dose of 7 × 108 EVs (corresponding to 7000 EVs/cell) was defined for future experiments.
Figure EV1. Purification, quantification, and characterization of bacterial EVs used in this study.
(A) Schematic protocol of the purification strategy used for EVs. (B) Mean size (left) and concentration (right) of Spy EVs (n = 15) characterized by nanoparticle tracker analysis (NTA). (C) Linear correlation between particles/mL (measured by NTA) and the amount of protein of the same batches (Bradford assay, n = 15). (D) LPS analysis of EV preparations separated by SDS PAGE followed by Pro-Q Emerald 300 staining. Smooth LPS standard from E. coli serotype O55:B5 with characteristic ladder pattern was used as positive control. (E) Spy EV preparation imaged by transmission electron microscopy (TEM, scale bar: 1 µm). (F) IL-1β and IL-6 released by human monocytes stimulated with increasing amounts of Spy EVs for 18 h. Bars represent the mean ± SD of six biological replicates. P < 0.0001 (for all comparisons). Data information: (F) One-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. ***P ≤ 0.001, n.s. not significant.
To obtain an unbiased view on the early (4 h) transcriptional response of monocytes to Spy EVs or infections with S. pyogenes, we performed RNA sequencing as outlined in Fig. 1A. Principle component analysis (PCA) showed that samples were clearly separated and clustered by treatment, indicating that donor variability had minor impact in our experimental setting (Fig. 1B).
Figure 1. S. pyogenes EVs induce a distinct expression profile in monocytes.
(A) Schematic summary of the study design. Primary monocytes were purified through density gradient followed by magnetic negative selection. Four hours after S. pyogenes infection or challenge with its EVs, RNA from monocytes was purified and processed for RNA sequencing. (B) Principal component analysis of gene transcription colored by treatment. (C) Scatterplot comparison of Log2 fold change (FC) (treatment vs untreated control, Unt) in transcript abundance for S. pyogenes-infected (x axis) or EV-treated samples (y axis). Dashed lines mark cut-off criteria of a log2 FC > |2|. Grey dots represent transcripts commonly regulated but with an FC < |2|. Blue dots denote commonly regulated transcripts with a FC ≥ |2|. Orange and purple dots indicate differentially expressed genes for either EVs or S. pyogenes, respectively. Dashed areas indicate four distinct gene groups which meet our cut-off criteria that share (2 and 4) or display unique (1 and 3) transcription patterns for both or either stimulus, respectively. Representative genes in each group are highlighted. Wald test was applied for statistical analyses. (D) Venn diagrams summarizing unique and overlapping up- or downregulated genes with FC ≥ |2|. The top three genes for each category are displayed. (E) Heatmap of twenty representative genes from each gene group in (D). Data information: (B–E) Data are representative of eight biological replicates. (C–E) Graphs include gene transcription after 4 h upon EV or S. pyogenes stimulation compared to untreated monocytes. Genes were defined as differentially regulated when FC ≥ |2| as compared to untreated cells and the adjusted P value was <0.01 after Benjamini-Hochberg correction for multiple comparisons.
Our transcriptomic datasets showed that 1107 and 445 genes are upregulated in response to both Spy EVs and bacterial cells in monocytes and BLaER1 cells, respectively (Figs. 1C,D and EV2A,B). Since primary monocytes respond more strongly than BLaER cells, we focused on our monocyte dataset for further analyses. Accordingly, pathway analysis of primary monocytes revealed that KEGG pathways and GO terms enriched for each treatment show a high similarity (Fig. EV2C,D). Among the commonly upregulated genes (Fig. 1C–E, group 2), we found cytokines (e.g., IL1A, IL1B and IL6), chemokines (e.g., CCL3, CCL4 and CXCL8) and growth factors (e.g., CSF3). In contrast, several immune receptors (e.g., TLR1, TLR6 and CCR2) and adhesion factors (e.g., PECAM1 and VCAN) were downregulated for both stimuli (Fig. 1C–E, group 4). We then analyzed the differentially expressed genes (DEGs) belonging specifically to one of the treatments. DEGs upon challenge with Spy EVs included the metalloreductase STEAP4, components of the Wnt signaling pathway (WNT5A), and, strikingly, the immune sensor CASP5 (Fig. 1C–E, group 1). Infection with S. pyogenes specifically upregulated an additional 1107 genes (Fig. 1C–E, group 3), including interferon and interferon-related genes (IRGs, e.g., IFNG, IFNB1 and CXCL10), growth factors (e.g., CSF1), and chemokines (e.g., CCL8), indicating that the sensing of whole bacteria induces responses remarkably distinct from those of EVs. Real-time quantitative PCR (RTqPCR) was used to validate representative candidates from each gene group (Fig. EV2E–H).
Figure EV2. RNA sequencing of monocytes: pathway analysis and RTqPCR of specific genes.
(A) Scatterplot comparison of Log2 fold change (FC) (treatment vs untreated control, Unt) in transcript abundance for S. pyogenes-infected (x-axis) or EV-treated samples (y-axis) in BLaER1 cells (related to Fig. 1). Grey dots represent transcripts commonly regulated but with an FC < | 2 | . Blue dots denote commonly regulated transcripts with a FC ≥ | 2 | . Orange and purple dots indicate differentially expressed genes for either Spy EVs or S. pyogenes, respectively. (B) Venn diagrams of differentially transcribed genes in BLaER1 cells after stimulation with Spy EVs or S. pyogenes for 4 h. Numbers indicate the total amount of genes for each category. (C, D) Top ten upregulated KEGG pathways (C) or GO terms (D) are shown for Spy EV-treated versus untreated monocytes and S. pyogenes-treated versus untreated monocytes. Solid lines indicate same pathway and dashed lines indicate a related pathway. (E–H) Quantitative real time PCR of selected genes. Floating bar plots displaying the negative ΔCt values of three biological replicates. The central line represents the mean, the borders represent the minimum and the maximum values. A dotted line indicates the mean expression of the housekeeping genes (GAPDH/TUBB). (E) Genes upregulated by Spy EV treatment. (WNT5A) P = 0.0029, P = 0.0004. (CASP5) P = 0.0028. (F) Genes upregulated by S. pyogenes and Spy EV treatments. (IL1A) P = 0.0003 (for both comparisons). (CCL4) P = 0.0014 (for both comparisons). (G) Genes upregulated by S. pyogenes treatment. (IFNG) P = 0.0025. (IFNB) P = 0.0308. (H) Genes downregulated for both S. pyogenes and Spy EV treatments. (CCR2) P = 0.0055 (Spy), P = 0.0010 (EVs). (PTGRFN) P = 0.0130 (for both comparisons). Data information: (E–H) One-way ANOVA with Holm–Šídák correction for multiple comparisons was applied for statistical analyses. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant.
Spy EVs induce a proinflammatory response
During S. pyogenes infection, inflammation facilitates bacterial clearance by innate immune cells but may also promote a potentially detrimental immune response. In mouse models, this balance is orchestrated by IL-1β and type I interferon signaling, which promote and repress inflammation, respectively (Castiglia et al, 2016). Accordingly, IL-1β inhibitors used to treat patients with autoimmune diseases increase the risk of developing invasive S. pyogenes infections by 330-fold, suggesting a central role for this cytokine in controlling S. pyogenes pathogenesis in humans (LaRock et al, 2016). Interestingly, while IL-1β and other members of the IL-1 cytokine family, such as IL-1α and IL-36γ, are upregulated by both S. pyogenes and its EVs, we observed expression of IFNβ, IFNγ, and IRGs (e.g., CXCL9, CXCL10, CXCL11) predominantly during bacterial infection (Dataset EV1; Figs. 1C,E and EV4G,H). Although RNA sequencing detected both IFNβ and -γ expression also in response to Spy EVs, they were only classified among the lowest 5% or 10% of the DEGs. In contrast, infection with S. pyogenes triggered a significantly higher expression of both cytokines, with IFNβ ranked among the lowest 50% and IFNγ among the highest 50% of the DEGs, indicating that only bacterial cells induce a robust IFN response in human monocytes.
Figure EV4. Cytokine controls and cell death of monocytes and BLaER1 cells: canonical inflammasome.
(A) IL-6 in supernatants from human monocytes either left untreated or preincubated with Ac-LEVD-CHO, MCC950, VX-765, or Ac-YVAD-CMK. Cells were then left unstimulated or challenged with S. pyogenes or its EVs for 18 h. Bars represent the mean ± SD of four biological replicates. P = 0.0011. (B, C) IL-6 released from BLaER1 WT, PYCARD KO (ASC−/−), NLRP3 KO clones 1 and 2 (NLRP3−/−), Caspase-1 KO clones 1 and 2 (CASP1−/−), and non-targeted control (NTC) cells stimulated with Spy EVs or infected with S. pyogenes for 18 h. Bars represent the mean ± SD of three biological replicates. (D) Box plot showing caspase and NLRP protein levels differentially expressed in BLaER1 cells (5 biological replicates) and primary human monocytes (4 biological replicates). Each dot represents the protein level of an individual sample. The boxplot displays the distribution of the data with the box representing the interquartile range (IQR) between the 25th (Q1) and 75th (Q3) percentiles. The line inside the box indicates the median (50th percentile). Whiskers extend to the smallest and largest values within 1.5 × IQR below Q1 and above Q3, respectively. Data points outside this range are shown individually as outliers. In total, 6472 proteins were quantified in at least 3 biological replicates of BLaER1 cells, and 6092 proteins were quantified in primary monocytes. (E) Immunoblots displaying GSDMD and IL-1β in human monocytes either left untreated or challenged with Spy EVs or S. pyogenes for 18 h. Representative of four biological replicates. Data information: (A–C) Two-way ANOVA with Holm–Šídák correction for multiple comparisons was applied for statistical analyses. **P ≤ 0.01, n.s. not significant.
Caspase-4/-5 are required for Spy EV-induced IL-1β release
Inflammasomes in innate immune and epithelial cells play an important role in bacterial sensing (Martinon et al, 2002; Hayward et al, 2018). In monocytes, IL-1β is secreted upon activation of the canonical, non-canonical and/or alternative inflammasome (Martinon et al, 2002; Kayagaki et al, 2011; Shi et al, 2014; Gaidt et al, 2016). Notably, we observed that Spy EVs treatment induced expression of the non-canonical inflammasome component, caspase-5, compared with untreated cells (log2FC: 5.45, Dataset EV1; Figs. 1C,D and EV4E). Caspase-4 and -5 and their murine homolog, caspase-11, have been shown to recognize intracellular LPS during Gram-negative infection, and are also activated in response to the Gram-positive species Staphylococcus aureus (S. aureus) and Listeria monocytogenes (L. monocytogenes) (Shi et al, 2014; Casson et al, 2015; Vanaja et al, 2016; Hara et al, 2018; Krause et al, 2019). Upregulation of caspase-5 upon EV treatment in the absence of a robust interferon signature (Figs. 1 and EV4) prompted us to investigate whether IL-1β might be induced differently by Spy EVs than by their parental cells.
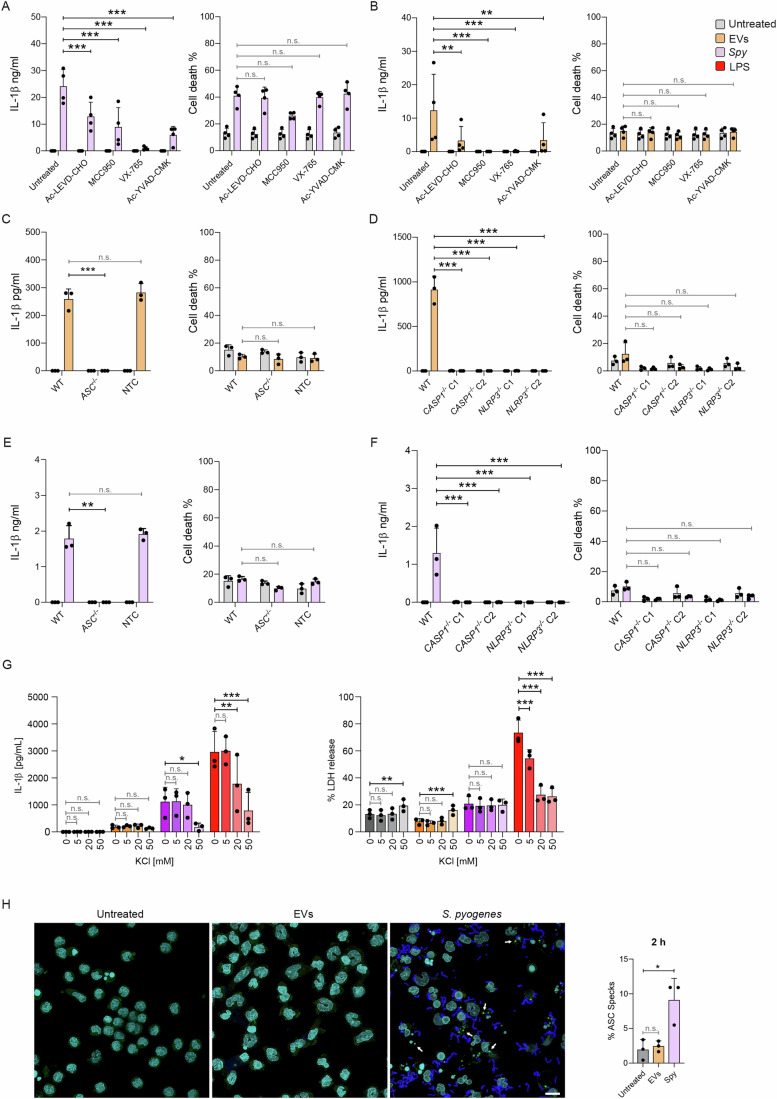
To determine the role of caspase-4 and -5 in Spy EV-induced IL-1β release, we used the chemical inhibitor Ac-LEVD-CHO, which blocks the action of these two caspases. Incubation of monocytes with the caspase-4/-5 inhibitor prior to the addition of Spy EVs or S. pyogenes decreased IL-1β secretion, albeit with a greater reduction in IL- 1β in response to EVs (Fig. 2A,B). In contrast, the levels of caspase-independent cytokines such as IL-6 and IL-1α remained unchanged. Activation of the non-canonical inflammasome can lead to pyroptotic cell death (Shi et al, 2015). However, cell death measured by secretion of lactate dehydrogenase (LDH) did not vary upon inhibition of caspase-4/-5 (Fig. 2A,B). To validate that Ac-LEVD-CHO only impairs the function of caspase-4/-5 and does not affect the expression of its genes, we measured the mRNA levels of caspase-4 and -5 as well as IL1B and IL6 using RTqPCR. No difference in the transcript levels for each target gene between untreated vs. Ac-LEVD-CHO-treated cells could be detected (Fig. EV3A), indicating that the observed reduction in IL-1β secretion upon inhibition of caspase-4/-5 does not originate from lower caspase-5 or pro-IL1β gene expression. In addition, we analyzed the amounts of pro-IL1β by immunoblotting. While the induction of pro-IL1β in response to Spy EVs was much lower than for bacterial cells, no difference was found between untreated and Ac-LEVD-CHO-treated cells (Fig. EV3B). Consistent with the fact that LPS is a major activator of caspase-4/-5, Ac-LEVD-CHO also significantly reduced the release of IL-1beta in response to LPS (Fig. EV3C). Finally, we detected significant cleavage of the caspase-4/-5 substrate Ac-LEVD-AFC in cell culture supernatants from monocytes infected with S. pyogenes and, to a lesser extent, in response to Spy EVs as well (Fig. 2C). Collectively, our data suggest that IL-1β release in response to Spy EVs and S. pyogenes requires caspase-4/-5 without inducing cell death.
Figure 2. S. pyogenes EVs are recognized by caspase-4 and caspase-5.
(A, B) IL-1β, IL-6, IL-1α, and LDH released by monocytes after stimulation with either Spy EVs (A) or intact S. pyogenes (B) for 18 h. Monocytes were either preincubated with the caspase-4/-5 inhibitor Ac-LEVD-CHO or left untreated. The percentage of LDH released from the positive control is shown as a measure of cell death. Bars represent the mean ± standard deviation (SD) of four biological replicates. (A) P = 0.0385, (B) P = 0.0036, (C) Caspase-4/5 substrate cleavage in supernatants from monocytes after stimulation with Spy EVs or intact S. pyogenes. Bars represent the mean ± SD of six biological replicates. P = 0.0139 (EVs_3.5 × 109), P = 0.0024 (Spy MOI 5). (D) IL-1β released by BLaER1 cells after stimulation with either Spy EVs (left panel) or S. pyogenes (right panel) for 18 h. BLaER1 cell lines: wild-type (WT), Caspase-4 knock-out (KO, CASP4−/−), Caspase-5 KO (CASP5−/−, clones 1 and 2) and non-targeting control (NTC). Bars represent the mean ± SD of three biological replicates. (Left panel) P = 0.0034, P < 0.0001, P = 0.003. (Right panel) P = 0.0239, P = 0.0131, P = 0.0131. (E) Immunoblot analysis of cleavage products for caspase-1/4/5 in cell culture supernatants from human monocytes & macrophages. Cells were either left untreated or infected with S. pyogenes strains M1T1 5448, M1T1 5448 AP, ISS3348, or ISS3348 Δslo at MOI 5 (covS-: covS inactivation). Representative of four biological replicates. (F) Immunoblot, RTqPCR & ELISA analysis of IL-1β released by monocytes after stimulation with Spy EVs in the presence or absence of BafA1 (100 nM) for 2 h or 18 h. A dotted line indicates the mean expression of the housekeeping genes (GAPDH/TUBB). Representative of four biological replicates. P = 0.0254. (G) Immunoblot analysis of LC3 levels in human monocytes after stimulation with Spy EVs in the presence or absence of BafA1 (100 nM). Bars represent the mean ± SD of four biological replicates. P = 0.0132 (30’), P = 0.0151 (90’). Data information: (ABDG) Two-way ANOVA with Holm–Šídák correction for multiple comparisons was applied for statistical analyses. (C) Multiple t tests with Holm–Šídák correction for multiple comparisons. (F) One-way ANOVA with Holm–Šídák correction for multiple comparisons was applied for statistical analyses. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant. Source data are available online for this figure.
Figure EV3. BLaER1 knock-out characterization.
