Abstract
In addition to the four chlorophylls (Chls) involved in primary charge separation, the photosystem II (PSII) reaction center polypeptides, D1 and D2, coordinate a pair of symmetry-related, peripheral accessory Chls. These Chls are axially coordinated by the D1-H118 and D2-H117 residues and are in close association with the proximal Chl antennae proteins, CP43 and CP47. To gain insight into the function(s) of each of the peripheral Chls, we generated site-specific mutations of the amino acid residues that coordinate these Chls and characterized their energy and electron transfer properties. Our results demonstrate that D1-H118 and D2-H117 mutants differ with respect to: (a) their relative numbers of functional PSII complexes, (b) their relative ability to stabilize charge-separated states, (c) light-harvesting efficiency, and (d) their sensitivity to photo-inhibition. The D2-H117N and D2-H117Q mutants had reduced levels of functional PSII complexes and oxygen evolution capacity as well as reduced light-harvesting efficiencies relative to wild-type cells. In contrast, the D1-H118Q mutant was capable of near wild-type rates of oxygen evolution at saturating light intensities. The D1-H118Q mutant also was substantially more resistant to photo-inhibition than wild type. This reduced sensitivity to photo-inhibition is presumably associated with a reduced light-harvesting efficiency in this mutant. Finally, it is noted that the PSII peripheral accessory Chls have similarities to a to a pair of Chls also present in the PSI reaction center complex.
Photosystem II (PSII) is a membrane-bound pigment-protein complex that catalyzes the light-driven oxidation of water and reduction of plastoquinone. The simplest functional PSII complex capable of charge separation is the reaction center complex. The PSII reaction center complex contains five polypeptides (Nanba and Satoh, 1987; Gounaris et al., 1990). The largest polypeptides (32 kD) are the D1 and D2 polypeptides, each of which has five transmembrane-spanning alpha helices. Sandwiched between the D1 and D2 polypeptides are the four chlorophylls (Chls) and two pheophytins (Pheos) involved in primary charge separation (for review, see Ruffle and Sayre, 1998). Three additional small (10 kD) polypeptides are present in the PSII reaction center complex. Two of these proteins, psbE and psbF, coordinate the redox active heme, cytochrome (Cyt) b559. This heme is not involved in primary charge separation but participates in a low quantum yield electron transfer cycle around PSII that protects the complex from photodamage (Buser et al., 1992; Stewart et al., 1998). The fifth protein component is the psbI protein, a small structural polypeptide that stabilizes the complex (Nanba and Satoh, 1987).
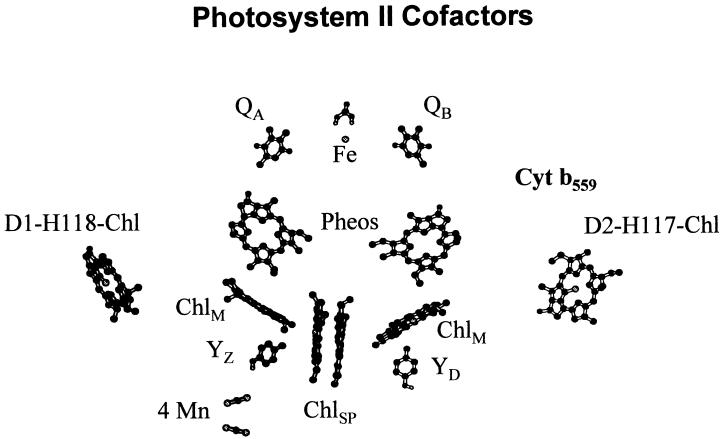
Amino acid sequence similarities between the D1 and D2 proteins and the analogous L and M subunits of the bacterial (Rhodopseudomonas viridis) photosynthetic reaction center indicated that the PSII reaction center was structurally analogous to the bacterial photosynthetic reaction center (Deisenhofer et al., 1984; Trebst, 1986; Sayre et al., 1986; Ruffle et al., 1992; Svensson et al., 1996; Xiong et al., 1998). Experimental analyses of the pigment composition of isolated PSII reaction center complexes indicated, however, that they contained two additional Chls not present in the bacterial photosynthetic reaction center (Kobayashi et al., 1990; Kurreck et al., 1997). Possible coordination sites for the additional pair of Chls in PSII were first proposed by Deisenhofer et al. (1984). They hypothesized that a pair of conserved and presumably symmetry-related His residues on the D1 (H118) and D2 (H117) proteins might coordinate Chls. These histidines are not conserved in the analogous L and M polypeptides of the bacterial reaction center (for review, see Ruffle and Sayre, 1998). Site-directed mutagenesis studies in Chlamydomonas reinhardtii and Synechocystis PCC6803, and the recently resolved 3.8-Å crystal structure of the Synechococcus elongatus PSII core complex, subsequently demonstrated that the D1-H118 and D2-H117 residues coordinate the additional Chls associated with the PSII D1 and D2 proteins (Hutchison and Sayre, 1995; Cua et al., 1998; Lince and Vermaas, 1998; Schweitzer et al., 1998; Ruffle et al., 1999; Johnston et al., 2000; Zouni et al., 2001). Unlike the Chls involved in primary charge separation, the Chls coordinated by the D1-H118 and D2-H117 residues are located on the exterior of the D1-D2 heterodimer approximately midway across the thylakoid membrane and hence are designated the peripheral accessory Chls (Fig. 1; Trebst, 1986; Sayre et al., 1986; Ruffle et al., 1992; Svensson et al., 1996; Xiong et al., 1998).
Figure 1.
Relative location of selected cofactors of the PSII reaction center model. Figure is redrawn from Zouni et al. (2001).
Two functions have been proposed for the PSII peripheral accessory Chls: (a) mediation of energy transfer from the proximal antennae complexes (CP43 and CP47) to P680, and (b) participation in a low quantum-yield electron transfer cycle around PSII that protects the complex from photo-inhibition (Koulougliotis et al., 1994; Hutchison and Sayre, 1995; Schweitzer and Brudvig, 1997; Cua et al., 1998; Lince and Vermaas, 1998; Ruffle et al., 1999; Johnston et al., 2000). We previously demonstrated that the Chl coordinated by the D2-H117 residue was involved in energy transfer to the primary donor, P680 (Johnston et al., 2000). Relative to wild type, we observed a shift in the 30-ps Chl fluorescence decay lifetime component to 10 ps in D2-H117 mutants consistent with an alteration in energy transfer from the Chl coordinated by this residue to P680 (Schelvis et al., 1994; Johnston et al., 2000).
One of the peripheral accessory Chls also has been proposed to participate in a low quantum yield electron transfer cycle around PSII (Thompson and Brudvig, 1988; Koulougliotis et al., 1994). This electron transfer cycle includes plastoquinone bound at the B binding site (QB), Cyt b559, ChlZ (the redox active Chl monomer coordinated by either the D1-H118 or D2-H117 residue), possibly (Car) carotenoid, and the primary donor, P680+ (Koulougliotis et al., 1994; Noguchi et al., 1994; Hanley et al., 1999; Vrettos et al., 1999). Under high-light intensities, both P680+ and doubly reduced QA may accumulate leading to damage and turnover of the D1 protein (photo-inhibition; Chen et al., 1992; Andersson and Barber, 1996; Napiwotzki et al., 1997; Gadjieva et al., 2000). The low quantum yield electron transfer cycle involving ChlZ is thought to protect PSII from photo-inhibitory damage by facilitating the reduction of long-lived P680+ states and the oxidation of over-reduced QA.
