Abstract
The Brassica napus gene, Cel16, encodes a membrane-anchored endo-1,4-β-glucanase with a deduced molecular mass of 69 kD. As for other membrane-anchored endo-1,4-β-glucanases, Cel16 consists of a predicted intracellular, charged N terminus (methionine1-lysine70), a hydrophobic transmembrane domain (isoleucine71-valine93), and a periplasmic catalytic core (lysine94-proline621). Here, we report the functional analysis of Δ1-90Cel16, the N terminally truncated Cel16, missing residues 1 through 90 and comprising the catalytic domain of Cel16 expressed recombinantly in the methylotrophic yeast Pichia pastoris as a soluble protein. A two-step purification protocol yielded Δ1-90Cel16 in a pure form. The molecular mass of Δ1-90Cel16, when determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, was about 130 kD and about 60 kD after enzymatic removal of N-glycans, fitting the expected molecular mass of 59 kD. Δ1-90Cel16 was highly N glycosylated as compared with the native B. napus Cel16 protein. Δ1-90Cel16 had a pH optimum of 6.0. The activity of Δ1-90Cel16 was inhibited by EDTA and exhibited a strong dependence on calcium. Δ1-90Cel16 showed substrate specificity for low substituted carboxymethyl-cellulose and amorphous cellulose. It did not hydrolyze crystalline cellulose, xyloglycan, xylan, (1→3),(1→4)-β-d-glucan, the highly substituted hydroxyethylcellulose, or the oligosaccharides cellotriose, cellotetraose, cellopentaose, or xylopentaose. Size exclusion analysis of Δ1-90Cel16-hydrolyzed carboxymethylcellulose showed that Δ1-90Cel16 is a true endo-acting glucanase.
The primary cell wall of dicot plants has been described as a network of cellulose microfibrils cross-linked by xyloglycan and reinforced by pectins (Carpita et al., 1996; Reiter, 1998). Plant growth involves the controlled action of many different cell wall-related enzymes on the wall architecture. Among others, this complex process involves the action of cellulose synthases (Turner and Somerville, 1997; Arioli et al., 1998; Burton et al., 2000; Fagard et al., 2000), xyloglucan endotransglucosylases (McQueen-Mason et al., 1993; Catala et al., 2000), expansins (Cosgrove, 1998, 2000), and endo-1,4-β-glucanases (EGases; Hayashi and Ohsumi, 1994; Wu et al., 1996; del Campillo, 1999; Catala et al., 2000). Most plant EGases (EC 3.2.1.4) have an endoplasmatic reticulum import signal peptide and are secreted to the periplasm where they modify the cell wall, whereas plant membrane-anchored EGases are type II integral membrane proteins predicted to be integrated in the plasma membrane and to act at the plasma membrane-cell wall interface (Brummell et al., 1997; Nicol et al., 1998). Because membrane-anchored EGases are expected to be associated with the plasma membrane, they probably do not have access to the majority of the cell wall, and so they probably do not function as cell wall-loosening enzymes. In Arabidopsis, there are at least 17 genes encoding secreted EGases and only three encoding membrane-anchored EGases. A mutation (KORRIGAN) in one of the membrane-anchored EGases, encoded by the Arabidopsis KOR gene, disrupts the correct assembly of the cellulose-hemicellulose network (Nicol et al., 1998). This results in the absence of stratified microfibrils in the inner part of the cell wall. Other results suggest that KOR plays a critical role during cytokinesis, more specifically during cell plate maturation (Zuo et al., 2000). A stronger mutant allele than the previously identified mutation in the KORRIGAN mutant causes the formation of aberrant cell plates, incomplete cell walls, and multinucleated cells, leading to severely abnormal seedling morphology (Zuo et al., 2000).
Brassica napus Cel16 is orthologous to KOR, and recently we have shown that there is no strict correlation between Cel16 expression and elongation in light-grown seedlings (Mølhøj et al., 2001a). In Arabidopsis, membrane-anchored EGases belong to a small gene family of three genes: KOR, KOR2, and KOR3 (Nicol et al., 1998; Zuo et al., 2000; Mølhøj et al., 2001a, 2001b). KOR and Cel16 are ubiquitously expressed membrane-anchored EGases, whereas KOR2 and KOR3 expression is restricted to specific cell types. KOR2 and KOR3 were shown to be differentially expressed in developing leaf trichomes and their support cells, respectively (Mølhøj et al., 2001b). Furthermore, KOR2 is expressed in the developing root hairs within the root differentiation zone, the basal region of leaves, and floral organs, whereas KOR3 is also expressed in the bundle sheath cells that surround the vascular bundle within the leaf mesophyll tissue (Mølhøj et al., 2001b). Although KORRIGAN shows a defect in non-tip-growing cells (Nicol et al., 1998), KOR2 seems at least partly to be expressed in tip-growing cells like trichomes and root hairs.