(A) Quantitative real time PCR of selected genes in human monocytes. Shown are mean ± SD of three biological replicates. A dotted line indicates the mean expression of the housekeeping genes (GAPDH/TUBB). (CASP5) P = 0.0013 (−), P = 0.0018 (+). (IL1B) P = 0.0006 (EVs for both comparisons), P = 0.0001 (Spy, for both comparisons). (IL6) P = 0.0001 (EVs for both comparisons), P < 0.0001 (Spy, for both comparisons). (B) Immunoblot analysis of pro-IL-1β in human monocytes. Cells were either left untreated or preincubated with the caspase-4/-5 inhibitor Ac-LEVD-CHO. Representative of three biological replicates. (C) IL-1β in response to LPS in supernatants from human monocytes either left untreated or preincubated with Ac-LEVD-CHO at 18 h. Shown are mean ± SD of three biological replicates. P < 0.0001 (D) Graphical representation of the differentiation protocol of BLaER1 cells as well as CRISPR-Cas9 mutagenesis of BLaER1 cells. The existence of frameshift mutations on each allele of the CASP4−/−, CASP5−/−, TLR2−/−, CASP1−/−, and NLRP3-/- clones was assessed using next generation sequencing. The guide RNAs used for genome editing are shown in the Reagents and Tools Table. Mass spectrometry analysis was used to confirm the absence of caspase-4 and caspase-5 from their respective KO clones compared to wild-type or non-targeted control (NTC) BLaER1 cells. (E–H) IL-6 and LDH released from BLaER1 WT, caspase-4 KO (CASP4−/−), caspase-5 KO clones 1 and 2 (CASP5−/− C1 or C2), and non-targeted cells (NTC). BLaER1 cells were left untreated or were stimulated for 18 h with either Spy EVs or S. pyogenes. The percentage of LDH released from the positive control is shown as a measure of cell death. Bars represent the mean ± SD of three biological replicates. Data information: (A, C, E–H) Two-way ANOVA with Holm–Šídák correction for multiple comparisons was applied for statistical analyses. **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant.
Human monocytes are short-lived, non-replicating cells (Patel et al, 2017), which complicates loss-of-function studies using engineering tools (e.g., CRISPR/Cas9). Furthermore, commonly used monocytic cell lines, such as THP-1, have been shown to only partially mimic the functions of primary cells (Gaidt et al, 2016, 2018). To overcome these limitations, we targeted genes in BLaER1 cells, which have been described to closely recapitulate human monocytic functions such as TLR and inflammasome signaling (Rapino et al, 2013; Gaidt et al, 2016). BLaER1 is an immortalized B cell line which, upon induction of the transcription factor C/EBPα, irreversibly differentiates into monocyte-like cells (Rapino et al, 2013). We have shown that BLaER1 cells respond similarly, albeit less pronounced, to Spy EVs and bacterial cells (Fig. EV2AB). To dissect the individual contributions of caspase-4 and -5 in Spy EV-induced IL-1β release, we generated gene knock-out (KO) BLaER1 cell clones using CRISPR-Cas9 (Fig. EV3D). BLaER1 CASP4−/− cells secreted less IL-1β than their wild-type counterparts upon Spy EVs challenge, while IL-1β secretion in CASP5−/− cells was almost abolished (Fig. 2D, left panel). A slight reduction in IL-1β production was also observed upon stimulation with live S. pyogenes (Fig. 2D, right panel). Consistent with our experiments in primary monocytes, IL-6 and LDH levels were unaltered in wild-type and CASP4−/− and CASP5−/− cells (Fig. EV3E–H). Non-targeted control (NTC, electroporated with a control guide RNA) cells showed similar secretion of IL-1β, IL-6 and LDH when compared to wild-type cells (Figs. 2D and EV3E–H). Immunoblot analysis of cell culture supernatants from human monocytes infected with different M1 serotypes of S. pyogenes revealed no difference in caspase-1/-4/-5 cleavage between wild-type and covS-inactivated (covS-) strains (Fig. 2E). In contrast, when monocytes were differentiated to macrophages, cleavage products for all three caspases were only observed when cells were infected with hypervirulent S. pyogenes harboring covS inactivating mutations leading to increased production of streptolysin O (Slo) (Sumby et al, 2006). Altogether, our experiments suggest that caspase-4 and caspase-5 contribute to IL-1β release in response to Spy EVs and intact S. pyogenes, which is independent of the virulence regulator CovS in monocytes but dependent on CovS in differentiated macrophages.
During inflammasome activation, mature IL-1β is typically secreted through Gasdermin D (GSDMD) pores. However, Spy EV stimulation of human monocytes did not result in cell death. Alternative secretion pathways for IL-1β through secretory lysosomes (Andrei et al, 1999; Semino et al, 2018), microvesicle shedding (MacKenzie et al, 2001), or autophagy (Iula et al, 2018) have been described in LPS-activated monocytes or neutrophils. Consistent with these previous findings, we found that Bafilomycin A (BafA1), a V-ATPase inhibitor shown to stimulate lysosomal exocytosis (Tapper and Sundler, 1995), promotes an increase in EV-induced IL-1β release from human monocytes (Fig. 2F) without affecting pro-IL-1β or caspase-5 expression. Finally, we also observed an increase in LC3-II levels and autophagic flux in human monocytes upon EV treatment (Fig. 2G), suggesting an increase in the formation of autophagosomes.
EV signaling through TLR2-MyD88 are required for cytokine production
Caspase-4/-5 are cytosolic receptors that do not bind directly to any component of Gram-positive lysates (Shi et al, 2014a). Consequently, signaling through other proteins acting upstream of caspase-4 and -5 may be the trigger for IL-1β production. TLRs are the most thoroughly characterized family of PRRs involved in S. pyogenes recognition (Gratz et al, 2008; Loof et al, 2008; Eigenbrod et al, 2015; Fieber et al, 2015). In particular, TLR2 signaling via the adaptor protein MyD88 is essential for generating an inflammatory response to S. pyogenes and other Gram-positive pathogens by promoting NFκB-dependent gene expression (Underhill et al, 1999; Loof et al, 2008). Yet, conflicting results have been published on the role of TLR2 in S. pyogenes recognition (Gratz et al, 2008; Fieber et al, 2015). Since LPS-induced activation of the alternative inflammasome in human monocytes has been linked to lipid A-induced TLR4 signaling, we investigated whether the lipid and lipoteichoic acid (LTA) sensing TLR2 and its adaptor protein MyD88 might coordinate the sensing of Spy EVs. Indeed, TLR2- or MyD88-deficiency in BLaER1 cells severely impaired both IL-1β and IL-6 secretion in response to Spy EVs challenge (Fig. 3A), implying a critical role for TLR2-MyD88 signaling in Spy EVs recognition. In contrast, sensing of S. pyogenes required MyD88, but not TLR2, to induce IL-6 and IL-1β (Fig. 3B), indicating a redundant role for other MyD88-coupled TLRs such as TLR1 or TLR4-9. To address this redundancy, we used a variety of small molecule inhibitors. Treatment of monocytes with TLR2/1 (TLR-IN-C29) or TLR2/6 (Cu-CPT22) inhibitors significantly reduced the release of Spy EV-induced IL-1β, but only minor effects were observed following infection with S. pyogenes (Fig. 3C). Notably, inhibition of the Receptor-Interacting serine/threonine-Protein Kinase 1 (RIPK1), the key kinase mediating the alternative inflammasome pathway upon LPS treatment, resulted in a reduction in IL-1β secretion only in Spy EV-treated cells, but not in those treated with S. pyogenes. In support of TLR2 appearing as the major sensor of Spy EVs, a TLR2 reporter cell-line (HEK-Blue) showed a dose-dependent activity in response to Spy EVs (Fig. 3D). We further examined the potential involvement of TLR4, TLR7, TLR8, and TLR9 in the sensing of intact S. pyogenes cells by means of small molecule inhibitor treatment. We found a role for intracellular TLR8 in mediating IL-1β secretion upon live S. pyogenes infection (Fig. 3E,F). Lastly, and in accordance with previous observations for LPS-stimulated monocytes (Gaidt et al, 2016), both S. pyogenes and its EVs required caspase-8 for IL-1β release (Fig. 3G). Together, we conclude that the activation of the inflammasome in response to Spy EVs is most likely initiated at the cell surface via TLR2, whereas S. pyogenes triggers IL-1β secretion predominantly through TLR8, thereby indicating that internalization of the bacteria is required to activate the inflammasome.
Figure 3. EV-dependent cytokine release requires Toll-like receptor 2-mediated signaling.
(A) IL-1β, IL-6, and LDH in supernatants of BLaER1 WT, MYD88 KO (MYD88−/−), Toll-like receptor 2 KO (TLR2−/−) and NTC cells stimulated with Spy EVs for 18 h. Bars represent the mean ± SD of three biological replicates. (left panel, IL-1β) P < 0.0001 (MYD88−/−), P < 0.0001 (TLR2−/−). (middle panel, IL-6) P = 0.0019 (MYD88−/−), P = 0.0040 (TLR2−/−). (B) IL-1β, IL-6, and LDH released by BLaER1 WT, MYD88−/−, TLR2−/−, and NTC cells after 18 h stimulation with S. pyogenes. Bars represent the mean ± SD of three biological replicates. (left panel, IL-1β) P = 0.0017 (MYD88−/−). (Middle panel, IL-6) P = 0.0016 (MYD88−/−). (A, B) The percentage of LDH released from the positive control is shown as a measure of cell death. (C) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with Ac-LEVD-CHO (caspase-4/5), GSK3145095 (RIPK1), TLR2-C29 (TLR2/1, TLR2/6), or CU-CPT22 (TLR2/6). Cells were then left unstimulated or challenged with Spy EVs or S. pyogenes for 18 h. Bars represent the mean ± SD of four biological replicates. (EVs, IL-1β) P = 0.0001, P = 0.0018, P = 0.0003, P = 0.0018. (EVs, IL-6) P = 0.0082. (Spy, IL-1β) P < 0.0001, P = 0.0437. (D) TLR2 activity in HEK-Blue TLR2 cells after stimulation with Spy EVs for 18 h. Baseline represents untreated cells. Bars represent the mean ± SD of four biological replicates. (7 × 108) P < 0.0001, (3.5 × 108) P < 0.0001, (7 × 107) P < 0.0001, (3.5 × 107) P = 0.0081. (E) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with TLR inhibitors CLI-095 (TLR4), M5049 (TLR7/8, 0,1 µM = TLR7, 1 µM = TLR8), or TLR9-IN-1 (TLR9). Cells were then left unstimulated or infected with S. pyogenes for 18 h. Bars represent the mean ± SD of four biological replicates. P = 0.0048. (F) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with the TLR8 inhibitor CU-CPT9a. Cells were then left unstimulated or infected with S. pyogenes for 18 h. Bars represent the mean ± SD of four biological replicates. P < 0.0001. (G) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with the caspase-8 inhibitor Ac-IETD-CHO. Cells were then left unstimulated or challenged with Spy EVs, S. pyogenes, or LPS for 18 h. Bars represent the mean ± SD of four biological replicates. (EVs) P = 0.0043, (Spy) P = 0.0075, (LPS) P = 0.0043. Data information: (A–C, E–G) Two-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (D) One-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant. Source data are available online for this figure.
The NLRP3 inflammasome coordinates EV-mediated secretion of IL-1β
The NLRP3 inflammasome is composed of three main proteins: Nucleotide-binding domain Leucine-rich Repeat family containing a Pyrin domain 3 (NLRP3), Apoptosis-associated Speck-like containing a Caspase recruitment domain (ASC), and caspase-1 (Martinon et al, 2002; Muñoz-Planillo et al, 2013). Previous studies have shown that the NLRP3 inflammasome coordinates IL-1β secretion during S. pyogenes infection (Harder et al, 2009; Lin et al, 2015; Valderrama et al, 2017) and a recent study established that S. aureus EVs are also recognized by this system (Wang et al, 2020). We therefore investigated whether, in addition to caspase-4/-5, the NLRP3 inflammasome contributed to IL-1β release in response to Spy EVs. To this end, we left monocytes untreated or treated them with the NLRP3 inhibitor MCC950, the caspase-1 inhibitor Ac-YVAD-cmk, the caspase-4/-5 inhibitor Ac-LEVD-CHO, or the caspase-1/-4/-5 inhibitor VX-765 prior to stimulation with S. pyogenes or Spy EVs. All inhibitors significantly reduced IL-1β for both S. pyogenes and its EVs (Fig. 4A,B). Release of IL-6 and LDH were comparable for all treatments, with the exception of increased IL-6 and decreased cell death upon inhibition of NLRP3 following infection with S. pyogenes (Figs. 4A,B and EV4A).
Figure 4. The NLRP3 inflammasome contributes to IL-1β and LDH release upon recognition of S. pyogenes EVs.
(A, B) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with Ac-LEVD-CHO, MCC950, VX-765, or Ac-YVAD-CMK. Cells were then left unstimulated or stimulated with either S. pyogenes (A) or Spy EVs (B) for 18 h. Bars represent the mean ± SD of four biological replicates. (A) P < 0.0001 (for all comparisons). (B) P = 0.0068, P = 0.0006, P = 0.0006, P = 0.0076. (C, D) IL-1β and LDH were measured in supernatants of BLaER1 WT, PYCARD KO (ASC-/-), Caspase-1 KO (CASP1−/− clone 1 and 2), NLRP3 KO (NLRP3−/− clone 1 and 2), and NTC cells after 18 h either left untreated or stimulated with Spy EVs. Bars represent the mean ± SD of three biological replicates. (C) P = 0.0010. (D) P < 0.0001 (for all comparisons). (E, F) IL-1β and LDH were measured in supernatants of BLaER1 WT, ASC−/−, CASP1−/− clone 1 and 2, NLRP3−/− clone 1 and 2, and NTC cells after 18 h either left untreated or stimulated with S. pyogenes. Bars represent the mean ± SD of three biological replicates. (E) P = 0.0016. (F) P = 0.0003 (for all comparisons). (G) IL-1β and LDH in supernatants from BLaER1 WT cells after 18 h, either left untreated or stimulated with Spy EVs, S. pyogenes, or LPS (200 ng/mL). Bars represent the mean ± SD of three biological replicates. (IL-1β) P = 0.0177 (Spy, 50 mM), P = 0.0017 (LPS, 20 mM), P < 0.0001 (LPS, 50 mM). (LDH) P = 0.0084 (Untreated, 50 mM), P = 0.0007 (EVs, 50 mM), P < 0.0001 (LPS, all comparisons). (H) Immunofluorescence staining and corresponding quantification results for ASC speck formation in human monocytes either left untreated or stimulated with Spy EVs or S. pyogenes for 2 h (scale bar: 10 µm). White arrows indicate ASC specks. Bars represent the mean ± SD of three biological replicates. P = 0.0103. Data information: (A–G) The percentage of LDH released from the positive control is shown as a measure of cell death. Two-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (H) One-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant. Source data are available online for this figure.
To further validate these results, we used BLaER1 monocytes lacking ASC (Vierbuchen et al, 2017), caspase-1, or NLRP3. Consistent with the pharmacological inhibition, the genetic deficiency of NLRP3, ASC, or CASP1 abolished IL-1β secretion in response to S. pyogenes or Spy EVs treatment (Fig. 4C–F), while IL-6 levels remained unchanged for all cell lines and conditions tested (Fig. EV4B,C). In addition, whereas the release of LPS-induced IL-1β and LDH was strongly dependent on K+ efflux, no effect of extracellular potassium could be detected for Spy EVs (Fig. 4G). In contrast, S. pyogenes-infected monocytes showed significantly reduced IL-1β secretion in the presence of 50 mM KCl, indicating that the reduction of intracellular K+ contributes to activation of the inflammasome triggered by S. pyogenes, but not by its derived EVs. Using immunofluorescence microscopy, we further analyzed ASC speck formation in human monocytes either treated with Spy EVs or infected with S. pyogenes. In accordance with the previously reported alternative inflammasome pathway, which lacks the typical features of classical NLRP3 inflammasome activation (Gaidt et al, 2016), we observed ASC specks and cleavage of GSDMD only in S. pyogenes-infected monocytes, but not in EV-treated cells (Figs. 4H and EV4E).
Since NLRP6 and NLRP7 have been reported to recognize LTA and acetylated lipoproteins (Khare et al, 2012; Hara et al, 2018), we also attempted to generate corresponding knockout cells for these proteins to study their involvement in inflammasome activation upon Spy EVs or S. pyogenes challenge. However, we were unable to detect expression of either sensor in BLaER1 cells or in primary monocytes (Fig. EV4D). Collectively, these results indicate that Spy EVs or S. pyogenes engage the NLRP3 inflammasome as well as caspase-4 and -5, but employ distinct upstream sensors and signaling cascades for the induction and release of IL-1β in monocytes.
Caspase-4/-5-Mediated Secretion of IL-1β is Triggered by Lipoteichoic Acid
LTA is a major component of the cell membrane of Gram-positive bacteria that is sensed by TLR2 (Schwandner et al, 1999; Takeuchi et al, 1999) and has been shown to induce caspase-11-mediated secretion of IL-1β in mice (Hara et al, 2018). Since we found that Spy EVs induced secretion of IL-1β depends on TLR2, we hypothesized that LTA might be the bacterial ligand triggering caspase-4/-5 signaling. The clinical isolate used in this study is highly encapsulated due to its inactive two-component system CovRS (Resch et al, 2016). Therefore, we first examined whether LTA was available on the surface of this bacterial strain and its EVs. Using immunogold-labelling of LTA and electron microscopy, we observed that this structural acid can indeed be detected on the surface of S. pyogenes and its EVs (Fig. 5A,B, white arrows). In addition, we used immunoblotting to confirm the presence of LTA in our EV preparations (Fig. 5C).
Figure 5. Lipoteichoic acid conserved in Gram-positive EVs promotes caspase-4/-5-dependent IL-1β secretion.