For ChlZ to be oxidized by P680+, however, it must be sufficiently close to either P680+ for direct electron transfer or to some potential intermediate electron carrier, such as a Car or Pheo (Noguchi et al., 1994). As hypothesized by Schelvis et al. (1994) the peripheral Chls coordinated by the D1-H118 and D2-H117 residues are sufficiently far removed (approximately 30 Å) from the Chl special pair (P680) to reduce the likelihood of their direct oxidation by P680+. Consistent with this prediction, it has been shown that Car radicals are generated under cryogenic conditions that lead to the photo-accumulation of ChlZ+ suggesting that Car may participate in ChlZ oxidation (Hanley et al., 1999). ChlZ+ subsequently is reduced by Cyt b559 (Buser et al., 1992). Whether Cyt b559 is the only cofactor to reduce ChlZ+ directly is unclear, however.
The most controversial issues regarding ChlZ are whether it is a single Chl species or two and if it is a single Chl species whether it is coordinated by the D1-H118 or the D2-H117 residue. There are several observations that suggest that ChlZ is coordinated by the D2-H117 residue. Pulsed EPR analyses of the magnetic dipole interactions between ChlZ+ and redox active Tyr-160 on the D2 protein (YD●) indicate that the distance between ChlZ+ and YD● is 29.4 Å (Shigemori et al., 1998). Consistent with this measurement, PSII models and the recent crystal structure indicate that the peripheral accessory Chl coordinated by the D2-H117 residue is about 26 to 30 Å from YD (Fig. 1; Ruffle et al., 1992; Svensson et al., 1996; Xiong et al., 1998; Zouni et al., 2001). In contrast, the peripheral accessory Chl coordinated by the D1-H118 residue is >50 Å from YD. In addition, the recent PSII crystal structure demonstrates that the Cyt b559 heme, the presumed electron donor to ChlZ+ (Schweitzer and Brudvig, 1997), is located adjacent to the peripheral accessory Chl coordinated by the D2-H117 residue and not the Chl coordinated by the D1-H118 residue (Schweitzer et al., 1998; Zouni et al., 2001). Last of all, the kinetics of ChlZ+-dependent quenching of the 695-nm low temperature Chl fluorescence emission band is substantially slower in D2-H117Q and D2-H117N mutants but is unaltered in the D1-H118Q mutant (J. Wang, D. Gosztola, S.V. Ruffle, C. Heman, M. Seibert, M.R. Wasielewski, R. Hille, T.L. Gustafson, and R.T. Sayre, unpublished data).
Evidence in support of ChlZ coordination by the D1-H118 residue comes from observations of mutation-induced alterations in the ChlZ+ vibrational spectra of cyanobacterial PSII complexes (Cua et al., 1998; Schweitzer et al., 1998; Stewart et al., 1998). Brudvig and coworkers demonstrated that the resonance Raman spectrum of ChlZ+ was perturbed in a cyanobacterial D1-H118Q mutant but was unaffected in a D2-H117Q mutant. It is noted, however, that the cryogenic conditions used to trap ChlZ+ could lead to the photo-accumulation of Car radicals that would affect the relaxation properties of ChlZ+ (Noguchi et al., 1994; Hanley et al., 1999).
In this report, we describe the effects of site-directed mutations of the peripheral accessory Chl ligands, D1-H118 and D2-H117, on PSII electron transfer processes, light-harvesting efficiency, and sensitivity to photo-inhibition. The results of these studies clearly demonstrate that the peripheral accessory Chls mediate energy transfer between the proximal PSII antennae complexes (CP43 and CP47) and the primary electron donor, P680, and reconfirm earlier observations that indicated that the peripheral accessory Chls play a critical role in regulating the sensitivity to photo-inhibition in the PSII complex. It also is evident that equivalent amino acid substitutions (Gln) at the D1-H118 and D2-H117 residues do not result in equivalent phenotypes. D2-H117 (Gln and Asn substitutions) mutants have reduced numbers of functional oxygen-evolving complexes and dramatically reduced light-harvesting efficiencies. In contrast, the D1-H118Q mutant has near wild-type numbers of functional oxygen-evolving complexes but has dramatically reduced sensitivity to photo-inhibitory light treatments. The reduced sensitivity to photo-inhibition in the D1-H118Q mutant is attributed to a reduction in its light-harvesting efficiency. Finally, we speculate on the conservation of structure and function between the PSII peripheral accessory Chls and the analogous connecting Chls (cC-Chls) of the PSI reaction center.
RESULTS
PSII Content and Activity
To characterize the function(s) of the peripheral accessory Chls conservative mutations were introduced at the D1-H118 and the D2-H117 residues. Asn and/or Gln substitutions were chosen to conserve potential Chl coordination sites and to minimize potential secondary structural perturbations. All chloroplast mutants were confirmed to be homoplasmic for the mutant gene by PCR and DNA sequence analysis.
Preliminary studies indicated that the D1-H118Q and the D2-H117N and D2-H117Q mutants were able to grow photosynthetically on medium lacking acetate although at reduced growth rates relative to wild type (data not shown). These results were in contrast to those obtained for the D1-H118R and D1-H118L mutants that were unable to grow photosynthetically and could not assemble a functional PSII complex (Hutchison and Sayre, 1995).
As shown in Table I, thylakoids from the D1-H118Q mutant had D1 protein levels that were essentially similar to wild type. In a similar manner, the functional manganese content of D1-H118Q PSII particles approximated that of wild type. To determine whether mutations of the peripheral accessory Chls ligands affected PSII-dependent electron transfer processes, we measured the rate of oxygen evolution at subsaturating light intensities (67% of saturation). At sub-saturating light intensities, differences in the efficiency of PSII electron transfer are more likely to be apparent than at saturating light intensities. As shown in Table I, oxygen evolution rates for the D1-H118Q mutant were only 60% of wild type. In a similar manner, the D2-H117Q and D2-H117N mutants had less than one-half the rate of oxygen evolution of wild type. The reduced rates of oxygen evolution in the D2-H117 mutants were correlated, however, with reduced functional manganese contents. Because the D1 protein content of thylakoids from light- and/or dark-grown D2-H117Q and D2-H117N mutants was nearly equivalent to wild-type thylakoids, however, the reduction in functional manganese content in these mutants could not be attributed to a loss of the D1 protein.
Table I.