The membrane-anchored EGases are of particular interest in the context of a function in cell wall assembly, but their substrate specificity has not yet been characterized. Like all plant-secreted EGases, membrane-anchored EGases belong to family 9 of the glycoside hydrolase families (Henrissat, 1991), characterized by an inverting hydrolyzing mechanism. Inverting glycoside hydrolases mediate an inversion of the anomeric configuration, thus leaving the product with the opposite stereochemistry to the substrate. Neither KOR nor Cel3 (Brummell et al., 1997), a tomato (Lycopersicon esculentum) homolog, is up-regulated by ethylene or auxin, which suggests that membrane-anchored EGases are functionally distinct from secreted EGases.
Thus, membrane-anchored EGases are good candidates for EGases involved in cellulose biosynthesis in plants. However, very little is known about the biochemical properties of membrane-bound EGases, and information on their substrate specificity would be especially valuable to clarify whether these enzymes could be involved in the biosynthesis of cellulose. Therefore, we have expressed the catalytic domain of Cel16 as a truncated soluble protein, Δ1-90Cel16, in the methylotrophic yeast Pichia pastoris, and, in the present paper, we describe the expression, purification, and characterization of Δ1-90Cel16, including its substrate specificity.
RESULTS
Primary Structure of Membrane-Anchored EGases
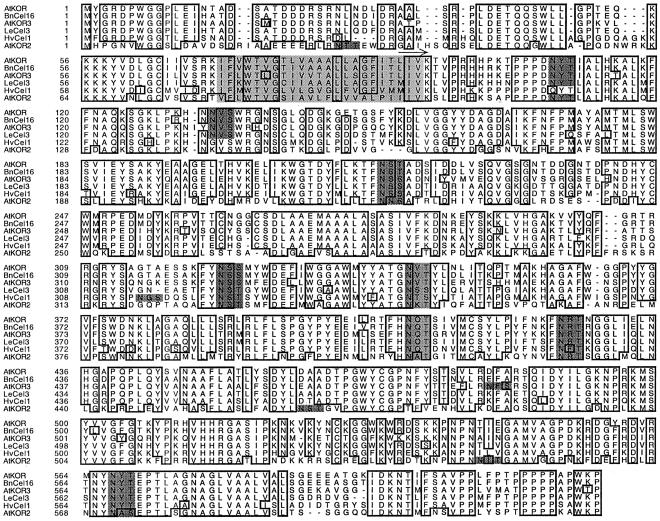
Membrane-anchored EGases have been highly conserved through plant evolution as homologous genes have been identified in both dicots and grasses. A multiple alignment of the predicted amino acid sequences of the six full-length membrane-anchored EGases found in the databases confirms that they share a relatively high degree of amino acid identity (Fig. 1), even in the cytoplasmic N terminus and the extreme, Pro-rich C terminus, indicating important functions of these domains. The six full-length membrane-anchored EGases known to date contain seven to 10 putative N-glycosylation sites, among which six are conserved (Fig. 1). These conserved putative N-glycosylation sites have also been found in partial sequences found in the databases encoding membrane-anchored EGases from soybean (Glycine max) (accession nos. AW666343 and AW704037), cotton (Gossypium hirsutum) (accession nos. BF271380, BF268560, and AW729417), rice (Oryza sativa) (accession no. C25232), maize (Zea mays) (accession no. AW065538), hybrid aspen (Populus sp.) (accession no. AI164537), Lotus japonicus (accession no. AW163991), and alfalfa (Medicago sativa) (accession no. AW692796; data not shown). The predicted amino acid sequences of all six full-length membrane-anchored EGases exhibit the diagnostic characteristics of EGases belonging to the glycoside hydrolase family 9 characterized by an inverting hydrolysis mechanism.
Figure 1.
Multiple alignment of deduced amino acid sequences of full-length plant membrane-anchored EGases. Abbreviations and Genbank accession nos. are as follows: Arabidopsis KOR (AtKOR, U37702), Arabidopsis KOR2 (AtKOR2, AC001229), Arabidopsis KOR3 (AtKOR3, AL078637), B. napus Cel16 (BnCel16, AJ242807), tomato Cel3 (LeCel3, U78526), and barley Cel1 (HvCel1, AB040769). Dots denote gaps to maximize alignment. Boxed residues are identical in at least five sequences. Dark-gray residues denote putative N-glycosylation sites among which six are conserved in the membrane-anchored EGase amino acid sequences. The core of the transmembrane domain is shown in light gray, and the catalytic domain of Cel16 expressed in P. pastoris is marked with an arrow above the sequence.