(A) S. pyogenes cells imaged by Scanning Electron Microscopy and immunogold staining (SEM, scale bar = 200 nm). (B) Immunogold-labelling of surface lipoteichoic acid from purified Spy EVs (LTA clusters indicated with white arrows, scale bar = 200 nm). (C) Immunoblot analysis of LTA in Spy EVs. (D) IL-1β in supernatants from monocytes either left untreated or preincubated with Ac-LEVD-CHO prior to the addition of S. pyogenes LTA after 18 h. Bars represent the mean ± SD of four biological replicates. P = 0.0181. (E) IL-1β in supernatants from BLaER1 cells either left untreated or preincubated with Ac-LEVD-CHO prior to the addition of S. pyogenes LTA after 18 h. Bars represent the mean ± SD of three biological replicates. P = 0.0177. (F) IL-1β release at 18 h from monocytes that were left untreated or treated with S. pyogenes LTA, either directly applied (LTA) or transfected using Lipofectamine LTX (LTx:LTA). Bars represent the mean ± SD of four biological replicates. (G) TLR2 activity in HEK-Blue TLR2 cells after stimulation with Spy LTA for 18 h. Baseline represents untreated cells. Bars represent the mean ± SD of four biological replicates. (H) Immunoblot, RTqPCR & ELISA analysis of IL-1β released by monocytes after stimulation with S. pyogenes LTA in the presence or absence of BafA1 (100 nM) for 2 h or 18 h. Bars represent the mean ± SD of four biological replicates. P = 0.0067. (I) Immunoblot analysis of LC3 levels in human monocytes after stimulation with S. pyogenes LTA in the presence or absence of BafA1 (100 nM). Bars represent the mean ± SD of four biological replicates. P = 0.0412 (30’), P = 0.0143 (60’), P = 0.0002 (90’). (J) IL-1β release at 18 h from monocytes either left untreated or preincubated with Ac-LEVD-CHO before the addition of EVs from S. aureus (left), S. agalactiae (middle), or B. subtilis (right). Bars represent the mean ± SD of four biological replicates. P = 0.0164 (S. aureus), P = 0.0099 (S. agalactiae). (K) IL-1β and IL-6 in supernatants from monocytes or BLaER1 cells either left untreated or stimulated with Spy EVs isolated −/+ LtaS-IN-1771 for 18 h. Bars represent the mean ± SD of three biological replicates. (Primary, IL-1β) P = 0.0003 (both comparisons). (Primary, IL-6) P < 0.0001 (both comparisons). (BLaER1, IL-1β) P = 0.0005, P = 0.0160, P = 0.0044. (BLaER1, IL-6) P = 0.0005, P = 0.0051, P = 0.0161. Data information: (D, E, I) Two-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (F, H, K) One-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (J) Statistical significance was assessed using paired t tests. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant. Source data are available online for this figure.
LTA mutants of Gram-positive bacteria have been reported to display severe growth defects (Schneewind and Missiakas, 2017), which is why we used purified LTA from S. pyogenes to test its role in monocyte activation. Consistent with our hypothesis, stimulation of monocytes or BLaER1 cells with S. pyogenes LTA induced IL-1β secretion, which was significantly reduced upon pretreatment with the caspase-4/-5 inhibitor peptide Ac-LEVD-CHO (Fig. 5D,E), while IL6 secretion and LDH release were not affected (Fig. EV5A,B). Moreover, the lipofectamine-mediated encapsulation and intracellular delivery of LTA into monocytes did not further increase IL-1β production (Fig. 5F) or affect IL-6 and LDH release (Fig. EV5C), suggesting that the caspase-4/-5-mediated signaling triggered by LTA most likely begins at the cell surface via TLR2, in line with the alternative inflammasome pathway described for various TLR ligands (Gaidt et al, 2016; Unterberger et al, 2023). Finally, to test whether LTA from other Gram-positive bacteria similarly induces caspase-4/5-dependent IL-1β secretion, we used LTA derived from S. aureus and, akin to S. pyogenes LTA, we observed a caspase-4/-5-dependent reduction in the release of IL-1β, but not IL-6 (Fig. EV5D). Of note, we also detected a significant upregulation of caspase-5 gene expression upon stimulation with both S. pyogenes and S. aureus LTA, whereas gene expression levels of caspase-4 remained unaffected (Fig. EV5E). In line with our previous results showing Spy EV-induced TLR2 activity in a reporter cell-line, LTA from both S. pyogenes and S. aureus induced TLR2 activity in a dose-dependent manner, albeit S. aureus LTA was considerably more potent and showed a measurable response at concentrations as low as 0.01 µg/mL (Figs. 5G and EV5F). As shown before for Spy EVs, LTA-induced IL-1β secretion is dependent on caspase-8 (Fig. EV5G). Furthermore, BafA1 increased IL-1β release in response to S. pyogenes LTA, suggesting a vesicular secretion mechanism for IL-1β as observed for Spy EVs (Fig. 5H). Since the turnover of autophagosomes was also significantly increased by S. pyogenes LTA (Fig. 5I), a positive role of autophagy during IL-1β secretion in human monocytes upon Spy EV or LTA encounter may be a plausible conclusion.
Figure EV5. Cytokine controls and cell death of monocytes and BLaER1 cells: role of LTA in EV-dependent monocyte activation.
(A) IL-6 and LDH release at 18 h from monocytes that were left untreated or incubated with Ac-LEVD-CHO before the addition of S. pyogenes LTA. Shown are mean ± SD of four biological replicates. P = 0.0069. (B) IL-6 and LDH release at 18 h from BLaER1 cells that were left untreated or incubated with Ac-LEVD-CHO prior to addition of purified S. pyogenes LTA. Shown are mean ± SD of three biological replicates. (C) IL-6 and LDH release at 18 h from monocytes that were left untreated or treated with S. pyogenes LTA, either directly applied (LTA) or transfected using Lipofectamine LTX (LTx:LTA). Shown are mean ± SD of four biological replicates. (D) IL-1β and IL-6 release at 18 h from monocytes that were left untreated or incubated with Ac-LEVD-CHO before the addition of S. aureus LTA. Shown are mean ± SD of four biological replicates. P = 0.0374. (E) Quantitative real time PCR of CASP5 and CASP4. Shown are mean ± SD of three biological replicates. A dotted line indicates the mean expression of the housekeeping genes (GAPDH/TUBB). P < 0.0001 (for all comparisons). (F) TLR2 activity in HEK-Blue TLR2 cells after stimulation with S. aureus LTA for 18 h. Baseline represents untreated cells. Bars represent the mean ± SD of four biological replicates. (G) IL-1β and IL-6 released by human monocytes that were either left untreated or preincubated with the caspase-8 inhibitor Ac-IETD-CHO. Cells were then left unstimulated or treated with S. pyogenes LTA for 18 h. Bars represent the mean ± SD of four biological replicates. P = 0.0066. (H) Mean size and particles/mL of S. pyogenes, S. aureus, B. subtilis, and S. agalactiae EVs (n = 2). (I) IL-6 and LDH release from monocytes either left untreated or preincubated with Ac-LEVD-CHO before the addition of EVs from S. aureus, S. agalactiae, or B. subtilis for 18 h. Shown are mean ± SD of four biological replicates. (J) Immunoblot analysis of LTA in bacterial pellets treated −/+ LtaS-IN-1771 and particles/mL of corresponding S. pyogenes EVs. Representative image of 2 biological replicates is shown. (K) S. pyogenes growth in the presence of LtaS-IN-1771 in THB media. Shown are mean ± SD of three biological replicates. (3 h) P = 0.0010, (4h-8h) P < 0.0001. Data information: (ABDEGK) Two-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (C) One-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. (I–H) Statistical significance was assessed using paired t tests. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant.
EVs from other Gram-positive bacteria induce IL-1β through caspase-4/-5
Since the synthesis of teichoic acids is moderately conserved among Gram-positive bacteria (Reichmann and Gründling, 2011; Brown et al, 2013), we asked whether EVs derived from other Gram-positive bacteria could trigger IL-1β production through caspase-4 and -5. In fact, monocytes treated with EVs from S. aureus, Streptococcus agalactiae, or Bacillus subtilis released less IL-1β when caspase-4/-5 was inhibited, while IL-6 secretion remained unaffected (Figs. 5J and EV5H,I), demonstrating that caspase-4/-5 are also involved in IL-1β release in response to EVs derived from other Gram-positive bacteria.
LTA-depleted EVs severely attenuate the IL-1β response of human monocytes
To determine whether surface ligands other than LTA are sensed upstream of caspase-4/-5 activation, we treated S. pyogenes with compound 1771, which was shown to inhibit LTA biosynthesis in S. aureus (Richter et al, 2013; Serpi et al, 2023) to produce LTA-depleted Spy EVs (Fig. EV5J). Consistent with its reported antimicrobial properties, compound 1771 significantly delayed the growth of S. pyogenes compared with DMSO-treated control cultures (Fig. EV5K). When monocytes were treated with LTA-depleted Spy EVs, their release of both IL-1β and IL-6 was significantly decreased (Fig. 5K). To a lesser extent, this phenotype was also reproduced in BLaER1 cells, indicating that LTA on Spy EVs is the predominant factor responsible for TLR and inflammasome activation.
Free LTA in EV-depleted supernatants recapitulates monocyte activation in response to EVs
In addition to EV-bound LTA, supernatants of S. pyogenes also contain free LTA that is released during bacterial growth. To compare the effects of EV-bound vs. free LTA on monocyte activation, we first measured the amount of LTA in double-filtered S. pyogenes supernatants before and after EV isolation (Fig. 6A). The LTA concentration in the remaining supernatants was reduced by ~45% after removal of EVs using ultracentrifugation. We then determined the amounts of cytokines and LDH released by human monocytes treated either with Spy EVs or with EV-depleted supernatant (SUP) (Fig. 6B). Notably, due to the presence of Slo in the EV-depleted SUPs from WT S. pyogenes, monocytes displayed high amounts of cell death and no significant release of IL1β or IL-6 could be observed. Therefore, we additionally isolated EVs and EV-depleted SUP from an S. pyogenes slo deletion strain. Increasing volumes of Δslo EV-depleted SUP significantly induced IL-1β and IL-6 secretion without affecting cell survival. Immunoblotting confirmed comparable levels of LTA in EV-depleted supernatants from WT and Δslo S. pyogenes (Fig. 6C). Interestingly, THB alone also led to significant IL-6 secretion by human monocytes, which is most likely attributable to the presence of beef heart peptones or high salt concentrations within the concentrated media. Furthermore, protein analysis using mass spectrometry revealed an 84% overlap between EVs and EV-depleted supernatants, suggesting that a high percentage of proteins encapsulated in EVs is also released into the media (Fig. 6D). Finally, as for monocytes treated with Spy EVs, stimulation with EV-depleted SUP prevented caspase-3 activation and induced phosphorylation of the MAP kinase (MAPK) p44/42 (ERK1/2) (Fig. 6D) indicative of monocyte survival pathways being activated.
Figure 6. Spy EVs isolated using size exclusion chromatography partially recapitulate the monocyte response against conventional EV preparations.
(A) LTA concentration in double-filtered S. pyogenes culture supernatants before and after ultracentrifugation. Bars represent the mean ± SD of four biological replicates. P = 0.0080. (B) IL-1β, IL-6, and LDH released by monocytes after 18 h stimulation with either Spy EVs, THB medium, or EV-depleted supernatants (SUP) derived from either S. pyogenes WT or Δslo mutant strains. The percentage of LDH released from the positive control is shown as a measure of cell death. Bars represent the mean ± standard deviation (SD) of four biological replicates. (IL-1β) P < 0.0001. (IL-6) P < 0.0001 (THB, all comparisons), P = 0.0020, P = 0.0002, P < 0.0001. (LDH) P < 0.0001 (all comparisons). (C) Immunoblot of Slo and LTA in EV-depleted supernatants from S. pyogenes WT (4 biological replicates) or Δslo mutant strains. THB media was used as negative control. (D) Venn diagram & immunoblots for EV-depleted supernatants. The Venn diagram shows protein expression in ultracentrifuged EV preparations (EV UC) vs. EV-depleted supernatants. Numbers indicate the total amount of proteins for each category. Immunoblots display caspase-3 cleavage and phospho-p44/42 MAPK in human monocytes after 18 h stimulation with either Spy EVs, THB medium, or EV-depleted supernatants (SUP) derived from either S. pyogenes WT or Δslo mutant strains. Representative of four biological replicates. (E) LTA and protein concentration in 28 fractions of double-filtered S. pyogenes culture supernatant fractionated using size exclusion chromatography. Bars represent the mean ± standard deviation (SD) of three biological replicates. Immunoblot shows LTA present in fractions 8–28. (F) Venn diagrams showing protein expression in ultracentrifuged EV preparations (EV UC) vs. pooled SEC fractions 8–16 (EVs) or 17–28 (soluble), respectively. (G) Concentration, size, and zeta potential of Spy EVs analyzed with the exoid instrument. Bars represent the mean ± SD of three biological replicates. (H) LTA concentration in EVs UC vs. EVs SEC (left panel). Bars represent the mean ± SD of four vs. two biological replicates. LDH (middle panel) and IL-6 (right panel) released by human monocytes treated with Spy EVs isolated using SEC for 18 h. Bars represent the mean ± SD of seven biological replicates with pooled EV fractions from 2 SECs (black circles: SEC1 F8-16; grey circles: SEC2 F8-15). P = 0.0299 (LDH), P = 0.0003 (IL-6). (I) TLR2 activity in HEK-Blue TLR2 cells after stimulation with Spy EVs isolated using SEC for 18 h. Baseline represents untreated cells. Bars represent the mean ± SD of eight biological replicates with pooled EV fractions from 2 SECs (black circles: SEC1 F8-16; grey circles: SEC2 F8-15). P = 0.0007, P = 0.0071. (J) Immunoblots displaying caspase-3 cleavage, phospho-p44/42 MAPK, and caspase-5 expression in human monocytes after 18 h stimulation with Spy EVs isolated using SEC. Representative of four biological replicates. Data information: (A, G, H) Statistical significance was assessed using unpaired t test. (B, H) Owo-way ANOVA was applied with Holm–Šídák correction for multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s. not significant. Source data are available online for this figure.
Spy EVs isolated using size exclusion chromatography (SEC) confirm that LTA is as driving factor for monocyte activation
Given the growing interest in EVs, isolation methodologies are constantly evolving and refined (Welsh et al, 2024). Since our EV preparations using ultracentrifugation most likely also contain non-vesicular extracellular particles (NVEP) such as pili, phages, or proteins, we attempted to isolate Spy EVs via size exclusion chromatography (SEC) as previously described for E. coli and other bacteria (Watson et al, 2021). In parallel to SEC, we processed the culture supernatants of S. pyogenes by ultracentrifugation to directly compare the EVs obtained. Figure 6E shows the LTA and protein concentration in sequential fractions, which increase successively. Using SDS-PAGE and TRPS, we identified Spy EVs enriched within the fractions 8-16 of SEC1, 8-15 of SEC2, and 8-13 of SEC3 (Appendix Fig. S2A). Characteristic blockage traces and size/concentration histograms of EVs UC and the pooled SEC fractions are summarized in Appendix Fig. S2B. Traces from individual SEC fractions are shown in Appendix Fig. S2C. Proteomic characterization revealed a 65% and 80% overlap between EVs derived from ultracentrifugation (EV UC) and SEC fractions 8-16 (EVs) and 17–28 (soluble), respectively. Since NTA analysis was not sensitive enough to detect particles in our SEC fractions, we used TRPS to determine the number, size, and zeta potential of our comparative UC and SEC-isolated EVs (Fig. 6G). We found negligible evidence of EV-aggregation after prolonged storage in PBS, supported by a unimodal size distribution (Appendix Fig. S3A). While the size of UC- and SEC-derived Spy EVs was comparable, the concentration of SEC EVs was one order of magnitude lower than that of the EV UC batches (1010 vs. 1011 particles/mL). The zeta-potential of EVs in PBS ranged from −16 to −24 mV (UC mean: −22 ± 2.0, SEC mean: −21 ± 3.3), which is commonly observed for EVs and indicates the presence of negatively charged cell-wall teichoic acid and membrane-anchored lipoteichoic acid (Rogers et al, 2023). Stimulation of human monocytes with SEC EVs significantly increased cell viability and IL-6 secretion (Fig. 6H). In addition, SEC EVs elicited pronounced TLR2 activity in HEK-Blue TLR2 reporter cells (Fig. 6I). In line with the lower particle and LTA concentration of SEC EVs compared to EVs UC (0.171 and 0.413 µg LTA per 1.9 × 108 particles for SEC1 and SEC2, respectively), the release of IL-1β was barely measurable (Appendix Fig. S3B). In fact, a dose response experiment with Spy LTA revealed that the minimal concentration of LTA required to achieve measurable IL-1β secretion was 0,5 µg/mL (Appendix Fig. S3C). However, we also detected inhibition of caspase-3 activation as well as the induction of p44/42 (ERK1/2) MAPK phosphorylation and caspase-5 protein expression upon SEC EV treatment. Conclusively, the SEC EV data corroborate the results of EV UC, with LTA being a major driver of monocyte activation and the subsequent inflammation in response to Spy EVs.
Discussion
Murine infection models have been used primarily to understand the role of specific receptors and downstream signaling mediators recognizing the strict human pathogen S. pyogenes (Watson et al, 2016). Although these models are extremely informative and enable the study of systemic infections and therapeutic interventions, it remains challenging to assess their contributions due to the intrinsic differences between the innate immune system of mice and humans (Heil et al, 2004; Hasan et al, 2005; Khare et al, 2012; Gaidt et al, 2016).
In the present study, we explored the response of primary human monocytes as well as monocyte-like BLaER1 cells to hypervirulent S. pyogenes and its EVs and identified mediators of recognition and downstream signaling events. Strikingly, only one report using murine macrophages has so far addressed the cellular response during S. pyogenes infection on a genome-wide level (Goldmann et al, 2007), and to our knowledge, no reports on primary human monocytes or macrophages have been published to date. Our RNA sequencing results revealed that responses triggered by S. pyogenes and its EVs strongly overlap, consistent with the observation that Spy EVs contain at least half of the proteins of the total proteome of their parental cells (Resch et al, 2016). Nonetheless, S. pyogenes and its EVs regulate the expression of distinct sets of genes in monocytes, indicating that unique components of intact bacteria or EVs convey a differential immune response.