Oxygen evolution, functional manganese, and D1 protein content of wild type and peripheral accessory Ch1 ligand mutants
| Strain | Growth Condition | Oxygen Evolution | Manganese Content (Mn/250 Chl) | D1 Content/Ch1 (Relative Mean Intensity) |
|---|---|---|---|---|
| μmol O2 mg Chl h−1 | ||||
| WT | Light | 225.7 ± 1.6 | 4.02 ± 0.10 | 100 |
| WT | Dark | 0 | 0 | 88 ± 3.8 |
| D1-H118Q | Light | 134 ± 5.6 (59) | 4.0 ± 0.1 (100) | ND |
| D2-H117N | Light | 105 ± 1.1 (47) | 2.54 ± 0.10 (63) | 85 ± 9.9 |
| D2-H117N | Dark | 0 | 0 | 84 ± 6.4 |
| D2-H117Q | Light | 125 ± 1.1 (55) | 2.11 ± 0.13 (52) | 88 ± 5.0 |
| D2-H117Q | Dark | 0 | 0 | 106 ± 12.5 |
Oxygen evolution was determined at sub-saturating light intensities using thylakoids. Functional manganese content was measured in PSII particles. D1 protein content was determined using thylakoids. Light intensity was 800 umol photons m2 s−1 (67% saturating light intensity for wild-type thylakoids). Data presented as mean ± se. Values in parentheses are percentages of wild type. ND, Not determined; WT, wild type.
Flash-Induced Chl Fluorescence Decay Kinetics
To gain further insights into the physical basis for the reduction in oxygen-evolving activity in the mutants, we characterized the Chl a fluorescence decay kinetics following a saturating flash. These measurements allow us to monitor the rate-limiting steps in electron transfer (QA → QB) as well as the back reaction from QA− to YZ● (dark grown) or the predominant S state of the water-oxidizing complex following a single flash (S2) state (light grown). For Chl fluorescence measurements with light-grown but dark-adapted wild-type cells, it is assumed that the PSII centers are all in the low Chl fluorescence state, S1P680QA. Immediately following a flash, the PSII complex advances to the high Chl fluorescence state, S2P680QA−, followed by the decay or loss of the high Chl fluorescent. The largest contribution to the Chl fluorescence decay kinetics (in the microsecond time scale) typically is electron transfer from QA− to the secondary electron acceptor, QB. Competing reactions including accelerated back reactions between QA− and an oxidized electron donor also contribute to the Chl fluorescence decay (Mamedov et al., 1998).
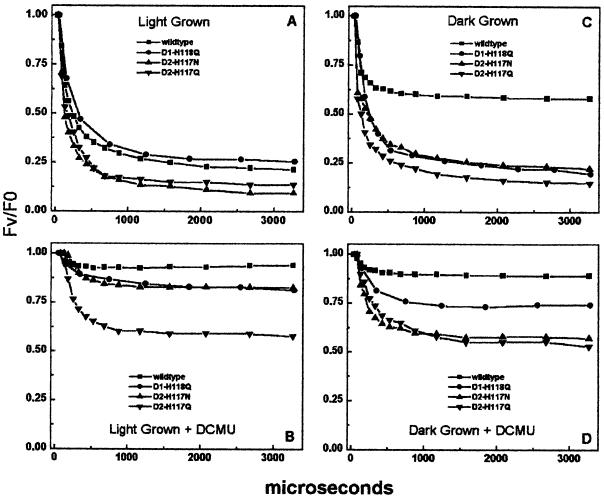
Figure 2 shows the Chl fluorescence decay kinetics of thylakoids isolated from dark- or light-grown cells excited in the presence or absence of DCMU (blocks QA to QB electron transfer). As shown in Figure 2A and summarized in Table II, the Chl fluorescence decay kinetics of the light-grown D2-H117 mutants were faster than wild type, indicating either a faster QA → QB electron transfer rate or an accelerated back reaction between QB and the S2 state of the water-splitting complex. In contrast, the Chl fluorescence decay kinetics of the D1-H118Q mutant were slightly slower than wild type, primarily due to the greater contribution of the slower Chl fluorescence decay lifetime component to the overall decay kinetics. To determine whether the back reaction between QA− and the water-splitting complex affected the Chl fluorescence decay kinetics in the mutants, we measured Chl fluorescence decay in dark-grown cells. These cells lack a tetra-manganese, water-oxidizing complex (Table I). Following a single turnover flash, the charge-separated state advances to the high Chl fluorescence state, YZ●P680QA−. Similar to light-grown cells, the Chl fluorescence decay kinetics of the dark-grown D2-H117Q, D2-H117N, and D1-H118Q mutants were faster than wild type with the exception of the D1-H118Q mutant, which had fewer resolvable lifetime components (Table II). However, the amplitude of the fastest Chl fluorescence decay lifetime component of the D1-H118Q mutant was greater than that of wild type (Fig. 2C, Table II). Thus, the decay of the P680/QA− high fluorescence state appears faster in the D1-H118Q mutant than in wild type.
Figure 2.
Chl fluorescence decay kinetics of wild-type, D1-H118Q, D2-H117N, and D2-H1117Q thylakoids following a single flash. A, Thylakoids from light-grown cells dark adapted for 10 min; B, A plus 10 μm 3-(3′,4′-dichloropheyl)-1,1-dimethylurea (DCMU); C, thylakoids from dark-grown cells dark adapted for 10 min; D, C plus 10 μm DCMU.
Table II.
Chl fluorescence decay kinetics lifetime and amplitude analysis of results presented in Fig. 2
| Relative Values | Light Grown
|
Light Grown + DCMU
|
Dark Grown
|
Dark Grown + DCMU
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | D1-H118Q | D2-H117N | D2-H117Q | WT | D1-H118Q | D2-H117N | D2-H117Q | WT | D1-H118Q | D2-H117N | D2-H117Q | WT | D1-H118Q | D2-H117N | D2-H117Q | |
| τ1 | 111 | 114 | 68 | 28 | 73 | 251 | 289 | 207 | 73 | 149 | 11 | 33 | 163 | 289 | 83 | 120 |
| % A1 | 64% | 45% | 70% | 55% | 24% | 20% | 30% | 53% | 57% | 70% | 93% | 72% | 77% | 35% | 56% | 39% |
| τ2 | 788 | 473 | 391 | 263 | – | 21,178 | – | 46,056 | 773 | 2,513 | 390 | 421 | – | – | 452 | 639 |
| % A2 | 22% | 37% | 22% | 37% | – | 47% | – | 28% | 7% | 15% | 5% | 19% | – | – | 14% | 28% |
| τ3 | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying | Nondecaying |
| % A3 | 14% | 17% | 8% | 8% | 76% | 33% | 70% | 19% | 36% | 15% | 2% | 9% | 23% | 65% | 30% | 33% |
Lifetimes (τ1/2) are indicated in microseconds and amplitudes (A) are expressed as a percentage of the total. Samples with no appreciable Chl fluorescence decay are indicated by –. WT, Wild type.
To eliminate the contribution of QA− → QB electron transfer from the overall Chl fluorescence decay kinetics, we measured Chl fluorescence decay kinetics in the presence of DCMU. The addition of DCMU blocks QA− → QB electron transfer leading to a long-lived high Chl fluorescence state (P680/QA−) in wild-type thylakoids. Under these conditions, the largest contribution to the Chl fluorescence decay is typically the back reaction from QA− to either YZ● (dark grown) or the S2 state (light grown). As shown in Figure 2, B and D, thylakoids from light- and dark-grown D2-H117Q cells exhibited accelerated Chl fluorescence decay kinetics relative to wild type in the presence of DCMU. We attribute this rapid decay of the high Chl fluorescence state in the D2-H117Q mutant to an accelerated back reaction between QA− and either YZ● (in dark-grown cells) or the S2 state (in light-grown cells).
It is significant that thylakoids from light-grown D2-H117N cells had substantially slower Chl fluorescence decay kinetics than dark-grown D2-H117N cells in the presence of DCMU (Fig. 2, B and D; Table II). Similar results were not observed in the D2-H117Q mutant. The light-grown D2-H117N cells lost only 17% of their maximal Chl fluorescence over a 3-ms period. These results suggest that the back reaction between QA− and the donor side (S2 state) was suppressed in the light-grown D2-H117N mutant cells but not in dark-grown cells (QA− → YZ●). One possible mechanism for this reduced rate of deactivation of the charge-separated state in light-grown D2-H117N cells is a rapid reduction of either the S2 state, YZ●, or P680+ by some alternate electron donor. The charge-separated state alternatively may be more stable in the D2-H117N mutant than in the D2-H117Q mutant.
In the presence of DCMU, the Chl fluorescence decay kinetics of light-grown D1-H118Q thylakoids were similar to light-grown wild-type or D2-H117N mutant cells. The Chl fluorescence decay (+DCMU) kinetics of dark-grown D1-H118Q mutant thylakoids, however, were intermediate between wild type and the D2-H117 mutants, indicative of a reduced back reaction between QA− and YZ● (relative to the D2-H117 mutants). Overall, these results suggest that the D1-H118Q mutation has fewer secondary effects on primary charge transfer processes relative to the D2-H117 mutations.
Stability of the S-State Cycle in D2-H117 Mutants
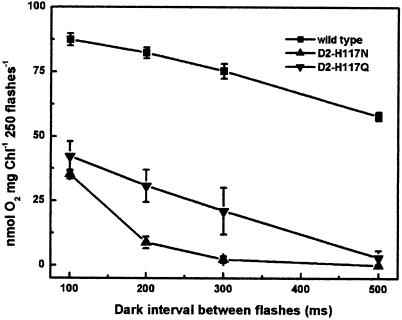
It was apparent from the Chl fluorescence decay kinetics of DCMU-treated D2-H117 mutants and wild-type thylakoids that the D2-H117 mutations affected PSII donor-side electron transfer processes. To further investigate these effects, we assessed the stability of the charge accumulating, water-splitting apparatus during S-state advancement. We measured the stability of the S states and their tendency to decay to lower S states by measuring oxygen production during a series of single-turnover flashes with varying dark intervals between each flash (Renger, 1972). The flash-dependent yield of oxygen typically decreases as the dark interval between flashes increases. This can be attributed to either a decay or reduction (electron donor) of the S-state complex (Mamedov et al., 1998). Factors that accelerate the decay of the S states include slowed electron transfer to P680+, charge recombination from QA−, reduction of YZ● or the S-state complex by alternate electron donors, or an unstable tetra-manganese complex. As shown in Figure 3, the oxygen yield of the D2-H117N mutant was significantly reduced, relative to wild type, as the time interval between flashes was increased. At a dark interval of 100 ms, only 41% of the wild-type oxygen yield was observed in the D2-H117N mutant and at dark intervals ≥ 300 ms, no oxygen was produced. In contrast, the rate of the dark interval-dependent loss of oxygen production per flash was similar for the D2-H117Q mutant and wild type (Fig. 3). Overall, these results indicate that advancement of the S states is inhibited in the D2-H117N mutant. We attribute this effect to either instability of the S-state complex or to the reduction of donor-side electron transfer cofactors by an alternate electron donor.
Figure 3.
The effect of varying dark intervals on the flash-dependent yield of oxygen. Oxygen yield of wild-type, D2-H117N, and D2-H117Q thylakoids following a series of 250 flashes with variable dark intervals between each flash.
Chl Fluorescence Decay Kinetics of D1-H118Q Reaction Center
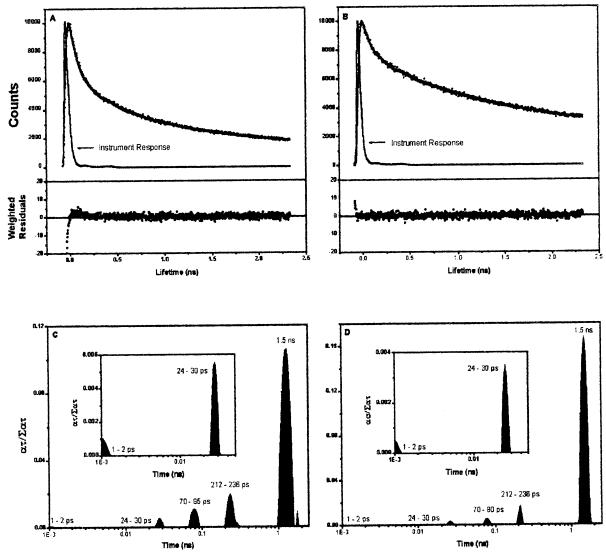
To assess the relative effects of mutagenesis of the D2-H117 and D1-H118 residues on energy transfer kinetics in reaction center particles lacking the proximal antennae complexes, we measured the Chl fluorescence decay kinetics of reaction center particles. It had been demonstrated previously that the 30-ps Chl fluorescence lifetime component attributed to energy transfer from the peripheral accessory Chl to P680 was altered in D2-H117N PSII reaction centers (Johnston et al., 2000). The mutation-induced alteration in energy transfer kinetics was attributed to a change in the distance or orientation of the peripheral accessory Chl relative to P680. To determine whether the D1-H118Q mutation had a similar effect on energy transfer kinetics, we analyzed the ultra-fast Chl fluorescence decay kinetics in D1-H118Q reaction center particles containing six Chls per two Pheos. As shown in Figure 4, the Chl fluorescence decay kinetics of the D1-H118Q mutant were substantially slower than those of wild type. Analysis of the distribution of Chl fluorescence decay lifetime amplitudes indicated that the Chl fluorescence decay was dominated by contributions from the slower lifetime components relative to wild type. Unlike the D2-H117N mutant, however, there was no major shift in any single Chl fluorescence lifetime component in the Chl fluorescence decay kinetics of D1-H118Q reaction center particles (Johnston et al., 2000). It is significant that the amplitudes of the slower Chl fluorescence decay lifetime components were greater for the D1-H118Q mutant than in wild type. These results indicate a reduced energy coupling efficiency between the peripheral accessory Chl and P680.
Figure 4.
Picosecond Chl fluorescence decay kinetics and exponential series distribution of lifetime components of wild-type and D1-H118Q mutant photosynthetic reaction centers. A, Chl fluorescence decay kinetics of wild-type reaction center particles; B, Chl fluorescence decay kinetics of D1-H118Q mutant reaction center particles; C and D, exponential series distribution of Chl fluorescence decay lifetime components for wild-type photosynthetic reaction centers and D1-H118Q mutant photosynthetic reaction centers, respectively.