In the broad spectrum of S. pyogenes clinical manifestations, inflammation plays a critical role in preventing bacterial invasion and limiting an excessive, devastating tissue-destructive immune response. It is well-established that interferons expressed during viral infections are beneficial to the host (Isaacs and Lindenmann, 1957). Notably, interferons produced during bacterial infections have been shown to play both protective and detrimental roles, depending on the bacterial species and the site of infection (Kovarik et al, 2016). In mice, the balance between protection and damage during S. pyogenes infection is orchestrated by IL-1β levels, a key immunoregulatory and proinflammatory cytokine matured by type I interferon-regulated multiprotein complexes, the inflammasomes (Castiglia et al, 2016). Similarly, studies with IL-1β inhibitors suggest a central role of IL-1β in controlling S. pyogenes dissemination in the human body (LaRock et al, 2016).
On one side of this balance, S. pyogenes triggers an interferon-mediated signature that is distinct from its EVs, as demonstrated here by unbiased RNA-Seq profiling. These results are consistent with a report showing that biopsies from patients with necrotizing streptococcal soft tissue infection (NSTI) displayed a recognizable IFN signature that was absent in polymicrobial NSTI’s (Thänert et al, 2019). On the other side, we found that IL-1β production in response to S. pyogenes or its EVs converge at the NLRP3 inflammasome, although different sensing and signaling routes are employed. Accordingly, several studies have demonstrated that NLRP3 is a key sensor of Gram-positive bacteria and a recent report indicated that it participates in the recognition of S. aureus EVs (Mariathasan et al, 2006; Harder et al, 2009; Wang et al, 2020).
In addition to the canonical inflammasome, two other systems, the non-canonical inflammasome and the alternative inflammasome, contribute to the production of IL-1 β in human monocytes (Shi et al, 2014; Gaidt et al, 2016), although little is known about the molecular interactions and crosstalk between the components of these three systems. In this respect, an important observation of this study is the identification of the non-canonical components of the inflammasome, caspase-4/-5, as mediators of IL-1β release induced by Spy EVs and to a lesser extent, by S. pyogenes. In most tissues, basal expression of caspase-5, like murine caspase-11, is low and transcription has been shown to be induced by LPS and IFN-γ (Lin et al, 2000; Casson et al, 2015; Viganò et al, 2015) through TLR/TRIF signaling, whereas caspase-4 is typically constitutively expressed (Shi et al, 2014). Consequently, we report a significant upregulation of caspase-5, but not caspase-4 upon challenge with Spy EVs or purified LTA. Although RNA sequencing revealed a slight increase in caspase-5 transcription in response to parental S. pyogenes cells, the expression level was considerably lower than in monocytes treated with Spy EVs and could not be validated by RT-qPCR. This could be explained by the differential sensing modes identified in monocytes, TLR8 for intact bacteria and TLR2 for Spy EVs, the former sensing bacterial RNA and predominantly activating interferon responses and the latter robustly activating NF-kB target genes such as caspase 5. Furthermore, several type III secretion effector proteins from Gram-negative bacterial pathogens have been demonstrated to actively suppress inflammasome activation (Galle et al, 2008; Brodsky et al, 2010; LaRock and Cookson, 2012; Kobayashi et al, 2013). Moreover, Gram-positive Streptococcus pneumoniae as well as Streptococcus oralis have been shown to prevent inflammasome assembly by introducing oxidative stress into host cells through the production of hydrogen peroxide (Erttmann and Gekara, 2019). Therefore, future work should address whether caspase-5 gene expression is regulated in a temporal- and redox-dependent manner during infection with S. pyogenes.
During Gram-negative bacterial infection, binding of LPS to caspase-4/-5/-11 results in Gasdermin D cleavage, cytosolic leakage, and pyroptotic cell death (Shi et al, 2015). In contrast, the activation of caspase-4/-5/-11 leading to IL-1β production by Gram-positive bacteria does not always result in cell death (Hara et al, 2018; Krause et al, 2019), which is in agreement with our findings as we did not observe a reduction in S. pyogenes-induced cytotoxicity when caspase-4/-5 were inhibited. Interestingly, a recent report demonstrates that S. pyogenes infection induces caspase-4 expression in human neutrophils, indicating that this system participates in S. pyogenes-induced IL-1β production in other cell types as well (Williams et al, 2021).
Although TLR2 has traditionally been linked to the recognition of Gram-positive bacteria (Schwandner et al, 1999; Takeuchi et al, 1999; Underhill et al, 1999), S. pyogenes infection of mice lacking TLR2 results in variable susceptibility compared to wild-type mice (Gratz et al, 2008; Fieber et al, 2015). Furthermore, TLR2 contributes to the sensing of bacterial EVs (Prados-Rosales et al, 2011; Kim et al, 2012; Shen et al, 2012; Choi et al, 2018) and mediates priming of the NLRP3 inflammasome during infection with S. aureus or S. pneumoniae in macrophages (Witzenrath et al, 2011; Wang et al, 2020). In monocytes, however, priming is not required for NLRP3 inflammasome activation, allowing direct release of IL-1β/IL-18 in response to TLR stimulation via the alternative inflammasome pathway (Netea et al, 2009; Viganò et al, 2015; Gaidt et al, 2016; Gritsenko et al, 2020). Here, we show that TLR2 is differentially involved in the sensing of Spy EVs and S. pyogenes: whereas EVs induced IL-6 and IL-1β via this receptor, TLR2 contributed only slightly to IL-1β secretion during bacterial infection. In fact, our results confirm previous findings that endolysosomal TLR8 is the primary receptor for S. pyogenes in human monocytes (Eigenbrod et al, 2015) that activates inflammatory responses.
We provide evidence that LTA, decorating the surface of S. pyogenes and its EVs, modulates caspase-4/-5-dependent responses. Noteworthily, S. pyogenes-derived LTA has already been described to prime the NLRP3 inflammasome in macrophages (Richter et al, 2021). Furthermore, we have shown that caspase-4/-5-dependent IL-1β released in response to EVs is conserved for a variety of Gram-positive species. Nevertheless, in contrast to LPS, it has been demonstrated that caspase-4/-5/-11 do not bind directly to any component present in lysates of several Gram-positive species (Shi et al, 2014), suggesting that other sensors act upstream of these enzymes. In line with this hypothesis, caspase-11 coordinates the sensing of cytosolic LTA from L. monocytogenes as a component of the NLRP6 inflammasome in mice (Hara et al, 2018). In addition, a newly identified sensor (NLRP7) that recognizes acylated Gram-positive lipoproteins, has been described in humans (Khare et al, 2012). Since we were unable to detect the expression of NLRP6 nor NLRP7 in monocytes, it remains to be elucidated whether any other cytosolic sensor acts upstream of caspase-4/-5 during encounters with Gram-positive bacteria. However, given that in our experimental set up, the cytosolic presence of LTA was not required to trigger caspase-4/-5 dependent IL-1β production, we believe that the upstream receptor(s) of this cascade are located on the cell surface. Since LPS stimulation of human monocytes has been reported to activate an alternative TLR4-linked and calcium-flux-dependent, one-step inflammasome pathway, it seems plausible that a similar mechanism might exist for TLR2 and LTA that potentially also involves caspase-4/-5. Indeed, TLR2 agonists have been implicated in the activation of the alternative inflammasome involving TRAF6, TAK1, and IKKβ in monocytes (Chen et al, 2023). Here, we found a dependence on TLR2 and the previously described alternative inflammasome component RIPK1 for IL-1β secretion solely for Spy EVs, indicating that inflammasome activation by intact S. pyogenes can be triggered by various sensor proteins. Consistent with these results, a previous report demonstrated that Spy EVs from a lgt mutant strain, characterized by a defect in lipoprotein modification, did not activate a TLR2-reporter cell line (Biagini et al, 2015). Furthermore, contrary to monocytes infected with S. pyogenes, monocytes stimulated with Spy EVs display hallmarks of the alternative inflammasome pathway (Gaidt et al, 2016) by lacking ASC speck formation or cleavage of GSDMD, and the resulting IL-1β secretion was also independent of potassium efflux. Similarly, previous research suggests that ASC specks are not required for caspase-1 activation in all instances (Nagar et al, 2021). While monocytes infected with S. pyogenes partially exhibit characteristics of the classical NLRP3 inflammasome, we found a comparable induction of caspase cleavage in both WT and a slo deletion strain, although Slo has been proven to be essential for inflammasome engagement in macrophages in providing signal 2 (Harder et al, 2009; Richter et al, 2021). In monocytes, however, signal 2 is dispensable and a TLR8 mediated one-step inflammasome activation process appears to take place. Moreover, both S. pyogenes and its EVs rely on caspase-4/-5 and -8 activities to facilitate IL-1β secretion. Caspase-4/-5 have been identified to facilitate LPS-induced IL-1β release in monocytes (Viganò et al, 2015), but how they fit into the alternative inflammasome pathway described by Gaidt et al, 2016 is currently unknown. Of note, we were unable to find evidence of caspase cleavage in response to Spy EVs. Given that the EV-induced IL-1β response is significantly weaker than that induced by S. pyogenes, this could be due to a sensitivity issue that prevents us from detecting caspase cleavage products. However, caspase activity does not always result in cleavage (Stennicke et al, 1999; Guey et al, 2014; Krause et al, 2018). In fact, initiator caspases (Muzio et al, 1998; Boatright et al, 2003; Pop et al, 2006), but also caspase-1 (Conos et al, 2016), have been reported to be activated by dimerization, with cleavage serving either to stabilize the dimer or to terminate protease activity (Boucher et al, 2018). In line with the aforementioned missing signs for classical inflammasome activation, it is plausible to assume that Spy EV induced-caspase activation is mild and does not necessarily result in cleavage. Finally, the discovery of additional players such as the tyrosine-protein kinase SYK as well as the adaptor protein SCIMP mediating inflammasome activation through TLR2/4 (Zewinger et al, 2020), suggests that there might not be a unique signaling pathway for the alternative inflammasome. Instead, some degree of exchangeability between signaling molecules might ensure the proper transmission of danger signals depending on their origin. Future studies are necessary to determine the exact sequence and molecules involved in caspase-1 activation and subsequent IL-1β release in response to S. pyogenes or its derived EVs.
The leaderless cytokine IL-1β is generally released during pyroptosis, requiring GSDMD-dependent pore formation (Evavold et al, 2018; Heilig et al, 2018). However, the absence of cell death in response to Spy EVs points towards a GSDMD-independent route of secretion. Notably, secretory lysosomes have been described to serve as non-classical pathway for IL-1β release upon LPS-induced alternative inflammasome activation in human monocytes (Andrei et al, 1999). Likewise, we observed elevated IL-1β levels in monocytes stimulated with BafA1 to excrete lysosomes. In addition, we noticed increased autophagosome formation and turn over in response to Spy EVs and LTA, indicating the induction of autophagic processes. Accordingly, autophagy has been shown to be important for monocyte survival and differentiation (Zhang et al, 2012). Since increased autophagy has also been reported to promote unconventional IL-1β secretion (Dupont et al, 2011), further studies should evaluate the molecular composition of the monocytes soluble and vesicular proteome following Spy EV treatment.
In this study, we titrated and evaluated a biologically relevant Spy EV dosage based on particle count (and not on protein content, as mostly reported in bacterial EV-host-cell-response experiments) to avoid excessive over-dosing. Moreover, because EV isolation techniques are continuously evolving, we set out to refine our EV-isolation procedures with the aim to localize LTA (being the primary agent for monocyte activation) to vesicular and non-vesicular/soluble entities. To this end, monocytes were incubated with EV-depleted supernatants as well as Spy EVs isolated using SEC, presumably depleted from potential non-vesicular secreted entities to a higher extend as compared to EVs UC. Nonetheless, the monocyte´s responses to SEC EVs were comparable to EVs UC. We conclude that the additional sample preparation steps are informative but increase the risk of contamination. Decent EV amounts are hardly feasible to obtain for encapsulated, non-hypervesiculating S. pyogenes strains and may not cover the entire size spectrum of EVs present in culture supernatants, as is the case with ultracentrifugation.
Monocytes are usually short-lived cells, that rapidly undergo apoptosis through caspase-3 activation in the absence of adequate survival signals (Fahy et al, 1999; Goyal et al, 2002). We provide evidence that cleavage of caspase-3 is abrogated when monocytes are stimulated with LTA derived from either Spy EVs (UC and SEC) or EV-depleted supernatants. In addition, we found phosphorylation of the MAP kinase p44/42 (ERK1/2), previously shown to promote monocyte survival and differentiation (Bianchi et al, 2007; Farzam-Kia et al, 2023). Interestingly, TLR2 has been demonstrated to induce selective autophagy in an ERK1/2-dependent manner (Anand et al, 2011; Chang et al, 2013; Lu et al, 2017). These reports are consistent with our findings of increased LC3-II turn over upon Spy EV and LTA treatment. Lastly, we observe that IL-6 and particularly IL-1β secretion in response to Spy EVs highly correlates with the LTA content in our EV preparations. Overall, we find free and vesicular Spy LTA promoting IL-1β release while inhibiting apoptotic and pyroptotic cell death mechanisms. Since LTA has been demonstrated to induce antibody secretion and memory B cells (Yi et al, 2024) as well as dendritic cell maturation (Wesa and Galy, 2001; Luft et al, 2002), we speculate that vesicular LTA may serve as a “booster” to establish a humoral immune response. As we have previously shown IgG-immune reactive epitopes on Spy EV components (Resch et al, 2016), LTA-deficient S. pyogenes may be useful for follow-up experiments to investigate whether other factors within Spy EVs exhibit immunomodulatory properties.
Altogether, our study highlights a new role for TLR2 and caspase-4/-5 in the recognition of Gram-positive EVs in human monocytes, balancing cell death, proteostasis, and autophagy mechanisms. Whether Spy EVs have an overall protective role during S. pyogenes infection or, on the contrary, exacerbate the disease in humans, remains to be shown. Given that no causative organism could be identified in 70% of sepsis cases (Paoli et al, 2018), it is possible that systemically distributed microbial products, such as EVs, may induce an inflammation overshoot after the parental strain has been eradicated by antibiotics or trigger flares of inflammation resulting in chronic inflammatory diseases. In fact, for Gram-negative bacteria, OMVs have already been shown to be sensed by caspase-11 in mice (Vanaja et al, 2016) leading to sepsis-like symptoms (Park et al, 2010; Svennerholm et al, 2017; Kim et al, 2018), endothelial cell activation (Soult et al, 2014; Kovarik et al, 2016), and even disseminated intravascular coagulation (DIC) (Wang et al, 2019; Peng et al, 2020). Notably, an OMV-producing Neisseria meningitidis strain was the causative agent in a fatal case of meningococcal septicemia (Namork and Brandtzaeg, 2002). In contrast, evidence for the pathophysiological role of EVs from Gram-positive pathogens is scarce. To date, only EVs from S. aureus have been implicated in the development of atopic dermatitis as well as asthma (Kim et al, 2012; Hong et al, 2014). However, the discovery of S. aureus EVs in house dust supports their persistence. Thus, bacterial EVs circulating in human body fluids or tissues after an overcome infection, may have unknown long-lasting immunomodulatory effects and may contribute to sterile inflammation processes. Strikingly, the expression of the metalloprotease STEAP4, which in our study was upregulated solely in response to Spy EVs, has been demonstrated to be associated with chronic inflammation in patients with obesity (Catalán et al, 2013), diabetes (Taylor et al, 2025), or inflammatory bowel disease (Xue et al, 2017). However, despite the identification of cross-reactive antibodies against S. pyogenes, that also recognize epitopes in heart and brain tissues, and are responsible for streptococcal post-infectious autoimmune sequelae (Cunningham, 2016), it is not known whether hypervesiculating strains or their EVs alone contribute to the emergence of these pathologies. Our study uncovers Spy EV constituent LTA as a critical factor for a non-toxic innate immune response, which may tip the balance in acute situations such as sepsis or have long-lasting effects triggering flares of low-level inflammation during chronic or recurring infections (where increased EV release is warranted). Therefore, future experiments should explore the role of Spy EVs in severe complications of rheumatic fever such as rheumatic heart disease or neurological disorders using in vivo models.