Light Intensity-Dependent Rates of Water Oxidation
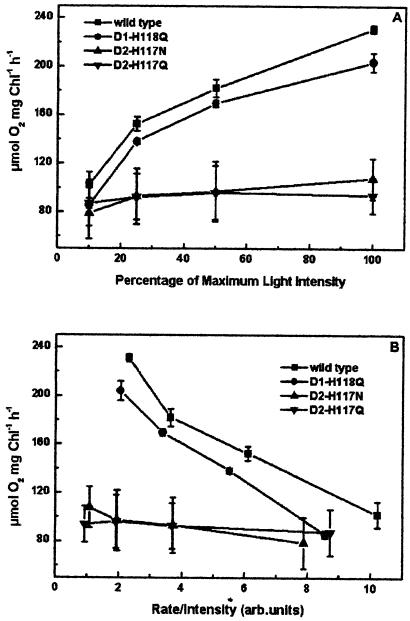
To determine whether light-harvesting efficiency was also altered in intact thylakoids of peripheral accessory Chl ligand mutants, oxygen evolution rates were measured as a function of light intensity using mutant and wild-type thylakoids. As shown in Figure 5, oxygen evolution rates for both D2-H117 mutants were light saturated at approximately 10% of the intensity that is light saturating for wild-type thylakoids. These results suggest that energy may be shared between nonfunctional (nonoxygen evolving) and functional PSII complexes in these mutants. In contrast to the D2-H117 mutants, the light intensity-dependent rates of oxygen evolution for the D1-H118Q mutant were only slightly reduced relative to wild type, indicative of only a partial reduction in light-harvesting efficiency.
Figure 5.
Light-dependent rates of oxygen evolution for wild-type and mutant thylakoids. The maximum light intensity used was 2,700 μmol photons m−2 s−1. Wild-type, D1-H118Q, D2-H117N, and D2-H117Q thylakoids.
Sensitivity to Photo-Inhibition
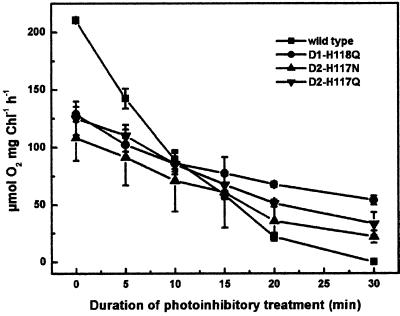
To determine the relative sensitivity of wild type and the D1 and D2 mutants to photo-inhibition, thylakoids were exposed to high-light intensities (1,000 μmol photons m−2 s−1 at 25°C) for periods up to 30 min followed by measurement of the residual rate of oxygen evolution in the presence of ρ-benzoquinone. As shown in Figure 6, both the D1-H118Q and the D2-H117 mutants were less sensitive to photo-inhibition than wild type. Following 30 min of exposure to high-light intensities, oxygen evolution was completely inhibited in wild-type thylakoids. Following a 30-min high-light exposure, there was only a 57% reduction of the oxygen evolution rate in the D1-H118Q mutant. The reduced sensitivity to photo-inhibitory light treatment in the D1-H118Q mutant is best accounted for by a reduction in light-harvesting efficiency. As previously indicated, light energy is less efficiently trapped in D1-H118Q PSII reaction centers.
Figure 6.
Sensitivity to photo-inhibition. Residual oxygen evolution activity following photo-inhibitory light treatment of thylakoids. Wild-type, D1-H118Q, D2-H117N, and D2-H117Q thylakoids.
In contrast, the relative sensitivity of the D2-H117Q and D2-H117N mutants to photo-inhibition was more similar to wild type. Following 30 min of photo-inhibitory light treatments, the D2-H117 mutants had 30% of the maximum rate of oxygen evolution. Although it is apparent that the D2-H117 mutants have a lower light-harvesting efficiency than the D1-H118Q mutant, the D2-H117 mutants also have fewer oxygen-evolving PSII complexes, thus accounting for their increased sensitivity to photo-inhibition relative to the D1-H118Q mutant.
DISCUSSION
We have demonstrated previously that the D1-H118 residue plays a critical role in the structural integrity of the PS II complex. Both a nonconservative (D1-H118L) and a conservative (D1-H118R) substitution of the peripheral accessory Chl ligand prevented accumulation of PSII complexes capable of charge separation. This outcome was presumably due to an inability to bind the peripheral accessory Chl coordinated at this site (Hutchison and Sayre, 1995; Ruffle et al., 1999).
To elucidate the functions of both peripheral accessory Chls it was necessary, however, to isolate D1-H118 and D2-H117 peripheral accessory Chl ligand mutants that had functional phenotypes. In this respect, the D1-H118Q mutant was more amenable to biochemical and biophysical analyses. The D1-H118Q mutant had nearly wild-type rates of oxygen evolution. Furthermore, analyses of Chl fluorescence decay kinetics in PSII reaction center particles and whole thylakoids indicated that the energy and electron transfer kinetics of the D1-H118Q mutants were similar to wild type. The D1-H118Q mutant, however, was substantially more resistant to photo-inhibitory light treatments than D2-H117 mutant or wild-type thylakoids. Photo-inhibitory light treatments that completely inhibited oxygen evolution in wild-type thylakoids reduced oxygen-evolving activity in the D1-H118Q mutant by only 50%. This effect could be attributed to either differences in light-harvesting efficiency or the operation of the ChlZ-mediated electron transfer pathway around PSII. In this study, we have focused on mutation-induced alterations in energy transfer efficiency. Analyses of the light-dependent rates of oxygen evolution in thylakoids of D1-H118Q mutants, as well as analyses of the Chl fluorescence decay kinetics in D1-H118Q PSII reaction centers, indicated a reduced efficiency for generating charge-separated states. The amplitudes of the longer lived Chl fluorescence decay lifetime components in isolated PSII reaction center complexes also were substantially greater than wild type, consistent with less efficient energy transfer in the D1-H118Q mutant. This reduced light-harvesting efficiency presumably accounts for the reduced sensitivity to photo-inhibitory damage in the D1-H118Q mutant. It is noted, however, that extrapolation from the Chl fluorescence decay kinetics of PSII reaction centers to the energy transfer processes taking place in intact thylakoids is speculative at best.
In marked contrast to the D1-H118Q mutant, oxygen evolution was light saturated at very low light intensities for the D2-H117Q and D2-H117N mutants. Although the D2-H117N mutant exhibited increasing, albeit low, rates of oxygen evolution with increasing light intensity, oxygen evolution rates for the D2-H117Q mutant did not increase at light intensities greater than 10% of saturating light intensity for wild type. In part, these results can be attributed to an impairment of oxygen evolution due to the reduced numbers of oxygen-evolving PSII complexes. However, the D2-H117 mutants also were less efficient at stabilizing a charge-separated state than wild type (Fig. 3). We observed an accelerated rate of charge recombination between QA− and the S2 state (light grown) or YZ● (dark grown) in the D2-H117Q mutant. The efficiency of charge separation in the D2-H117N mutant also was impaired but by other means. From analysis of the Chl fluorescence decay kinetics of thylakoids, as well as the reduced dark stability of the S-state complex (in a flash train), it was apparent that advancement of the S-state complex was inhibited in the D2-H117N mutant. Inhibition of advancement of the S-state complex is likely due to electron donation by some unidentified alternate electron donor. The identity of this alternate donor currently remains to be determined, although one intriguing possibility is that electron donation to P680+ from ChlZ may be affected in the D2-H117N mutant. Overall, it is apparent that the inhibition of donor-side processes associated with water oxidation is greater in the D2-H117 mutants than in the D1-H118Q mutant.
It is interesting that an identical mutation to the D2-H117N mutant described here has been generated in cyanobacteria. In contrast to the C. reinhardtii D2-H117N mutant, the cyanobacterial D2-H117N mutant had light-harvesting efficiencies (measured as light-dependent rates of oxygen evolution) that were nearly identical to wild type (Lince and Vermaas, 1998). An analysis of the amino acid sequences flanking the D2-H117 residues in C. reinhardtii and Synechocystis PCC6803 indicates that the amino acid residues adjacent to the H117 residue are conserved between the two species so local structural differences in the D2 proteins are not likely to account for the observed phenotypic differences. However, Chl-protein interactions between the peripheral accessory Chl coordinated by the D2-H117 residue and the adjacent proximal antennae complex (presumably CP47) may be differentially affected in the C. reinhardtii and cyanobacterial mutants (Buchel et al., 2000). These potential differences in quaternary organization of the chloroplastic and cyanobacterial PSII complexes may account for the different phenotypes between equivalent mutations in different organisms.
Finally, it is apparent from a comparison of the pigment organization in the PSI and PSII crystal structures that the PSI reaction center complex has a pair of Chls that are structurally analogous to the PSII peripheral accessory Chls (Schubert et al., 1998; Zouni et al., 2001). The C2 symmetry-related cC-Chls of the PSI complex occupy positions similar to those of the peripheral accessory Chls of PSII. The cC-Chls also have been proposed to mediate energy transfer from the proximal antennae Chls, bound to the N-terminal portions of the PsaA and/or PsaB proteins, to the Chls (P700) that participate in primary charge transfer (eC3, eC2, and eC1; Schubert et al., 1998). The pigments closest (15 Å, center to center) to the cC-Chls are the eC3 Chls, which include the first stable electron acceptor (A0) of PSI. The PSI reaction center eC3-Chls are structurally analogous to the Pheos of PSII.
From the emerging PSII crystal structure it has been determined that the center-to-center distance between the peripheral accessory Chls and the Pheos is 24 Å. Although the accessory Chl monomers of the PSII reaction center also are about 24 Å from the peripheral accessory Chls, the orientation of the Chl monomer and Pheo macro-cycle rings is apparently less favorable for Förster-mediated energy transfer than the apparent orientation between the peripheral accessory Chls and Pheos (Zouni et al., 2001). These results suggest that energy transfer from the peripheral accessory Chls to the primary donor may involve Pheo.
The structural relationships between the Pheos and the peripheral accessory Chls also may have implications for electron transfer processes involving ChlZ. Only the Pheo proximal to the peripheral accessory Chl coordinated by the D1-H118 residue participates in primary charge transfer (Dorlet et al., 2001). The peripheral accessory Chl located adjacent to the peripheral accessory Chl coordinated by the D2-H117 residue is not involved in primary charge transfer. As previously argued, it is likely that the Chl coordinated by the D2-H117 residue is the redox active Chl, ChlZ. Because ChlZ is spatially removed from the Pheo that participates in primary charge transfer direct electron transfer from Pheo− to ChlZ+ is less likely to occur than if ChlZ were adjacent to the Pheo involved in primary charge transfer. This separation of electron transfer pathway cofactors (linear versus cyclic) reduces the possibility of short-circuiting linear electron flow in PSII by ChlZ. In summary, these results demonstrate that the two peripheral accessory Chls have nonidentical functions that are optimized for efficient linear electron transfer, light harvesting, and protection from photo-inhibition.
MATERIALS AND METHODS
Generation of Mutants
Site-directed mutations were introduced into the Chlamydomonas reinhardtii psbA (encodes the D1 protein) and psbD (encodes the D2 protein) genes using the method of Kunkel et al. (1987) or the Quik-Change kit (Stratagene, La Jolla, CA) to generate His to Gln and Asn (D1-H118Q, D2-H117Q, and D2-H117N) substitutions, as well as silent nucleotide changes to introduce diagnostic restriction endonuclease recognition sites. For the D1 mutant, a modified psbA gene was generated in plasmid pBA155 (Minagawa and Crofts, 1994) and introduced into the psbA deletion mutant, CC-741, using a particle inflow gun (Finer et al., 1992; Hutchison et al., 1996). Transformants were selected on the basis of spectinomycin and streptomycin resistance conferred by the aadA gene (Goldschmidt-Clermont, 1991). For selection of psbD mutant transformants, an aadA gene from plasmid pBA155 was introduced 400 bp upstream of the 5′ end of psbD gene cloned into pUC18. The mutated psbD genes were transformed into wild-type C. reinhardtii (CC-2137) using a helium inflow particle gun (Finer et al., 1992; Hutchison et al., 1996). Primary heteroplasmic transformants were selected at low-light intensities (8–15 μmol photons m−2 s−1) on solid tris-acetate phosphate (TAP) medium (Harris, 1989) containing 100 μg spectinomycin mL−1 and 50 μg ampicillin mL−1 to inhibit bacterial growth. A secondary screening for psbD homoplasmic mutants was carried out by transferring the heteroplasmic transformants to TAP media containing 50 μg streptomycin mL−1 and 50 μg ampicillin mL−1. Transformants then were screened for the appropriate diagnostic restriction sites by Southern-blot analysis. Selection was alternated between the two antibiotics until homoplasmy was achieved as determined by Southern-blot analysis. All mutations were confirmed by DNA sequencing of PCR-derived fragments (ABI Prism, Perkin-Elmer, Wellesley, MA) using total Chlamydomonas sp. genomic DNA as a template.
Thylakoid Preparation
C. reinhardtii cultures were grown either in the dark or at low-light intensities (8–15 μmol photons m−2 s−1) in liquid TAP medium with 25 μg ampicillin mL−1 (Harris, 1989; Roffey et al., 1994). Mutant cell cultures had 30 μg spectinomycin mL−1 added to the media. Cultures were harvested when they achieved a density of 1 to 2 × 106 cells mL−1. Thylakoids were prepared by passing cells (1 mg Chl mL−1) in buffer A {300 mm sorbitol, 20 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.5, and 2 mm MgCl2}, twice through a Bio-Neb Cell disrupter (Glas-Col, Terre Haute, IN) using nitrogen gas (110 psi) as a carrier. Membranes were harvested by centrifugation at 40,000g for 20 min and unbroken cells were removed from the membrane fraction by centrifugation at 1,200g for 30 s in buffer B (400 mm Suc, 20 mm HEPES, pH 7.5, 5 mm MgCl2, and 5 mm EDTA). Thylakoids were harvested following centrifugation at 11,000g for 20 min and resuspended in buffer B at >1.0 mg Chl mL−1. Light-scattering particles were removed from thylakoids by centrifugation over a Suc pad (2 m Suc, 20 mm HEPES, pH 7.5, 5 mm EDTA, and 5 mm MgCl2) at 100,000g for 30 min prior to optical measurements. Thylakoids were finally resuspended in buffer B at >1.0 mg Chl mL−1. All steps were carried out in darkness at 4°C. Membranes were used immediately or frozen at 77 K in the dark at a Chl concentration of >1.0 mg mL−1 and stored at −80°C.