Methods
Reagents and tools table
| Reagent/resource | Reference or source | Identifier or catalog number | ||
|---|---|---|---|---|
| Experimental models | ||||
| BLaER1 wild-type | Prof. Thomas Graf (Rapino et al, 2013) | N/A | ||
| BLaER1 TLR2−/− | Prof. Holger Heine, this study | N/A | ||
| BLaER1 MYD88−/− |
Prof. Holger Heine (Vierbuchen et al, 2017) |
N/A | ||
| BLaER1 ASC−/− |
Prof. Holger Heine (Vierbuchen et al, 2017) |
N/A | ||
| BLaER1 CASP4−/− | Prof. Holger Heine, this study | N/A | ||
| BLaER1 Non-targeting cells | This study | N/A | ||
| BLaER1 CASP5−/− clone 1 and 2 | This study | N/A | ||
| BLaER1 CASP1−/− clone 1 and 2 | This study | N/A | ||
| BLaER1 NLRP3−/− clone 1 and 2 | This study | N/A | ||
| HEK-Blue™ hTLR2 Cells | Invivogen | hkb-htlr2 | ||
| ISS3348, M1 serotype Streptococcus pyogenes | G. Teti, University of Messina, Italy | N/A | ||
| M1T1 5448, M1 serotype Streptococcus pyogenes | M. Walker, University of Queensland, Australia | N/A | ||
| M1T1 5448 AP, M1 serotype Streptococcus pyogenes | M. Walker, University of Queensland, Australia | N/A | ||
| ISS3348 Δslo, M1 serotype Streptococcus pyogenes | This study | N/A | ||
| SA113, Staphylococcus aureus | ATCC | 35556 | ||
| 168, Bacillus subtilis | ATCC | 23857 | ||
| NEM316, Streptococcus agalactiae | S. Dramsi, Institute Pasteur, France | N/A | ||
| Recombinant DNA | ||||
| pUC19Ωlox71-PermAM/B-ermAM/B-lox66 | Lab collection | pEC454 | ||
| pEC85ΩPgyrA-cre | Lab collection | pEC455 | ||
| pRO1600/ColE1, bla | 10.1093/nar/gks1119 | pEC801 | ||
| pEC801Ωslo(up)-lox71-PermAM/B-ermAM/B-lox66-slo(down) | This study | pEC2898 | ||
| Antibodies | ||||
| Anti-S. pyogenes LTA | QEDBio | 15711 | ||
| Anti-Streptolysin O (Hemolytic streptococcus) antibody | Abcam | ab188539 | ||
| Gold-conjugated goat anti-mouse IgG | Jackson Immuno | 115-215-146 | ||
| IL-1β (D3U3E) rabbit mAb | Cell Signaling | 12703 | ||
| Cleaved-IL-1β (Asp116) (D3A3Z) rabbit mAb | Cell Signaling | 83186 | ||
| HSP90 | Proteintech | 13171-1-AP | ||
| anti-Caspase-1 (p20) (human), mAb | Adipogen | AG-20B-0048-C100 | ||
| Anti-Caspase-4 (Human) mAb | Medical & Biological Laboratories Co., LTD. | M029-3 | ||
| Anti-Caspase-5 (Human) mAb | Medical & Biological Laboratories Co., LTD. | M060-3 | ||
| LC3A/B (D3U4C) XP® Rabbit mAb | Cell Signaling | 12741S | ||
| Caspase-3 (D3R6Y) Rabbit mAb | Cell Signaling | 14220 | ||
| Cleaved Caspase-3 (Asp175) (5A1E) | Cell Signaling | 9664 | ||
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb | Cell Signaling | 4370 | ||
| Gasdermin D (E8G3F) Rabbit mAb | Cell Signaling | 97558 | ||
| Cleaved Gasdermin D (Asp275) (E7H9G) Rabbit mAb | Cell Signaling | 36425 | ||
| GAPDH (14C10) Rabbit mAb | Cell Signaling | 2118 | ||
| ASC/TMS1 Monoclonal antibody | Proteintech | 67494-1-Ig | ||
| Anti-Streptococcus pyogenes Group A Carbohydrate antibody | Abcam | ab9191 | ||
| Amersham ECL rabbit IgG, HRP-lined whole Ab | GE Healthcare | NA934-1ML | ||
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | 7076S | ||
| Cy™2 AffiniPure™ Donkey Anti-Mouse IgM | Jackson ImmunoResearch | 715-225-140 | ||
| Cy™5 AffiniPure™ Donkey Anti-Goat IgG (H + L) | Jackson ImmunoResearch | 705-175-147 | ||
| Oligonucleotides and other sequence-based reagents | ||||
| Primers for qRT-PCR | Sigma-Aldrich | N/A | ||
| Gene | Oligo code | Sequence | F/R | |
| TUBB | OLEC11016 | TGGACTCTGTTCGCTCAGGT | F | |
| TUBB | OLEC11017 | TGCCTCCTTCCGTACCACAT | R | |
| GAPDH | OLEC11889 | ACTTTGGTATCGTGGAAGGACT | F | |
| GAPDH | OLEC11890 | GTAGAGGCAGGGATGATGTTCT | R | |
| IL1A | OLEC10990 | AGATGCCTGAGATACCCAAAACC | F | |
| IL1A | OLEC10991 | CCAAGCACACCCAGTAGTCT | R | |
| IL1B | OLEC11897 | CCAGTGAAATGATGGCTTATTAC | F | |
| IL1B | OLEC11898 | CTGTAGTGGTGGTCGGAGATT | R | |
| IL6 | OLEC10966 | ACTCACCTCTTCAGAACGAATTG | F | |
| IL6 | OLEC10967 | CCATCTTTGGAAGGTTCAGGTTG | R | |
| CCL4 | OLEC10978 | CTGTGCTGATCCCAGTGAATC | F | |
| CCL4 | OLEC10979 | TCAGTTCAGTTCCAGGTCATACA | R | |
| CASP5 | OLEC10996 | TTCAACACCACATAACGTGTCC | F | |
| CASP5 | OLEC10997 | GTCAAGGTTGCTCGTTCTATGG | R | |
| CASP4 | OLEC10998 | CAAGAGAAGCAACGTATGGCA | F | |
| CASP4 | OLEC10999 | AGGCAGATGGTCAAACTCTGTA | R | |
| WNT5A | OLEC11000 | ATTCTTGGTGGTCGCTAGGTA | F | |
| WNT5A | OLEC11001 | CGCCTTCTCCGATGTACTGC | R | |
| IFNG | OLEC10986 | TCGGTAACTGACTTGAATGTCCA | F | |
| IFNG | OLEC10987 | TCGCTTCCCTGTTTTAGCTGC | R | |
| IFNB1 | OLEC10988 | ATGACCAACAAGTGTCTCCTCC | F | |
| IFNB1 | OLEC10989 | GGAATCCAAGCAAGTTGTAGCTC | R | |
| PTGFRN | OLEC11006 | CCTGCAACGTCAGTGACTATG | F | |
| PTGFRN | OLEC11007 | AGTCCGCCTTAACAGGATCTC | R | |
| CCR2 | OLEC11008 | TACGGTGCTCCCTGTCATAAA | F | |
| CCR2 | OLEC11009 | TAAGATGAGGACGACCAGCAT | R | |
| Guide RNAs | Thermo Fisher Scientific | N/A | ||
| Target gene | Provider | Guide RNA target sequence | Primer sequence (5’-3’) | |
| Non-targeting | A35526, Thermo Fisher | AAAUGUGAGAUCAGAGUAAU | n/a | |
| TLR2 | Designed with Benchling software | TGTCCATATTTCCCACTCTC |
F: TGTAATTCCGGATGGTTGTGC R: CCTGAAACAAACTTTCATCGGTG |
|
| CASP1_C1 | CRISPR917229_SGM, Thermo Fisher | GACAGTATTCCTAGAAGAAC |
F: CCATGCATACTCTTGAAGCAGC R: GCACTCTCTCATGGCAAGTTTG |
|
| CASP1_C2 | CRISPR937503_SGM, Thermo Fisher | GGCATCTGCGCTCTACCATC |
F: CATTTAGTTTCACCCCGCTCC R: ACCATACACATGTACCTGCCC |
|
| CASP4 | Designed with Benchling software | TGCAGCTCATCCGAATATGG |
F: TGGGGATTAGGAGGGGACGATT R: ATGGTTACTGTCATCCCCACCC |
|
| CASP5_C1 | CRISPR641087_SGM, Thermo Fisher | ACACGATGTTCTGACATTGA |
F: TAACACTGCATGGGCCTTGG R: TGTGTGTTATTCGCTGGAGACA |
|
| CASP5_C2 | CRISPR718889_SGM, Thermo Fisher | GGGGCTCACTATGACATCGT |
F: TTTCATTTGCTAGTGCATGTGCG R: TAGAGGTTAGGGAAGGTGGCA |
|
| NLRP3_C1 | CRISPR1098152_SGM, Thermo Fisher | CGCTAATGATCGACTTCAAT |
F: CTCATCCGTGTGCCGTGTTC R: CACTATGTGCACTTGGCCACAA |
|
| NLRP3_C2 | CRISPR1098154_SGM, Thermo Fisher | GATCGCAGCGAAGATCCACA |
F: CTCATCCGTGTGCCGTGTTC R: CACTATGTGCACTTGGCCACAA |
|
| Primers for slo deletion | Sigma-Aldrich | N/A | ||
| Purpose | Code | Sequence 5’–3’ | F/R | Usage * |
| Generation of slo deletion strain | ||||
| pEC801 linearisation | OLEC9943 | CTGCAGGCATGCAAGCTTGCG | F | PCR |
| OLEC9944 | TCTAGAGGATCCCCGGGTACCGAG | R | PCR | |
| slo upstream fragment | OLEC12497 | ACCCGGGGATCCTCTAGAAAGTAACAATATGTATAAGGTGCCAAAG | F | PCR |
| OLEC12498 | TGTATGCTATACGAACGGTAGTCCTTCATACCTTTTTATCATTCTAAAATG | R | PCR | |
| slo downstream fragment | OLEC12499 | GCATACATTATACGAACGGTAGACTGGTTCAAGAGGTTCGTCAAG | F | PCR |
| OLEC12500 | AAGCTTGCATGCCTGCAGAGTTGGGGTCAAATCAGTCCAA | R | PCR | |
| lox71-PermAM/B-ermAM/B-lox66 | OLEC1943 | TACCGTTCGTATAGCATACATTATACGAAGTTATCCGTAGCGGTTTTCAAAATTTGCAACC | F | PCR |
| OLEC1932 | TACCGTTCGTATAATGTATGCTATACGAAGTTATTTATTTCCTCCCGTTAAATAATAGATAACTATTAAA | R | PCR | |
| PCR ligation | OLEC11417 | ACCCGGGGATCCTCTAGA | F | LM-PCR |
| OLEC11418 | AAGCTTGCATGCCTGCAG | R | LM-PCR | |
| Validation of locus | OLEC12281 | CATTGCTCCAAATGGTGACC | F | SEQ |
| OLEC12282 | GAGATTTCCAGCCTTCATTATAACC | R | SEQ | |
| Verification of virulence regulator gene integrity in mutant strains | ||||
| covRS | OLEC4856 | TCGCTAGAAGACTATTTGACCAT | F | PCR, SEQ |
| OLEC4867 | AAGACATCGCGATTGACAGT | R | PCR, SEQ | |
| OLEC3609 | GGCTATGTTCAAGTCTTTCATG | F | SEQ | |
| OLEC3610 | CCAAATAACTCAACAACTAGTAGC | R | SEQ | |
| mga | oliRN172 | AGTTGACTAACCAATTGATCTACGCCTTTT | F | PCR, SEQ |
| OLEC290 | TTAACCTCTGTTTGATTCGC | R | PCR, SEQ | |
| ropB | OLEC4854 | AGCGACTATCATCCGAAACAT | F | PCR, SEQ |
| OLEC4855 | GCCCTGGAGCTGTTGAGATA | R | PCR, SEQ | |
| *LM-PCR PCR-mediated ligation, SEQ sequencing | ||||
| Chemicals, enzymes and other reagents | ||||
| Recombinant Human IL-3 | Peprotech | 200-03 | ||
| Recombinant Human M-CSF | Peprotech | 300-25 | ||
| Recombinant β-Estradiol | Sigma-Aldrich | E2257-1MG | ||
| Cas9 TrueCutV2 | Thermo Fisher Scientific | A36498 | ||
| S. pyogenes LTA | Sigma-Aldrich | L3140-5MG | ||
| S. aureus LTA | Invivogen | tlrl-pslta | ||
| Bafilomycin A1 | Enzo lifesciences | BML-CM110-0100 | ||
| Ac-YVAD-cmk | Invivogen | inh-yvad | ||
| MCC950 | Invivogen | inh-mcc | ||
| VX-765 | Invivogen | inh-vx765i-1 | ||
| Ac-LEVD-CHO | Enzo lifesciences | ALX-260-065 | ||
| Ac-LEVD-AFC | Enzo lifesciences | ALX-260-084 | ||
| EasySep Human Monocyte Isolation Kit | Stemcell | 19359 | ||
| Human GM-CSF | Miltenyi Biotec | 130-093-866 | ||
| LDH-Cytotoxicity Assay Kit II | Abcam | ab65393 | ||
| P3 Primary Cell 4D-Nucleofector Kit | Lonza | V4XP-3012 | ||
|
Power SYBR™ Green RNA-to-CT™ 1-Step Kit |
Thermo Fisher Scientific | 4367659 | ||
| Human IL-6 ELISA | R&D Systems | DY206 | ||
| Human IL-1β ELISA | R&D Systems | DY201 | ||
| Human IL-1α ELISA | R&D Systems | DY200 | ||
| Pro-QTM Emerald 300 Lipopolysaccharide Gel Stain Kit | Thermo Fisher Scientific | P20495 | ||
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | Thermo Fisher Scientific | 34580 | ||
| GSK3145095 | Selleckchem | S8845 | ||
| TLR2-IN-C29 | Selleckchem | S6597 | ||
| Cu-CPT22 | Selleckchem | S8677 | ||
| CU-CPT9a | Invivogen | inh-cc9a | ||
| CLI-095 | Invivogen | tlrl-cli95-4 | ||
| M5049 | Invivogen | inh-m5049 | ||
| TLR9-IN-1 | Hycultec | HY-148045 | ||
| Ac-IETD-CHO | Cayman Chemical | 27100 | ||
| HEK-Blue™ Selection | Invivogen | hb-sel | ||
| Normocin® | Invivogen | ant-nr-1 | ||
| HEK-Blue™ Detection | Invivogen | hb-det2 | ||
| Hoechst 33342 Solution (20 mM) | Thermo Fisher Scientific | 62249 | ||
| LTA (Lipoteichoic Acid) ELISA Kit | Biocat | AKR-5153-CB | ||
| TRPS Calibration Particles | Izon | CPC100, CPC200 | ||
| Nanopores | Izon | NP200, NP150, NP100 | ||
| Izon Reagent kit | Izon | RK3 | ||
| Software | ||||
| GraphPad Prism v10.0 | GraphPad | http://www.graphpad.com | ||
| NTA 3.1 analytical software | Malvern Panalytical | https://www.malvernpanalytical.com | ||
| Benchling gRNA design tool | Benchling | https://www.benchling.com | ||
| ICE Analysis | Synthego webtool | https://ice.synthego.com | ||
| DIA-NN 1.7.12 | Demichev et al, 2020 | N/A | ||
| DEseq2 (version 1.26.0.) | Love et al, 2014 | N/A | ||
| STAR (version 2.5.2a) | Dobin et al, 2013 | N/A | ||
| RNASeQC (version 2.1.0) | DeLuca et al, 2012 | N/A | ||
| QuPath | Bankhead et al, 2017 | https://qupath.github.io/ | ||
| Exoid Control Suite (v1.4.0.88) Izon Data Suite v1.0.2.32 v.3.1) | Izon | https://support.izon.com/how-can-i-get-the-latest-software-release | ||
| Other | ||||
| Buffy coats | German Red Cross Blood Transfusion Service | N/A | ||
| 8 Well Chamber, removable | ibidi | 80841 | ||
| qEV1 Columns 70 nm | IZON | IC1-70 | ||
| Nalgene™ Rapid-Flow™ Sterile Disposable Filter Units with PES Nylon Membrane | Thermo Fisher Scientific | 166-0045 | ||
| Amicon Ultra 15-30 kDa MWCO | Merck, Millipore | UFC9030 | ||
| Amicon Ultra 4-10 kDa MWCO | Merck, Millipore | UFC201024 | ||
Materials availability
BLaER1 knock-out cell lines newly generated for this study in our laboratory can be requested directly from the corresponding authors. The availability of the TLR2, MYD88, ASC, and CASP4 BLaER1 knock-out cell lines is limited due to an MTA agreement. These cell lines can be requested directly from the laboratories where they were generated.
Bacterial strains
S. pyogenes ISS3348 (G. Teti, University of Messina, Italy), M1T1 5448, M1T1 5448 AP (both M. Walker, University of Queensland, Australia), and S. agalactiae NEM316 (S. Dramsi, Institute Pasteur, France) were grown in Todd Hewitt Broth (THB, BD) at 37 °C with 5% CO2 without shaking. Trypticase soy agar (TSA, BD) supplemented with 3% sheep blood (Xebio Diagnostics) was used as a solid medium. B. subtilis 168 (ATCC 23857) and S. aureus SA113 (ATCC 35556) were grown in Brain Heart Infusion agar or broth (BHI, Sigma-Aldrich) at 37 °C with shaking.
Eukaryotic cells: BLaER1 cells
BLaER1 wild-type cells were a gift from Thomas Graf (Center for Genomic Regulation, Barcelona, Spain) (Rapino et al, 2013). BLaER1 ASC−/−, MYD88−/−, CASP4−/−, and TLR2−/− knockout cells were kindly provided by Holger Heine (German Center for Lung Research, Borstel, Germany). BLaER1 CASP1−/−, CASP5−/−, and NLRP3−/− were generated during this study. Cells were maintained in RPMI 1640 medium with Glutamax (Thermo Fisher Scientific) supplemented with 1% penicillin–streptomycin (P/S, Sigma-Aldrich), 10% FBS and 1% sodium pyruvate (both from Thermo Fisher Scientific) at 37 °C in a humidified atmosphere containing 5% CO2. Sex of the cell lines was not a consideration in this study. All cell lines were routinely tested for mycoplasma contamination with the MycoAlertTM Mycoplasma Detection Kit (Lonza).
Eukaryotic cells: primary monocytes
Human primary monocytes were purified from buffy coats from the German Red Cross Blood Transfusion Service upon ethical approval of the local authorities. For the isolation, buffy coats were diluted 1:1 in PBS and centrifuged (300 × g, 10 min, room temperature). The upper fraction was removed to reduce the final presence of platelets and the remainder was layered and separated in Pancoll density gradients (1.077 g/ml, Pan Biotech; 800× g, 20 min, room temperature, without break). Peripheral Blood Mononuclear Cells (PBMCs) were collected from the interphase and washed three times (500 × g, 10 min, 20 °C) with PBS containing 2% FBS and 0.1% Ethylenediaminetetraacetic acid (EDTA, Thermo Fisher Scientific). PBMCs were counted with CASY (OMNI Life Science) and resuspended at 50 × 106 cells/ml for monocyte purification. Classical monocytes (CD14+CD16−) were separated using the EasySep Human Monocyte Isolation Kit (Stemcell) according to the manufacturer’s instructions. Monocytes were maintained in RPMI 1640 medium with Glutamax supplemented with 10% FBS at 37 °C in a humidified atmosphere containing 5% CO2. In some experiments, monocytes were allowed to differentiate into adherent macrophages by prolonged cultivation (7 days) in the presence of 100 ng/mL GM-CSF. Sex of the primary cells was not considered in this study.
Eukaryotic cells: HEK-Blue TLR2 cells
HEK-Blue TLR2 cells (Invivogen) were maintained in DMEM medium supplemented with 1% penicillin–streptomycin (P/S, Sigma-Aldrich), 10% FBS, 100 µg/mL Normocin, and 1× HEK-Blue selection antibiotic mix at 37 °C in a humidified atmosphere containing 5% CO2.