Measurement of Electron Transfer Reactions
Oxygen evolution measurements were carried out according to Roffey et al. (1994) using a Hanstech CB1 oxygen electrode. The assay medium included 200 μm ρ-benzoquinone, 1 mm potassium ferricyanide, and 30 mm methylamine chloride in buffer A. For the determination of flash-dependent rates of oxygen evolution, the time interval between saturating flashes (250 flashes) was varied between 100 and 500 ms.
Manganese Determination
EDTA-washed “Berthold, Babcock, and Yocum-type” PSII-enriched membranes were prepared for manganese determination by induction-coupled plasma-mass spectroscopy, as described by Roffey et al. (1994).
D1 Protein Abundance
Thylakoids (0.25 mg Chl mL−1) were solubilized at 90°C for 10 min in sample loading buffer (50 mm Tris, pH 7.5, 2% [w/v] SDS, 20 mm EDTA, 2% [v/v] β-mercaptoethanol, 20% [v/v] glycerol, and 0.001% [w/v] bromphenol blue) and the proteins were separated by SDS-PAGE (12% [w/v] acrylamide; Laemmli, 1970). Western-blot analyses were carried out following the method of Towbin and Gordon (1984) as described in the Immuno-Blot assay kit procedure (Bio-Rad, Richmond, CA). The D1 antibody was raised against a synthetic peptide corresponding to residues 225 to 234 of the spinach D1 protein sequence (Sayre et al., 1986). The antigen-antibody complex was detected using an alkaline phosphatase-linked goat anti-rabbit secondary antibody. Quantification of the colorimetric reaction was determined by densitometry and titrated to confirm the linearity of the response.
Chl a Fluorescence Decay Kinetics of Thylakoids
Chl a fluorescence decay kinetics of thylakoids (10 μg Chl mL−1) in buffer A were recorded on a fluorometer (Occam Technologies, Cincinnati) at various time intervals (35 μs to 6 ms) following a 5-μs xenon actinic pulse, in the presence or absence of 10 μm DCMU. Samples were dark adapted for 10 min before recording. The Chl fluorescence decay kinetic lifetime components were determined using Microcal Origin software.
Chl a Fluorescence Decay Kinetics of Reaction Center Complex
PSII reaction center isolation from wild type and the D1-H118Q mutant, Chl fluorescence decay kinetic measurements (using a time-correlated single photon counting apparatus), and Chl fluorescence decay fits were conducted as described earlier (Johnston et al., 2000) using PSII reaction centers with six Chls per two Pheos (Eijckelhoff and Dekker, 1995).
Photo-Inhibition Studies
Thylakoids from light-grown cells were exposed to a photo-inhibitory light treatment (1,000 μmol photons m−2 s−1 at 25°C) in buffer A for up to 30 min at 20 μg Chl mL−1. Treated samples were assayed for oxygen evolution as described above.
Footnotes
This research was supported by the Department of Energy and Ohio State University (grants to T.L.G. and R.T.S.).
This paper is dedicated to Dr. George M. Cheniae.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010245.
LITERATURE CITED
- Andersson B, Barber J. Baker NR, ed, Advances in Photosynthesis (5): Photosynthesis and Environment. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. Mechanisms of photodamage and protein degradation during photoinhibition of photosystem II; pp. 101–121. [Google Scholar]
- Buchel C, Morris E, Barber J. Crystallization of CP43, a chlorophyll binding protein of PSII: an electron microscopy analysis of molecular packing. J Struct Biol. 2000;131:181–186. doi: 10.1006/jsbi.2000.4294. [DOI] [PubMed] [Google Scholar]
- Buser CA, Diner BA, Brudvig GW. Photooxidation of cytochrome b559 in oxygen-evolving photosystem II. Biochemistry. 1992;31:11449–11459. doi: 10.1021/bi00161a025. [DOI] [PubMed] [Google Scholar]
- Chen GX, Kazimir J, Cheniae GM. Photoinhibition of hydroxyalamine-extracted photosystem II membranes: studies of the mechanism. Biochemistry. 1992;31:11072–11083. doi: 10.1021/bi00160a017. [DOI] [PubMed] [Google Scholar]
- Cua A, Stewart DH, Brudvig GW, Bocian DF. Selective resonance Raman scattering from chlorophyll Z in photosystem II via excitation into the near-infrared absorption band of the cation. J Am Chem Soc. 1998;120:4532–4533. [Google Scholar]
- Deisenhofer J, Epp O, Miki K, Huber R, Michel H. X-ray structure analysis of a membrane protein complex: electron density map at 3 Å resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- Dorlet P, Xiong L, Sayre RT, Un S. High-field EPR study of the pheophytin anion radical in wild type and D1–E130 mutants of photosystem II in Chlamydomonas reinhardtii. J Biol Chem. 2001;276:22313–22316. doi: 10.1074/jbc.M102475200. [DOI] [PubMed] [Google Scholar]
- Eijckelhoff C, Dekker JP. Determination of the pigment stoichiometry of the photochemical reaction center of photosystem II. Biochim Biophys Acta. 1995;1231:21–28. [Google Scholar]
- Finer JJ, Vain P, Jones MW, McMullen MD. Development of the particles inflow gun for DNA deliver to plant cells. Plant Cell Rep. 1992;11:323–328. doi: 10.1007/BF00233358. [DOI] [PubMed] [Google Scholar]
- Gadjieva R, Eckert HJ, Renger G. Photoinhibition as a function of the ambient redox potential in Tris-washed PSII membrane fragments. Photosynth Res. 2000;63:237–248. doi: 10.1023/A:1006427408083. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris K, Chapman DJ, Booth P, Crystall B, Giorgi LB, Klug DR, Porter G, Barber J. Comparison of the D1/D2/Cytochrome b559 reaction center complex of photosystem 2 isolated by two different methods. FEBS Lett. 1990;265:88–92. doi: 10.1016/0014-5793(90)80890-u. [DOI] [PubMed] [Google Scholar]
- Hanley J, Deligiannakis Y, Pascal A, Faller P, Rutherford AW. Carotenoid oxidation in photosystem II. Biochemistry. 1999;38:8189–8195. doi: 10.1021/bi990633u. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. New York: Academic Press; 1989. [Google Scholar]
- Hutchison RS, Roffey RA, Sayre RT. Chloroplast transformation. In: Andersson B, Salter AH, Barber J, editors. Molecular Genetics of Photosynthesis. Oxford, UK: IRL Press; 1996. pp. 180–196. [Google Scholar]
- Hutchison RS, Sayre RT. Site-specific mutagenesis at histidine 118 of the photosystem II D1 protein of Chlamydomonas reinhardtii. In: Mathis P, editor. Photosynthesis: From Light to Biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 471–474. [Google Scholar]
- Johnston H, Wang J, Ruffle SV, Sayre RT, Gustafson TL. Fluorescence decay kinetics of wild type and D2–H117N mutant photosystem II reaction centers isolated from Chlamydomonas reinhardtii. J Phys Chem B. 2000;104:4777–4781. [Google Scholar]