Bacterial EVs: isolation and characterization
S. pyogenes and S. agalactiae EVs were isolated from late-logarithmic phase cultures in THB (OD600 nm: 0.4 in an Epoch 2, Agilent microplate reader). B. subtilis and S. aureus EVs were isolated from late-logarithmic phase cultures in BHI (OD600 nm: 0.8). 450 mL bacterial cultures were first centrifuged (9300 × g, 10 min, 10 °C) using a JLA 10.500 rotor and fitting tubes (both from Beckman Coulter), and cell free supernatants were centrifuged again to remove remaining cell debris (18,600 × g, 15 min, 10 °C). The supernatants were filtered twice through 0.22 µm polyethylsulfone membranes (Thermo Fisher Scientific) and EVs purified by ultracentrifugation at 175,000 × g (42,000 rpm) for 4 h at 10 °C using a Ti45 rotor with 70 ml polycarbonate-fitting tubes and an Optima XPN 100 ultracentrifuge. EV-pellets were resuspended and merged in phosphate-buffered saline (PBS, Thermo Fisher Scientific), centrifuged again at 175,000 × g for 3 h at 10 °C to concentrate and remove non-EV soluble proteins. Each final batch was resuspended in 1 ml of PBS and characterized using the NTA Nano Sight 300 device (NS300, Malvern Panalytical). The EV concentration (particles/mL) and size distribution of the particles in suspension (nm) were determined using the NTA 3.1 analytical software for data analysis (Malvern Panalytical) and revealed an average concentration for Spy EVs of ~7 × 1011 EVs/ batch. In addition, size, concentration, and zeta-potential were determined by tunable resistance pulse sensing (TRPS) using the Exoid (ECS v1.4.0.88) using appropriate nanopores (100, 150 and 200 nm) and calibration beads (CPC100, CPC200 and TKP200). 0.22 µm filtered PBS was used as the electrolyte and sample diluent. Size, concentration, or zeta potential were measured at 3 pressures (P1, P2, P3) or 3 voltages (V1, V2, V3) according to Izon-recommendations. Post-acquisition data processing was performed within the Izon Control Suite (v.3.1) in multi-and single pressure or charge analyses modules for size-concentration or zeta determination, respectively. Size-concentration histograms are provided in Appendix Fig. S2B.
For LTA-depleted EVs, S. pyogenes cultures were grown in the presence of 5 µM LtaS-IN-1771 (Richter et al, 2013) until reaching OD600 nm: 0.4 and further processed as described above.
Bacterial EVs: size exclusion chromatography (SEC) and characterization
A batch of double-filtered S. pyogenes supernatant was split into half to compare Spy EVs purified by conventional ultracentrifugation or size exclusion chromatography (SEC). For SEC, the supernatant was concentrated using an Amicon Ultra-15 centrifugal filter with a molecular weight cut-off 30 kDa (Merck, Millipore). In total, 1 mL of the concentrate were loaded onto a qEV 70 nm column and 28 fractions of 0.5 mL including void volume (3.5 ml) were collected in PBS. Fractions containing Spy EVs were identified by separation on a 12% SDS-PAGE and SYPRO Ruby protein gel staining. The EV containing fractions were pooled and concentrated using an Amicon Ultra-4 centrifugal filter device (MWCO 10 kDa). PBS only was concentrated using an Amicon Ultra-4 centrifugal filter device as well to serve as negative control. Spy EVs size, concentration, and zeta potential were determined as described above with the Exoid. SDS-Page protein profiles of SEC, size-concentration histograms, and IL-1β response to SEC are provided in Appendix Figs. S2 and S3.
EV-depleted supernatants
EV-depleted supernatants were concentrated after EVs have been removed using ultracentrifugation using an Amicon Ultra-15 centrifugal filter device (MWCO 10 kDa). The final volume of the concentrated supernatant was about 3.2 times higher than that of the Spy EV batches. THB media alone was concentrated as well and used as control in monocyte experiments.
Streptolysin O deletion
The chromosomal deletion of slo in S. pyogenes ISS348 was performed using the Cre-lox recombination approach as previously described (Le Rhun et al, 2017). The up- and downstream flanking regions of slo were amplified from S. pyogenes ISS3348 genomic DNA and ligated to the lox71-PermAM/B-ermAM/B-lox66 cassette by PCR using the primers listed in Reagents and Tools Table. The slo(up)-lox71-PermAM/B-ermAM/B-lox66-slo(down) fragment was then integrated into pEC801 using Gibson Assembly® Master Mix generating pEC2898. S. pyogenes ISS3348 competent cells were adjusted to an OD620 of 1.6 in 100 µL and incubated with 16.5 µg of Cfr42I-linearized pEC2898 on ice for 50 min before electroporation in a 0.1 cm electrode gap cuvette (1.5 kV, 400 Ω and 25 μF pulse; BioRad Gene Pulser Xcell electroporation system). After recovery in THB for 3 h, mutants were selected for two passages on TSA plates with 3% blood and 3 µg/mL erythromycin, cultured in THB without antibiotics, and flash-frozen in liquid nitrogen in THB with 20% glycerol. The obtained strains were validated by Sanger sequencing using OLEC12281 and OLEC12282.
To remove the ermAM/B resistance marker, competent cells of ISS3348 Δslo::lox71-ermAM/B-lox66 were adjusted to an OD620 of 2 in 100 µL and incubated with 500 ng of pEC455 on ice for 1 h. After electroporation at 1.5 kV, 400 Ω and 25 μF, cells were recovered for 2–3 h in THB. Transformants were selected on TSA plates with 3% blood and 300 µg/mL kanamycin and then grown in THB with 300 µg/mL kanamycin. To lose the Cre-expressing pEC455, transformants were then passaged four times in the absence of kanamycin, and strains susceptible to both kanamycin and erythromycin were finally cultured in THB and flash-frozen in liquid nitrogen in THB with 20% glycerol. The marker-less deletion strain of slo was validated by Sanger sequencing using primers targeting the slo locus and the virulence regulators mga, covRS and ropB (Reagents and Tools Table).
Transdifferentiation of BLaER1 cells
BLaER1 cells were transdifferentiated into monocyte-like cells for 7 days in six-well plates by seeding 106 cells/well in 3 mL of RPMI 1640 medium with Glutamax, 1% P/S, 10% FBS, 1% sodium pyruvate, 10 ng/mL of human recombinant IL-3, 10 ng/mL of human recombinant Macrophage Colony-stimulating Factor (both from PeproTech) and 100 nM of β-Estradiol (Sigma-Aldrich). Half of the medium was exchanged on days 2 and 5 with fresh medium containing the same supplements. On day 7, cells were harvested, counted with CASY and resuspended in fresh medium without cytokines or P/S at a final density of 1 × 106 cells/mL. Cells were seeded according to the experiment and maintained in incubators at 37 °C in a humidified atmosphere containing 5% CO2.
Construction of BLaER1 knock-outs
Guide RNAs (gRNA) designed with CRISPR gRNA design tool (Benchling) or commercially predesigned were selected for high on-target and low off-target scores (Reagents and Tools Table). Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) editing of BLaER1 cells was performed using the P3 Primary Cell 4D-Nucleofector Kit with strips (Lonza) according to the manufacturer’s protocol. Briefly, 1 µL of Cas9 TrueCutV2 (Thermo Fisher Scientific) + 1.6 µL of gRNA (100 µM) were complexed for 15 min at room temperature (RT). The complexed ribonucleoproteins were added to 106 cells previously resuspended in 20 µL of kit buffer and the mix was transferred to an electroporation cuvette. The Amaxa 4D-Nucleofector (Lonza, program EH-140) was used for electroporation and cells were transferred to pre-warmed medium and incubated in a 37 °C with 5% CO2. The whole bulk of targeted or non-targeted cells (control) was single-cell sorted in 96-well plates using a Sony SH800S Cell Sorter and individual clones were grown at 37 °C with 5% CO2. Genomic DNA was extracted with the NucleoSpin Tissue Kit (Macherey Nagel), PCR amplified (primers listed in the Reagents and Tools Table) and sent for Sanger sequencing (Microsynth). Upon confirmation of the presence of frameshift insertions/deletions (ICE Analysis Webtool Synthego), individual clones were stored in liquid nitrogen.
Infection assays and cell treatments
Infection assays were performed with S. pyogenes from late-logarithmic phase cultures in THB (OD600: 0.4). Bacteria were centrifuged (6000 × g, 3 min, 20 °C), washed with PBS and passed ten times through a 27 Gauge syringe insert (NeoLab) in a 1 mL syringe (VWR) to separate the cocci chains. Human monocyte assays were performed either in 96-well plates with 105 cells or 48-well plates with 106 cells per well. Cells were stimulated with a Multiplicity of Infection (MOI) of 5 or 7 × 108 particles of Spy EVs per 105 cells. Infection of BLaER1 cells was performed in 24 well plates with 106 cells per well. BLaER1 cells were stimulated with a MOI of 1 or 3.5 × 1010 particles of EVs. For experiments with EVs from other Gram-positive species, the volume of EVs was adjusted to add the same number of particles as for Spy EVs. Experiments with SEC EVs were performed in 48-well plates with 106 cells per well. Cells were stimulated with 9.8 × 106, 9.8 × 107, or 1.9 × 108 SEC EVs corresponding to 1, 10, or 20 µL of the SEC EV batch.
Upon bacterial or EV challenge, plates were centrifuged (300 × g, 5 min, 20 °C) for synchronization. For samples collected overnight (18 h post-treatment), 1% P/S was added after 1 h to inhibit bacterial replication and limit cell death. After centrifugation of the plates (500 × g, 5 min, 20 °C), the supernatants were collected at the designated time points and used for Lactate dehydrogenase assay (LDH). The remaining supernatants were stored at −20 °C until used for Enzyme-linked immunosorbent assay (ELISA). For immunoblotting experiments of cell culture supernatants, the FBS concentration of the media was reduced to 2%.
Purified S. pyogenes LTA (Sigma-Aldrich) or S. aureus LTA (Invivogen) was used at 1 µg/mL. LPS-B5 (lipopolysaccharide from E. coli serotype 055:B5, Invivogen) was used at 200 ng/mL. All chemical inhibitors were added 1 h prior to the addition of S. pyogenes, bacterial EVs or purified LTA. The inhibitors Ac-LEVD-CHO (Caspase-4/-5, Enzo Lifesciences), Ac-YVAD-cmk (caspase-1), Ac-IETD-CHO (caspase-8), MCC950 (NLRP3), and VX-765 (caspase-1/-4/-5) were used at a final concentration of 20 µM. GSK3145095 (RIPK1) was used at 2.5 µM. TLR2 inhibitors TLR2-IN-C29 and Cu-CPT22 were used at 50 µM and 10 µM. Final concentrations for other TLR inhibitors were: CLI-095 (TLR4; 0.05; 0.5; 5 µM), M5049 (TLR7/8; 0.1 and 1 µM), TLR9-IN-1 (TLR9; 5 µM), and Cu-CPT9a (TLR8; 5 µM).
RNA isolation
Human monocytes and BLaER1 cells were seeded at 1 × 106 cells/mL in six-well plates. Cells were left untreated or infected with S. pyogenes or treated with Spy EVs (7 × 109 or 7 × 1010 particles for monocytes or BLaER1 cells, respectively). Then, cells were collected by centrifugation (500 × g, 10 min, RT), and RNA was extracted with Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Samples were concentrated and DNAse-treated using the RNA Clean & Concentrator kit (Zymo Research). RNA quality was assessed with Agilent 2100 Bioanalyzer (Agilent Technologies) and a cut-off of RIN > 8 was established.
Library preparation and RNA sequencing
The Sequencing Core Facility at the Max Planck Institute for Molecular Genetics (Berlin, Germany) prepared cDNA libraries using the KAPA Stranded mRNA-Seq Kit (Roche) according to the manufacturer’s instructions. Monocyte and BLaER1 datasets were sequenced in single-end mode (75 base pairs) on a HiSeq 4000 instrument (Illumina) or a NextSeq 500 instrument (Illumina), respectively.
RNA sequencing: data processing
Fastq files from the Illumina sequencing were aligned with STAR (Dobin et al, 2013) against a combined reference containing the GRCh38 human genome reference (Schneider et al, 2017) and the S. pyogenes reference genome NC_002737.2 (Ferretti et al, 2001). Gene expression was quantified by counting overlapping features reads using RNASeQC 2.1.0 (DeLuca et al, 2012) against the Gencode v29 human gene annotation (Frankish et al, 2019). Normalization and differential gene expression (DE) were performed using the DEseq2 package (Love et al, 2014). Pathway analysis was performed using GAGE (Luo et al, 2009) and GO term enrichment analysis was generated using The Gene Ontology resource applying Bonferroni correction for multiple testing (Ashburner et al, 2000; Mi et al, 2019; Carbon et al, 2021). Data were visualized using Python programming environment using the matplotlib (John D. Hunter and Dept. of Computer Science, Univ. of Utah, Salt Lake, 2007), seaborn (Waskom, 2021), and bokeh (https://bokeh.org) libraries. Venn diagrams were generated using https://www.interactivenn.net/.
Gene expression analysis: quantitative PCR
The Power SYBR™ Green RNA-to-CT™ 1-Step Kit (Thermofisher Scientific) was used according to the manufacturer’s instructions. RNA samples were measured in a QuantStudio 5 qPCR machine (Thermo Fisher Scientific). Relative expression was calculated as ΔCt: Ct gene of interest-Ct housekeeping reference gene. All qPCR primers used in this study were selected from PrimerBank (Wang and Seed, 2003) and are summarized in the Reagents and Tools Table. For relative quantification, samples were normalized to TUBB and GAPDH.
LDH release assay and ELISA
Release of lactate dehydrogenase (LDH) was quantified using the Cytotoxicity Assay Kit II (Abcam) according to the manufacturer’s instructions. The percentage of cytotoxicity was calculated as follows:
Human IL-1α, IL-1β and IL-6 (R&D Systems) were measured by ELISA according to the manufacturer’s protocol. LTA was measured using the LTA ELISA Kit (Biocat) according to the manufacturer’s instructions.
Caspase-4/5 substrate cleavage assay
Monocytes were incubated in Hank’s balanced salt solution (HBSS) supplemented with 1% non-essential amino acids, 1% bovine serum albumin, 1% Glutamax, and 10 mM HEPES. At 20 h post-treatment, cell culture supernatants were harvested and cellular debris was removed by centrifugation at 500× g. 100 µL of the cell culture supernatant were mixed with 100 µL of caspase assay buffer (20 mM HEPES [pH7,4], 0,1 M NaCl, 10% Glycerol, 5 mM DTT) containing a final concentration of 250 µM Ac-LEVD-AFC in a 96-well black plate followed by incubation at 37 °C in a humidified atmosphere containing 5% CO2 for 2 h. The fluorescence was measured using a Cytation 5 microplate reader (Agilent BioTek) at ex/em 400/505 nm. All readings were normalized to untreated control wells.
TLR2 activity assay
HEK-Blue TLR2 cells were seeded in 96-well plates with 280,000 cells/200 µL of HEK-Blue Detection medium per well. Cells were stimulated for 18 h and production of the reporter gene secreted embryonic alkaline phosphatase (SEAP) upon TLR2 activation was monitored by absorbance measurement at 620 nm.
Immunoblot
In total, 30 μl of EVs were boiled with 6 µL of SDS-PAGE buffer (6×, Thermo Fisher Scientific) for 5 min at 95 °C. To determine the presence of LTA in LtaS-IN-1771-treated EV preparations, bacterial pellets of 50 mL cultures were boiled in PBS with 6× SDS-PAGE Buffer. The volume for LtaS-IN-1771-treated cultures was normalized to the OD600 measured for the corresponding DMSO-treated control culture for each time point. Protein extraction from monocytes was performed using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The resulting protein pellet was air-dried and dissolved in lysis buffer (2 M Thiourea, 8 M Urea, 0.065 M CHAPS, 0.13 M DTT, 0.079 M Tris). The Bradford method was used to determine protein concentrations. Equal amounts of protein were separated by 12% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. Membranes were incubated overnight with antibodies against LTA (#15711; QEDBio), pro-IL1B (#12703; Cell Signaling), and HSP90 (13171-1-AP; Proteintech). Corresponding secondary antibodies conjugated with horseradish peroxidase and in combination with SuperSignal West Pico PLUS Chemiluminescent Substrate (#34580; Thermo Fisher Scientific) were used to visualize bands on a Fusion FX imaging system (Vilber).
MS-based proteomics—monocytes
Cells were lysed and denatured in 4% SDS dissolved in PBS and processed according to the SP3 method (Hughes et al, 2019), digested with trypsin (SERVA) at a 1:50 enzyme-to-protein ratio and peptides desalted using OASIS HLB 96-well cartridges (Waters, #186001828BA). All samples were analyzed using a 90-min gradient and data-independent acquisition (DIA) on an Orbitrap Exploris 480 (Thermo Fisher Scientific) that was coupled to an RSLC nano UPLC (Thermo Fisher Scientific).
The MS was operated in DIA mode for single-injection quantitative measurements of individual samples with the following settings: 45k MS1 resolution, MS1 scan range 390–1010 m/z, 15k MS2 resolution, AGC target of 1000%, maximum injection time 22 ms, fixed normalized collision energy of 30. In all, 12 m/z precursor isolation windows were used in a staggered window pattern with optimized window placements from 394.4 to 1006.7 m/z.
DIA data were analyzed by DIA-NN 1.7.12 (Demichev et al, 2020) using the human fasta-database (UP000005640_9606, downloaded on 190111, 20412 entries). N-terminal methionine excision enabled. All data was filtered to 1% FDR. Cross-run normalization was enabled. Oxidation (M) was used as variable modification, and carbamidomethylation (C) was used as fixed modification. Default parameters were used unless specified otherwise. The unique genes table was used to obtain quantitative data on caspase-4 and caspase-5 protein abundance.
MS-based LPS analysis
All solvents and water used throughout all experiments were Fisher (Waltham, MA) Optima LC/MS grade. Intact LPS-B5 (lipopolysaccharide from E. coli serotype 055:K59(B5)H−, Source strain: ATCC 12014; CDC 5624-50 [NCTC 9701] from InvivoGen) was dissolved in a solution of 50% 2-propanol containing 10 mM ammonium acetate at an estimated LPS concentration of 1 μg/μL.
The LPS was directly infused via an ESI source at a flow rate of 5 μL min−1 into an Orbitrap Exploris 480 (Thermo Fisher Scientific) mass spectrometer. The mass spectrometer was operated in positive ionization mode with a capillary potential of 3 kV. MS1 (600–1800 m/z) and MS2 (100–1800 m/z) scans were acquired at 60 K resolution. Selected precursor ions were fragmented by stepped Higher Energy Collision Dissociation (@15,30,45,50%). LPS composition assignments and annotations are based on the chemical structure of LPS from E. coli as reported previously (Pupo et al, 2021). The supernatant obtained from the “bacterial EVs: isolation and quantification” step was also treated similarly to determine potential LPS contamination. MS/MS spectra are provided in Appendix Fig. S1.
MS-based proteomics—EVs
Data-independent acquisition (DIA)
EV samples were lysed and denatured in 4% SDS in PBS and processed following the SP3 protocol (Hughes et al, 2019) as described above. Label-free DIA analyses of peptides were acquired over 120 min by an Orbitrap Exploris 480 (Thermo Scientific) coupled to a 3000 RSLC nano UPLC (Thermo Scientific) from 1 µg of peptides based on Pierce™ Quantitative Peptide Assays & Standards (23275). Samples were loaded on a pepmap trap cartridge (300 µm i.d. x 5 mm, C18, Thermo) with 2% acetonitrile, 0.1% TFA at a flow rate of 20 µL/min. Peptides were separated over a 25 cm analytical column (PepSep C18, 75 µm I.D., 1.5 µm). Solvent A consists of 0.1% formic acid in water. Elution was carried out at a constant flow rate of 250 nL/min within 120 min. Initially, a two-step linear gradient was applied: 3–30% solvent B (0.1% formic acid in 80% acetonitrile) within 70.5 min, 30–45% solvent B within 13 min, followed by column washing and equilibration. The column was kept at a constant temperature of 50 °C.
The MS was operated in DIA mode for single-injection quantitative measurements of individual samples with the following settings: 60k MS1 resolution, MS1 scan range 375–1600 m/z, 15k MS2 resolution, MS2 scan range 120–1600 m/z, Normalized AGC target of 1000%, maximum injection time 54 ms, and fixed normalized collision energy of 30. In total, 12 m/z precursor isolation windows with optimized window placements from 400.4319 to 1210.8024 m/z.
DIA data analysis
Raw data analysis was performed using Spectronaut® (Biognosys AG, Zurich, Switzerland) version 19.7.250203.62635 in directDIA+ deep mode with reviewed UniProt databases (S. pyogenes serotype M1 – version 2024_02_05, 1,690 canonical entries). Methionine oxidation and Acetyl (Protein N-term) were set as a variable, and carbamidomethylation on cysteine residues was used as a static modification. The FDR for PSM-, peptide-, and protein-level was set to 0.01. All tolerances were set to dynamic for pulsar searches. All downstream data processing was carried out in R.
Scanning electron microscopy
Ten µl of sample were layered on 12 mm polylysinated coverslips and washed with PBS. For immunogold labelling, samples were blocked briefly with a BSA/cold water fish gelatin solution (Sigma-Aldrich), and reactions were incubated O/N with a 1:500 mouse anti-LTA antibody (QEDBio) diluted in 1% BSA at 4 °C. After three washes with PBS, the samples were incubated with gold-conjugated goat anti-mouse IgG (Jackson Immuno) for 1 h, washed four times with PBS and fixed in 2.5% glutaraldehyde (Electron Microscopy Sciences). Samples were then fixed in 0.5% osmium-tetroxide (Polysciences Inc), tannic acid (Sigma-Aldrich) and again in osmium-tetroxide. The coverslips were dehydrated in a graded ethanol series, dried in carbon dioxide at critical point and vacuum-coated with 3 nm carbon-platinum (Bal-Tec). Imaging was performed using a LEO 1550 scanning-electron microscope (Zeiss) at a 20 kV acceleration voltage. For topographic analysis, secondary electrons were detected with the in-lens secondary electron (SE) detector, while immunogold labelling was imaged using a detector for backscattered electrons (BSE).
Transmission electron microscopy
Overall, 10 µl of sample was applied to freshly glow discharged carbon-film-coated copper grids and allowed to adsorb for 6 min. After three washes with distilled water, the grids were contrasted with 4% phospho-tungstic-acid / 1% trehalose, touched on filter paper and air-dried. The grids were examined in a LEO 906 (Zeiss AG, Oberkochen) electron microscope operated at 100 kV and images were recorded with a Morada (SIS-Olympus, Münster) digital camera.
ASC speck formation
Monocytes were grown in removable 8-well chambers in slide format (ibidi). After treatment, cells were fixed with 4% PFA in PBS overnight at RT, permeabilized with 0.5% Triton X-100 in PBS for 1 min at RT, and non-specific binding was blocked for 30 min using a blocking buffer containing 3% normal donkey serum, 3% cold water fish gelatin, 1% BSA, and 0.05% Tween20 in PBS. Samples were incubated for 1 h at 37 °C with primary antibodies diluted in block buffer (mouse anti-ASC, 1:500 and goat anti-S. pyogenes Group A Carbohydrate antibody 1:1000). After washing, samples were incubated with the fluorophore-conjugated secondary antibodies a-mouse-Cy™2 and a-goat- Cy™5 both at 15 µg/mL. Hoechst 33342 at 5 µg/mL was added for DNA staining for 45 min at 37 °C. After washing, the removable silicon chambers were removed, and coverslips were mounted using Mowiol. Slides were imaged with a Zeiss AxioScan Z1 slide scanner (Zeiss, Jena, Germany) at ×20 magnification. Images were quantified using QuPath (Bankhead et al, 2017). Selected samples were imaged in more detail using an SP8 confocal microscope (Leica). Image series were prepared as collapsed confocal stacks using Arivis software.
Statistical analysis
Statistical tests were performed with GraphPad Prism 10.0.0 software, except for the RNA sequencing data. All quantitative data represent mean and standard deviation (SD). Figure legends indicate the number of biological replicates. Student’s t test was applied for pairwise comparisons. One-way or two-way ANOVA with Holm–Šídák’s multiple comparisons test were used for comparisons with more than two groups or for comparisons between groups with more than one variable, respectively. A P value of <0.05 or <0.01 (RNAseq) was defined as statistically significant. No blinding procedures were implemented in any of the experiments presented.
Supplementary information
Acknowledgements
We thank Karin Hahnke for measuring RNA concentrations with the Bioanalyzer and Dominik Rigo for measuring the protein content of EVs using the Bradford assay. We acknowledge the Max Planck Institute for Molecular Genetics for library preparation and RNA sequencing of the monocyte and BLaER1 samples. We thank Thomas Graf for providing BLaER1 wild-type cells. We acknowledge Knut Finstermeier for assistance with data analysis. We are much obliged to Thomas F Wulff for assistance in generating the S. pyogenes slo deletion strain. We are grateful to the members of the Charpentier Lab for their critical reading and discussion of the manuscript. This work was supported by the Max Planck Society (to EC), the German Research Foundation (DFG, Leibniz Prize to EC), Louis-Jeantet Foundation (Louis-Jeantet Price to EC), and the Tang Prize Foundation (Tang Prize to EC).
Expanded view
Author contributions
Kathrin Krause: Conceptualization; Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; Project administration; Writing—review and editing. Sandra Franch Arroyo: Conceptualization; Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; Project administration; Writing—review and editing. Matteo Ugolini: Investigation; Methodology. Tonya Kueck: Investigation; Methodology. Timothy J Sullivan: Formal analysis; Visualization. Eric J C Gálvez: Formal analysis; Visualization. Matthias Muenzner: Formal analysis; Investigation. Christian Goosmann: Investigation; Visualization. Volker Brinkmann: Formal analysis; Investigation; Visualization. Christian K Frese: Formal analysis; Investigation. Kathirvel Alagesan: Formal analysis; Investigation; Visualization. Tim Vierbuchen: Resources. Holger Heine: Resources. Ulrike Resch: Conceptualization; Resources; Formal analysis; Supervision; Investigation; Writing—review and editing. Leif E Sander: Resources; Supervision; Funding acquisition; Writing—review and editing. Emmanuelle Charpentier: Conceptualization; Supervision; Funding acquisition; Writing—review and editing.
Source data underlying figure panels in this paper may have individual authorship assigned. Where available, figure panel/source data authorship is listed in the following database record: biostudies:S-SCDT-10_1038-S44319-025-00558-7.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets produced in this study are available in the following databases: The code developed to analyze the RNA sequencing datasets of monocytes and BLaER1 cells has been deposited at https://github.com/MPUSP/human_RNA_seq_pipeline/releases/tag/v1.0.0. The raw RNA sequencing datasets are available under the NCBI-BioProject accession number: PRJNA753844. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al, 2022) partner repository (https://www.ebi.ac.uk/pride/profile/reviewer_pxd046570) with the dataset identifier PXD046570. Confocal images for ASC speck formation were deposited on BioImage Archive (Accession number: S-BIAD1725).
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-025-00558-7.
Disclosure and competing interests statement
The authors declare no competing interests.
Footnotes
These authors contributed equally: Kathrin Krause, Sandra Franch Arroyo.
Contributor Information
Kathrin Krause, Email: krause@mpusp.mpg.de.
Ulrike Resch, Email: ulrike.resch@meduniwien.ac.at.
Emmanuelle Charpentier, Email: research@emmanuelle-charpentier.org.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44319-025-00558-7.
References
- Anand PK, Tait SWG, Lamkanfi M, Amer AO, Nunez G, Pagès G, Pouysségur J, McGargill MA, Green DR, Kanneganti T-D (2011) TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J Biol Chem 286(50):42981–42991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A (1999) The secretory route of the leaderless protein interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol Biol Cell 10(5):1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed]
- Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG et al (2017) QuPath: open source software for digital pathology image analysis. Sci Rep 7(1):16878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini M, Garibaldi M, Aprea S, Pezzicoli A, Doro F, Becherelli M, Taddei AR, Tani C, Tavarini S, Mora M et al (2015) The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol Cell Proteom 14(8):2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G, Montecucco F, Bertolotto M, Dallegri F, Ottonello L (2007) Immune complexes induce monocyte survival through defined intracellular pathways. Ann N Y Acad Sci 1095:209–219 [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR et al (2003) A unified model for apical caspase activation. Mol Cell 11(2):529–541 [DOI] [PubMed] [Google Scholar]
- Boucher D, Monteleone M, Coll RC, Chen KW, Ross CM, Teo JL, Gomez GA, Holley CL, Bierschenk D, Stacey KJ et al (2018) Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J Exp Med 215(3):827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Palm NW, Sadanand S, Ryndak MB, Sutterwala FS, Flavell RA, Bliska JB, Medzhitov R (2010) A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7(5):376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados-Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13(10):620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP, Walker S (2013) Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M (2005) The global burden of group A streptococcal diseases. Lancet Infect Dis 5(11):685–694 [DOI] [PubMed] [Google Scholar]
- Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E et al (2021) The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49(D1):D325–D334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson CN, Yu J, Reyes VM, Taschuk FO, Yadav A, Copenhaver AM, Nguyen HT, Collman RG, Shin S (2015) Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc Natl Acad Sci USA 112(21):6688–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia V, Piersigilli A, Ebner F, Janos M, Goldmann O, Damböck U, Kröger A, Weiss S, Knapp S, Jamieson AM et al (2016) Type I interferon signaling prevents IL-1β-driven lethal systemic hyperinflammation during invasive bacterial infection of soft tissue. Cell Host Microbe 19(3):375–387 [DOI] [PubMed] [Google Scholar]
- Catalán V, Gómez-Ambrosi J, Rodríguez A, Ramírez B, Rotellar F, Valentí V, Silva C, Gil MJ, Salvador J, Frühbeck G (2013) Six-transmembrane epithelial antigen of prostate 4 and neutrophil gelatinase-associated lipocalin expression in visceral adipose tissue is related to iron status and inflammation in human obesity. Eur J Nutr 52(6):1587–1595 [DOI] [PubMed] [Google Scholar]
- Cecil JD, O’Brien-Simpson NM, Lenzo JC, Holden JA, Singleton W, Perez-Gonzalez A, Mansell A, Reynolds EC (2017) Outer membrane vesicles prime and activate macrophage inflammasomes and cytokine secretion in vitro and in vivo. Front Immunol 8:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-P, Su Y-C, Hu C-W, Lei H-Y (2013) TLR2-dependent selective autophagy regulates NF-κB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell Death Differ 20(3):515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Yu S, Gao Y, Li J, Wang X, Wei B, Meng G (2023) TRAF6-TAK1-IKKβ pathway mediates TLR2 agonists activating “one-step” NLRP3 inflammasome in human monocytes. Cytokine 169:156302 [DOI] [PubMed] [Google Scholar]
- Choi EJ, Lee HG, Bae IH, Kim W, Park J, Lee TR, Cho EG (2018) Propionibacterium acnes-derived extracellular vesicles promote acne-like phenotypes in human epidermis. J Investig Dermatol 138(6):1371–1379 [DOI] [PubMed] [Google Scholar]
- Conos SA, Lawlor KE, Vaux DL, Vince JE, Lindqvist LM (2016) Cell death is not essential for caspase-1-mediated interleukin-1β activation and secretion. Cell Death Differ 23(11):1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW (2016) Post-Streptococcal Autoimmune Sequelae: Rheumatic Fever and Beyond. In: Ferretti JJ, Stevens DL, Fischetti VA (eds) Streptococcus pyogenes: basic biology to clinical manifestations. University of Oklahoma Health Sciences Center, Oklahoma City, OK, p 703–733
- Deatheragea BL, Cooksona BT, Deatherage BL, Cookson BT (2012) Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immunity 80:1948–1957 [DOI] [PMC free article] [PubMed]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, Getz G (2012) RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28(11):1530–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M (2020) DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods 17(1):41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 30(23):4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrod T, Pelka K, Latz E, Kreikemeyer B, Dalpke AH (2015) TLR8 senses bacterial RNA in human monocytes and plays a nonredundant role for recognition of Streptococcus pyogenes. J Immunol 195(3):1092–1099 [DOI] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, Feldman MF (2014) Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5(2):e00909–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erttmann SF, Gekara NO (2019) Hydrogen peroxide release by bacteria suppresses inflammasome-dependent innate immunity. Nat Commun 10(1):3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC (2018) The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity 48(1):35–44.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy RJ, Doseff AI, Wewers MD (1999) Spontaneous human monocyte apoptosis utilizes a caspase-3-dependent pathway that is blocked by endotoxin and is independent of caspase-1. J Immunol 163(4):1755–1762 [PubMed] [Google Scholar]
- Farzam-Kia N, Moratalla AC, Lemaître F, Levert A, Da Cal S, Margarido C, Carpentier Solorio Y, Arbour N (2023) GM-CSF distinctly impacts human monocytes and macrophages via ERK1/2-dependent pathways. Immunol Lett 261:47–55 [DOI] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S et al (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA 98(8):4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieber C, Janos M, Koestler T, Gratz N, Li XD, Castiglia V, Aberle M, Sauert M, Wegner M, Alexopoulou L et al (2015) Innate immune response to Streptococcus pyogenes depends on the combined activation of TLR13 and TLR2. PLoS ONE 10(3):e0119727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Luoma S, Orench-Rivera N, Feeley EM, Haldar AK, Yamamoto M, Kanneganti TD, Kuehn MJ, Coers J (2017) Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. mBio 8(5):e01188–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J et al (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47(D1):D766–D773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AAB, Cooper MA, Graf T, Hornung V (2016) Human monocytes engage an alternative inflammasome pathway. Immunity 44(4):833–846 [DOI] [PubMed] [Google Scholar]
- Gaidt MM, Rapino F, Graf T, Hornung V (2018) Modeling primary human monocytes with the trans–differentiation cell line BLaER1. In: Dominic De Nardo, Christine M. De Nardo (eds) Methods in molecular biology. Humana Press Inc., pp 57–66 [DOI] [PubMed]
- Galle M, Schotte P, Haegman M, Wullaert A, Yang HJ, Jin S, Beyaert R (2008) The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase‐1 mediated IL‐1β maturation. J Cell Mol Med 12(5a):1767–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann O, Von Köckritz-Blickwede M, Höltje C, Chhatwal GS, Geffers R, Medina E (2007) Transcriptome analysis of murine macrophages in response to infection with Streptococcus pyogenes reveals an unusual activation program. Infect Immun 75(8):4148–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Wang Y, Graham MM, Doseff AI, Bhatt NY, Marsh CB (2002) Monocyte survival factors induce Akt activation and suppress caspase-3. Am J Respir Cell Mol Biol 26(2):224–230 [DOI] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CAL, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM (2002) Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA 99(21):13855–13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz N, Siller M, Schaljo B, Pirzada ZA, Gattermeier I, Vojtek I, Kirschning CJ, Wagner H, Akira S, Charpentier E et al (2008) Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J Biol Chem 283(29):19879–19887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko A, Yu S, Martin-Sanchez F, Diaz-del-Olmo I, Nichols EM, Davis DM, Brough D, Lopez-Castejon G (2020) Priming is dispensable for NLRP3 inflammasome activation in human monocytes in vitro. Front Immunol 11:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guey B, Bodnar M, Manié SN, Tardivel A, Petrilli V (2014) Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc Natl Acad Sci USA 111(48):17254–17259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim S, Il et al (2011) Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS ONE 6(11):e27958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, Núñez G (2018) The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell 175(6):1651–1664.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder JJ, Franchi L, Muñoz-Planillo R, Park J-H, Reimer T, Núñez G (2009) Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-κB activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol 183(9):5823–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, Tancredi S, Guiet C, Brière F, Vlach J, Lebecque S et al (2005) Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J Immunol 174(5):2942–2950 [DOI] [PubMed] [Google Scholar]
- Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF (2011) Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 286(2):1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JA, Mathur A, Ngo C, Man SM (2018) Cytosolic recognition of microbes and pathogens: inflammasomes in action. Microbiol Mol Biol Rev 82(4):e00015–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S (2004) Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303(5663):1526–1529 [DOI] [PubMed] [Google Scholar]
- Heilig R, Dick MS, Sborgi L, Meunier E, Hiller S, Broz P (2018) The Gasdermin-D pore acts as a conduit for IL-1β secretion in mice. Eur J Immunol 48(4):584–592 [DOI] [PubMed] [Google Scholar]
- Hong S-W, Choi E-B, Min T-K, Kim J-H, Kim M-H, Jeon SG, Lee B-J, Gho YS, Jee Y-K, Pyun B-Y et al (2014) An important role of α-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS ONE 9(7):e100499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J (2019) Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc 14(1):68–85 [DOI] [PubMed] [Google Scholar]
- Hynes W, Sloan M (2016) Secreted extracellular virulence factors. In: Ferretti JJ, Stevens DL, Fischetti VA (eds) Streptococcus pyogenes: basic biology to clinical manifestations. University of Oklahoma Health Sciences Center, Oklahoma City, OK, p 327–357