- Kobayashi M, Maeda H, Watanabe T, Nakane H, Satoh K. Chlorophyll a and β-carotene content in the D1/D2/cytochrome b559 reaction center complex from spinach. FEBS Lett. 1990;260:138–140. [Google Scholar]
- Koulougliotis D, Innes JB, Brudvig GW. Location of chlorophyll Z in photosystem II. Biochemistry. 1994;33:11814–11822. doi: 10.1021/bi00205a018. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kurreck J, Liu B, Napiwotzki A, Sellin S, Eckert HJ, Eichler HJ, Renger G. Stoichiometry of pigments and radical pair formation under saturating pulse excitation in D1/D2/cyt b559 preparations. Biochim Biophys Acta. 1997;1318:307–315. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lince MT, Vermaas W. Association of His117 in the D2 protein of photosystem II with a chlorophyll that affects excitation: energy transfer efficiency to the reaction center. Eur J Biochem. 1998;256:595–602. doi: 10.1046/j.1432-1327.1998.2560595.x. [DOI] [PubMed] [Google Scholar]
- Mamedov F, Sayre RT, Styring S. Involvement of histidine 190 on the D1 protein in electron/proton transfer reactions on the donor side of photosystem II. Biochemistry. 1998;37:14245–14256. doi: 10.1021/bi980194j. [DOI] [PubMed] [Google Scholar]
- Minagawa J, Crofts AR. A robust protocol for site-directed mutagenesis of the D1 protein in Chlamydomonas reinhardtii: a PCR spliced psbA gene in a plasmid conferring spectinomycin resistance was introduced into a psbA deletion strain. Photosynth Res. 1994;42:121–131. doi: 10.1007/BF02187123. [DOI] [PubMed] [Google Scholar]
- Nanba O, Satoh K. Isolation of a Photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b559. Proc Natl Acad Sci USA. 1987;84:109–112. doi: 10.1073/pnas.84.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiwotzki A, Bergmann A, Decker K, Legall H, Eckert HJ, Eichler HJ, Renger G. Acceptor side photoinhibition in PS II: on the possible effects of the functional integrity of the PS II donor side on photoinhibition of stable charge separation. Photosynth Res. 1997;52:199–213. [Google Scholar]
- Noguchi T, Mitsuka T, Inoue Y. Fourier-transform infrared spectrum of the radical cation of β-carotene photoinduced in photosystem II. FEBS Lett. 1994;356:179–182. doi: 10.1016/0014-5793(94)01263-6. [DOI] [PubMed] [Google Scholar]
- Renger G. Studies on the mechanism of the water oxidation in photosynthesis. Eur J Biochem. 1972;27:259–269. doi: 10.1111/j.1432-1033.1972.tb01835.x. [DOI] [PubMed] [Google Scholar]
- Roffey RA, Kramer DA, Govindjee, Sayre RT. Lumenal side histidine mutations in the D1 protein of photosystem II affect donor side electron transfer in Chlamydomonas reinhardtii. Biochim Biophys Acta. 1994;1185:257–270. doi: 10.1016/0005-2728(94)90240-2. [DOI] [PubMed] [Google Scholar]
- Ruffle SV, Donnelly D, Blundell TL, Nugent JHA. A three-dimensional model of the Photosystem II reaction center of Pisum sativum. Photosynth Res. 1992;34:287–300. doi: 10.1007/BF00033446. [DOI] [PubMed] [Google Scholar]
- Ruffle SV, Hutchison RS, Sayre RT. Mutagenesis of the symmetry related H117 residue in the photosystem II D2 protein of Chlamydomonas: implications for energy transfer from accessory chlorophylls. In: Garab G, editor. Photosynthesis: Mechanism and Effects. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 1013–1016. [Google Scholar]
- Ruffle SV, Sayre RT. Functional analysis of photosystem II. In: Goldshmidt-Cleremont M, Merchant S, Rochaix JD, editors. Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 287–322. [Google Scholar]
- Sayre RT, Andersson B, Bogorad L. The topology of a membrane protein: the orientation of the 32 kD Qb-binding chloroplast thylakoid membrane protein. Cell. 1986;47:601–608. doi: 10.1016/0092-8674(86)90624-0. [DOI] [PubMed] [Google Scholar]
- Schelvis JPM, van Noort PI, Aartsma TJ, van Gorkom HJ. Energy transfer, charge separation and pigment arrangement in the reaction center of photosystem II. Biochim Biophys Acta. 1994;1184:242–250. [Google Scholar]
- Schubert W-D, Klukas O, Saenger W, Witt HT, Fromme P, Krauβ N. A common ancestor for oxygenic and anoxygenic photosynthetic systems: a comparison based on the structural model of photosystem I. J Mol Biol. 1998;280:297–314. doi: 10.1006/jmbi.1998.1824. [DOI] [PubMed] [Google Scholar]
- Schweitzer RH, Brudvig GW. Fluorescence quenching by chlorophyll cations in photosystem II. Biochemistry. 1997;36:11351–11359. doi: 10.1021/bi9709203. [DOI] [PubMed] [Google Scholar]
- Schweitzer RH, Melkozernov AN, Blankenship RE, Brudvig GW. Time-resolved fluorescence measurements of photosystem II: the effect of quenching by oxidized chlorophyll Z. J Phys Chem B. 1998;102:8320–8326. [Google Scholar]
- Shigemori K, Hara H, Kawamori A, Akabori K. Determination of distances from tyrosine D to QA and chlorophyllZ in photosystem II studied by “2+1” pulsed EPR. Biochim Biophys Acta. 1998;1363:187–198. doi: 10.1016/s0005-2728(97)00100-x. [DOI] [PubMed] [Google Scholar]
- Stewart DH, Cua A, Chisholm DA, Diner BA, Bocian DF, Brudvig GW. Identification of hisitidine 118 in the D1 polypeptide of photosystem II as the axial ligand of chlorophyll Z. Biochemistry. 1998;37:10040–10046. doi: 10.1021/bi980668e. [DOI] [PubMed] [Google Scholar]
- Svensson B, Etchebest C, Tuffery P, van Kan P, Smith J, Styring S. A model for the photosystem II reaction center core including the structure of the primary donor P680. Biochemistry. 1996;35:14486–14502. doi: 10.1021/bi960764k. [DOI] [PubMed] [Google Scholar]
- Thompson LK, Brudvig GW. Cytochrome b559 may function to protect photosystem II from photoinhibition. Biochemistry. 1988;27:6653–6658. doi: 10.1021/bi00418a002. [DOI] [PubMed] [Google Scholar]
- Towbin H, Gordon J. Immunoblotting and dot immunobinding: current status and outlook. J Immunol Methods. 1984;72:313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- Trebst A. The topology of plastoquinone and herbicide binding polypeptides of photosystem II in the thylakoid membrane. Z Naturforsch. 1986;41:240–245. [Google Scholar]
- Vrettos JS, Stewart DH, de Paula JC, Brudvig GW. Low-temperature optical and resonance Raman spectra of a carotenoid cation radical in photosystem II. J Phys Chem B. 1999;102:6403–6406. [Google Scholar]
- Xiong J, Subramanian S, Govindjee A knowledge-based three dimensional model of the Photosystem II reaction center of Chlamydomonas reinhardtii. Photosynth Res. 1998;56:229–254. [Google Scholar]
- Zouni A, Witt H-T, Kern J, Fromme P, Kraub N, Saenger W, Orth P. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]