- Isaacs A, Lindenmann J (1957) Virus interference. I. The interferon. Proc R Soc Lond Ser B - Biol Sci 147(927):258–267 [DOI] [PubMed] [Google Scholar]
- Iula L, Keitelman IA, Sabbione F, Fuentes F, Guzman M, Galletti JG, Gerber PP, Ostrowski M, Geffner JR, Jancic CC et al (2018) Autophagy mediates interleukin-1β secretion in human neutrophils. Front Immunol 9:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John D, Hunter and Dept. of Computer Science, Univ. of Utah, Salt Lake, U (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9(3):90–95 [Google Scholar]
- Kaparakis-Liaskos M, Ferrero RL (2015) Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15(6):375–387 [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Walle L, Vande, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S et al (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479(7371):117–121 [DOI] [PubMed] [Google Scholar]
- Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C (2012) An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36(3):464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee J, Park K-S, Hong S-W, Gho YS (2018) Drug repositioning to alleviate systemic inflammatory response syndrome caused by Gram-negative bacterial outer membrane vesicles. Adv Healthc Mater 7(13):e1701476 [DOI] [PubMed] [Google Scholar]
- Kim MR, Hong SW, Choi EB, Lee WH, Kim YS, Jeon SG, Jang MH, Gho YS, Kim YK (2012) Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy: Eur J Allergy Clin Immunol 67(10):1271–1281 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogawa M, Sanada T, Mimuro H, Kim M, Ashida H, Akakura R, Yoshida M, Kawalec M, Reichhart J-M et al (2013) The Shigella OspC3 effector inhibits caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe 13(5):570–583 [DOI] [PubMed] [Google Scholar]
- Kovarik P, Castiglia V, Ivin M, Ebner F (2016) Type I interferons in bacterial infections: a balancing act. Front Immunol 7(7):652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Caution K, Badr A, Hamilton K, Saleh A, Patel K, Seveau S, Hall-Stoodley L, Hegazi R, Zhang X et al (2018) CASP4/caspase-11 promotes autophagosome formation in response to bacterial infection. Autophagy 14(11):1928–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Daily K, Estfanous S, Hamilton K, Badr A, Abu Khweek A, Hegazi R, Anne MN, Klamer B, Zhang X et al (2019) Caspase‐11 counteracts mitochondrial ROS‐mediated clearance of Staphylococcus aureus in macrophages. EMBO Rep 20(12):1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Cookson BT (2012) The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 12(6):799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Todd J, LaRock DL, Olson J, O’Donoghue AJ, Robertson AAB, Cooper MA, Hoffman HM, Nizet V (2016) IL-1b is an innate immune sensor of microbial proteolysis. Sci Immunol 1(2):eaah3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rhun A, Lécrivain A-L, Reimegård J, Proux-Wéra E, Broglia L, Della Beffa C, Charpentier E (2017) Identification of endoribonuclease specific cleavage positions reveals novel targets of RNase III in Streptococcus pyogenes. Nucleic Acids Res 45(5):2329–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-Y, Choi D-Y, Kim D-K, Kim J-W, Park JO, Kim S, Kim S-H, Desiderio DM, Kim Y-K, Kim K-P et al (2009) Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9(24):5425–5436 [DOI] [PubMed] [Google Scholar]
- Lei B, Mackie S, Lukomski S, Musser JM (2000) Identification and immunogenicity of group {A} {Streptococcus} culture supernatant proteins. Infect Immun 68(12):6807–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Beasley FC, Keller N, Hollands A, Urbano R, Troemel ER, Hoffman HM, Nizet V (2015) A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective interleukin 1β-dependent macrophage immune response. mBio 6(2):e00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XY, Choi MSK, Porter AG (2000) Expression analysis of the human caspase-1 subfamily reveals specific regulation of the CASP5 gene by lipopolysaccharide and interferon-γ. J Biol Chem 275(51):39920–39926 [DOI] [PubMed] [Google Scholar]
- Loof TG, Goldmann O, Medina E (2008) Immune recognition of Streptococcus pyogenes by dendritic. Cells 76(6):2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Xie D, Chen Y, Tian E, Muhammad I, Chen X, Miao Y, Hu W, Wu Z, Ni H et al (2017) TLR2 mediates autophagy through ERK signaling pathway in Mycoplasma gallisepticum-infected RAW264.7 cells. Mol Immunol 87:161–170 [DOI] [PubMed] [Google Scholar]
- Luft T, Jefford M, Luetjens P, Hochrein H, Masterman K-A, Maliszewski C, Shortman K, Cebon J, Maraskovsky E (2002) IL-1β enhances CD40 ligand-mediated cytokine secretion by human dendritic cells (DC): a mechanism for T cell-independent DC activation1. J Immunol 168(2):713–722 [DOI] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinforma 10(1):161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A (2001) Rapid secretion of interleukin-1β by microvesicle shedding. Immunity 15(5):825–835 [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM (2006) Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440(7081):228–232 [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell 10(2):417–426 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47(D1):D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G (2013) K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38(6):1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Aikawa C, Nozawa T, Nakatake A, Sakamoto K, Kikuchi T, Nakagawa I (2021) Biological effect of Streptococcus pyogenes-released extracellular vesicles on human monocytic cells, induction of cytotoxicity, and inflammatory response. Front Cell Infect Microbiol 11:711144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM (1998) An induced proximity model for caspase-8 activation. J Biol Chem 273(5):2926–2930 [DOI] [PubMed] [Google Scholar]
- Nagar A, Rahman T, Harton JA (2021) The ASC speck and NLRP3 inflammasome function are spatially and temporally distinct. Front Immunol 12:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ñahui Palomino RA, Vanpouille C, Costantini PE, Margolis L (2021) Microbiota–host communications: bacterial extracellular vesicles as a common language. PLoS Pathog 17(5):e1009508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namork E, Brandtzaeg P (2002) Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 360(9347):1741 [DOI] [PubMed] [Google Scholar]
- Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K et al (2014) Evolutionary pathway to increased virulence and epidemic group {A} {Streptococcus} disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA 111(17):E1768–E1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Nold-Petry CA, Nold MF, Joosten LAB, Opitz B, Van Der Meer JHM, Van De Veerdonk FL, Ferwerda G, Heinhuis B, Devesa I et al (2009) Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 113(10):2324–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya-Abril A, Prados-Rosales R, Mcconnell MJ, Martín-Peña R, González-Reyes JA, Jiménez-Munguía I, Gómez-Gascón L, Fernández J, Luque-García JL, García-Lidón C et al (2014) Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteom 106(4):46–60 [DOI] [PubMed] [Google Scholar]
- Orench-Rivera N, Kuehn MJ (2016) Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536 [DOI] [PMC free article] [PubMed]
- Paoli CJ, Reynolds MA, Sinha M, Gitlin M, Crouser E (2018) Epidemiology and costs of sepsis in the United States-an analysis based on timing of diagnosis and severity level. Crit Care Med 46(12):1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-S, Choi K-H, Kim Y-S, Hong BS, Kim OY, Kim JH, Yoon CM, Koh G-Y, Kim Y-K, Gho YS (2010) Outer membrane vesicles derived from Escherichia coli induce systemic inflammatory response syndrome. PLoS ONE 5(6):e11334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, Bigley V, Flavell RA, Gilroy DW, Asquith B et al (2017) The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 214(7):1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence MA, Rooijakkers SHM, Cogen AL, Cole JN, Hollands A, Gallo RL, Nizet V (2010) Streptococcal inhibitor of complement promotes innate immune resistance phenotypes of invasive M1T1 group A Streptococcus. J Innate Immun 2(6):587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Gao M, Liu Y, Qiu X, Cheng X, Yang X, Chen F, Wang E (2020) Bacterial outer membrane vesicles induce disseminated intravascular coagulation through the caspase-11-gasdermin D pathway. Thrombosis Res 196:159–166 [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M et al (2022) The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50(D1):D543–D552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS (2006) The apoptosome activates caspase-9 by dimerization. Mol Cell 22(2):269–275 [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B et al (2011) Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Investig 121(4):1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo E, van der Ley P, Meiring HD (2021) Nanoflow LC-MS method allowing in-depth characterization of natural heterogeneity of complex bacterial lipopolysaccharides. Anal Chem 93(48):15832–15839 [DOI] [PubMed] [Google Scholar]
- Rapino F, Robles EF, Richter-Larrea JA, Kallin EM, Martinez-Climent JA, Graf T (2013) C/EBPα induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep 3(4):1153–1163 [DOI] [PubMed] [Google Scholar]
- Reichmann NT, Gründling A (2011) Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett 319:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Nyunt Wai S, Charpentier E (2016) A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 7(6):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Monteleone MM, Cork AJ, Barnett TC, Nizet V, Brouwer S, Schroder K, Walker MJ (2021) Streptolysins are the primary inflammasome activators in macrophages during Streptococcus pyogenes infection. Immunol Cell Biol 99(10):1040–1052 [DOI] [PubMed] [Google Scholar]
- Richter SG, Elli D, Kim HK, Hendrickx APA, Sorg JA, Schneewind O, Missiakas D (2013) Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for Gram-positive bacteria. Proc Natl Acad Sci USA 110(9):3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A (2010) Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci USA 107(44):19002–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers NMK, McCumber AW, McMillan HM, McNamara RP, Dittmer DP, Kuehn MJ, Hendren CO, Wiesner MR (2023) Comparative electrokinetic properties of extracellular vesicles produced by yeast and bacteria. Colloids Surf B Biointerfaces 225:113249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D (2017) Lipoteichoic acid synthesis and function in gram-positive bacteria. In: Otto Geiger (ed) Biogenesis of fatty acids, lipids and membranes. Springer International Publishing, pp 1–18
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D et al (2017) Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27(5):849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ (1999) Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J Biol Chem 274(25):17406–17409 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ (2015) Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13(10):605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino C, Carta S, Gattorno M, Sitia R, Rubartelli A (2018) Progressive waves of IL-1β release by primary human monocytes via sequential activation of vesicular and gasdermin D-mediated secretory pathways. Cell Death Dis 9(11):1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpi M, Pertusati F, Morozzi C, Novelli G, Giannantonio D, Duggan K, Vittorio S, Fallis IA, De Luca L, Williams D (2023) Synthesis, molecular docking and antibacterial activity of an oxadiazole-based lipoteichoic acid inhibitor and its metabolites. J Mol Struct 1278:134977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK (2012) Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12(4):509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526(7575):660–665 [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F (2014) Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514(7521):187–192 [DOI] [PubMed] [Google Scholar]
- Soult MC, Dobrydneva Y, Wahab KH, Britt LD, Sullivan CJ (2014) Outer membrane vesicles alter inflammation and coagulation mediators. J Surg Res 192(1):134–142 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS (1999) Caspase-9 can be activated without proteolytic processing. J Biol Chem 274(13):8359–8362 [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM (2006) Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2(1):e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerholm K, Park K-S, Wikström J, Lässer C, Crescitelli R, Shelke GV, Jang SC, Suzuki S, Bandeira E, Olofsson CS et al (2017) Escherichia coli outer membrane vesicles can contribute to sepsis induced cardiac dysfunction. Sci Rep 7(1):17434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S (1999) Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11(4):443–451 [DOI] [PubMed] [Google Scholar]
- Tapper H, Sundler R (1995) Bafilomycin A1 inhibits lysosomal, phagosomal, and plasma membrane H(+)-ATPase and induces lysosomal enzyme secretion in macrophages. J Cell Physiol 163(1):137–144 [DOI] [PubMed] [Google Scholar]
- Taylor BE, Howell SJ, Lee C, Taylor Z, Barber K, Taylor PR (2025) Diabetes-mediated STEAP4 enhances retinal oxidative stress and impacts the development of diabetic retinopathy. Antioxidants 14(2):205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thänert R, Itzek A, Hoßmann J, Hamisch D, Madsen MB, Hyldegaard O, Skrede S, Bruun T, Norrby-Teglund A, Oppegaard O et al (2019) Molecular profiling of tissue biopsies reveals unique signatures associated with streptococcal necrotizing soft tissue infections. Nat Commun 10(1):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatsaronis JA, Walker MJ, Sanderson-Smith ML (2014) Host responses to group A Streptococcus: cell death and inflammation. PLoS Pathog 10(8):e1004266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens J, Vergauwen G, Van Deun J, Geeurickx E, Dhondt B, Lippens L, De Scheerder MA, Miinalainen I, Rappu P, De Geest BG et al (2020) Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 69:191–193 [DOI] [PMC free article] [PubMed]
- Uhlmann J, Rohde M, Siemens N, Kreikemeyer B, Bergman P, Johansson L, Norrby-Teglund A (2016) LL-37 triggers formation of Streptococcus pyogenes extracellular vesicle-like structures with immune stimulatory properties. J Innate Immun 8(3):243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A (1999) The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401(6755):811–815 [DOI] [PubMed] [Google Scholar]
- Unterberger S, Mullen L, Flint MS, Sacre S (2023) Multiple TLRs elicit alternative NLRP3 inflammasome activation in primary human monocytes independent of RIPK1 kinase activity. Front Immunol 14:1092799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama JA, Riestra AM, Gao NJ, Larock CN, Gupta N, Ali SR, Hoffman HM, Ghosh P, Nizet V (2017) Group A streptococcal M protein activates the NLRP3 inflammasome. Nat Microbiol 2(10):1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VAK (2016) Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165(5):1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O’Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC (2014) Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13(5):2420–2432 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Bang C, Rosigkeit H, Schmitz RA, Heine H (2017) The human-associated archaeon Methanosphaera stadtmanae is recognized through its RNA and induces TLR8-dependent NLRP3 inflammasome activation. Front Immunol 8:1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viganò E, Diamond CE, Spreafico R, Balachander A, Sobota RM, Mortellaro A (2015) Human caspase-4 and caspase-5 regulate the one-step non-canonical inflammasome activation in monocytes. Nat Commun 6(1):8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Liu Y, Qiu X, Tang Y, Wang H, Xiao X, Chen F, Lu B (2019) Bacteria-released outer membrane vesicles promote disseminated intravascular coagulation. Thromb Res 178:26–33 [DOI] [PubMed] [Google Scholar]
- Wang X, Eagen WJ, Lee JC (2020) Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proc Natl Acad Sci USA 117(6):3174–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B (2003) A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 31(24):e154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M (2021) seaborn: statistical data visualization. J Open Source Softw 6(60):3021 [Google Scholar]
- Watson DC, Johnson S, Santos A, Yin M, Bayik D, Lathia JD, Dwidar M (2021) Scalable isolation and purification of extracellular vesicles from Escherichia coli and other bacteria. J Vis Exp 2021(176):10.3791/63155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ME, Neely MN, Caparon MG (2016) Animal models of Streptococcus pyogenes infection. In: Ferretti JJ, Stevens DL, Fischetti VA (eds). Streptococcus pyogenes: basic biology to clinical manifestations. University of Oklahoma Health Sciences Center, Oklahoma City, OK, p 497–521
- Welsh JA, Goberdhan DCI, O’Driscoll L, Buzas EI, Blenkiron C, Bussolati B, Cai H, Di Vizio D, Driedonks TAP, Erdbrügger U et al (2024) Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles 13(2):e12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesa AK, Galy A (2001) IL-1β induces dendritic cells to produce IL-12. Int Immunol 13(8):1053–1061 [DOI] [PubMed] [Google Scholar]
- Williams JG, Ly D, Geraghty NJ, McArthur JD, Vyas HKN, Gorman J, Tsatsaronis JA, Sluyter R, Sanderson-Smith ML (2021) Streptococcus pyogenes M1T1 variants induce an inflammatory neutrophil phenotype including activation of inflammatory caspases. Front Cell Infect Microbiol 10:596023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzenrath M, Pache F, Lorenz D, Koppe U, Gutbier B, Tabeling C, Reppe K, Meixenberger K, Dorhoi A, Ma J et al (2011) The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. J Immunol 187(1):434–440 [DOI] [PubMed] [Google Scholar]
- Woith E, Fuhrmann G, Melzig MF (2019) Extracellular vesicles—connecting kingdoms. Int J Mol Sci 20:5695 [DOI] [PMC free article] [PubMed]
- Xue X, Bredell BX, Anderson ER, Martin A, Mays C, Nagao-Kitamoto H, Huang S, Győrffy B, Greenson JK, Hardiman K et al (2017) Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc Natl Acad Sci USA 114(45):E9608–E9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hwang I, Lee E, Shin SJ, Lee EJ, Rhee JH, Yu JW (2020) Bacterial outer membrane vesicle-mediated cytosolic delivery of flagellin triggers host NLRC4 canonical inflammasome signaling. Front Immunol 11:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X-Y, Hou X-R, Huang Z-X, Zhu P, Liu B-Y (2024) Immunization with a peptide mimicking lipoteichoic acid induces memory B cells in BALB/c mice. BMC Infect Dis 24(1):371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Kim OY, Gho YS (2014) Extracellular vesicles as emerging intercellular communicasomes. BMB Rep 47(10):531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewinger S, Reiser J, Jankowski V, Alansary D, Hahm E, Triem S, Klug M, Schunk SJ, Schmit D, Kramann R et al (2020) Apolipoprotein C3 induces inflammation and organ damage by alternative inflammasome activation. Nat Immunol 21(1):30–41 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Morgan MJ, Chen K, Choksi S, Liu Z (2012) Induction of autophagy is essential for monocyte-macrophage differentiation. Blood 119(12):2895–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets produced in this study are available in the following databases: The code developed to analyze the RNA sequencing datasets of monocytes and BLaER1 cells has been deposited at https://github.com/MPUSP/human_RNA_seq_pipeline/releases/tag/v1.0.0. The raw RNA sequencing datasets are available under the NCBI-BioProject accession number: PRJNA753844. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al, 2022) partner repository (https://www.ebi.ac.uk/pride/profile/reviewer_pxd046570) with the dataset identifier PXD046570. Confocal images for ASC speck formation were deposited on BioImage Archive (Accession number: S-BIAD1725).
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44319-025-00558-7.