Expression and Purification of Δ1-90Cel16
A PCR fragment encoding a truncated Δ1-90Cel16 protein (Fig. 1) was cloned in the pPICZαA P. pastoris expression vector and integrated into the P. pastoris genome by transformation. The vector pPICZαA contains the N terminus signal sequence of Saccharomyces cerevisia α-factor to allow entry into the secretory pathway. About 30 transformants were tested for expression levels in the following way: Transformants were grown under expression inducing conditions (methanol) over a period of 4 d. Aliquots of the culture medium were taken out every 24 h and the level of recombinant protein in the culture medium was estimated in dot blots incubated with an anti-Cel16 serum. The highest expressing transformant, T4, seemed to secrete Δ1-90Cel16 at highest levels already after 24 h following the induction with methanol (data not shown). Therefore, the time course of the expression of Δ1-90Cel16 in T4 was further analyzed at 4, 8, and 12 h after induction and compared with the expression level after 24 h (data not shown). The overall expression level of ΔCel16 was at its maximum at 24 h and the following large-scale cultures therefore were harvested at 24 h after induction.
To monitor purification, the enzymatic activity of Δ1-90Cel16 was measured viscometrically using carboxymethylcellulose (CMC4M) as substrate. CMC4M contains approximately four carboxymethyl groups per 10 anhydro-Glc units. The purification of Δ1-90Cel16 to homogeneity was achieved with ultrafiltration, cation exchange chromatography, and concentration. Following ultrafiltration, the active retentate was loaded onto an SP-Sepharose column. Elution was carried out stepwise, with 300, 500, and finally 1,000 mm NaCl in the equilibration buffer. Δ1-90Cel16 eluted during the 500 mm NaCl elution step. Active fractions were pooled and concentrated 6-fold using a Centricon Plus-20 (Biomax-100) centrifugal filter device. A summary of the purification steps is shown in Table I.
Table I.
Purification of Δ1-90 Cel16 from P. pastoris culture broth
| Step | Protein | Activity | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| mgb | unitsc | units/mg−2 | % | fold | |
| Concentrated Crude Extracta | 4,450 | 235 | 0.05 | 100 | 1 |
| SP-Sepharose | 0.55 | 2.21 | 4.02 | 0.9 | 80.4 |
| Centricon | 0.44 | 1.66 | 3.78 | 0.7d | 75.6 |
The crude protein extract (2 L) was concentrated to 100 mL using a Millipore Minitan II.
Protein was measured spectrophotometrically using an extinction coefficient of 1 and 2.01 for the concentrated crude extract and the pure protein, respectively.
One unit is defined as released reducing ends after 12-h incubation using CMC4M as substrate.
Due to an abundant EGase secreted by P. pastoris, the apparent yield of Δ1-90Cel16 was very low.
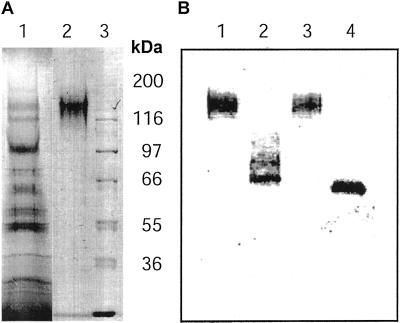
SDS-PAGE revealed the presence of a single protein band with an apparent molecular mass of approximately 130 kD in the purified Δ1-90Cel16. This was about 70 kD more than the expected molecular mass of 59 kD for the truncated protein. This band was furthermore recognized by an anti-Cel16 serum, thus confirming its identity (Fig. 2, A and B).
Figure 2.
SDS-PAGE and western analysis of the purified recombinant Δ1-90Cel16. Proteins were separated on a 10% (w/v) polyacrylamide gel by SDS-PAGE and stained with Coomassie Brilliant Blue (A) or transferred onto a polyvinylidene difluoride membrane and probed with a Cel16 antiserum (B). Lane A1, Crude extract from P. pastoris culture broth after ultrafiltration; lane A2, purified Δ1-90Cel16; lane A3, molecular mass protein marker; lane B1, purified Δ1-90Cel16 incubated with buffer; lane B2, purified Δ1-90Cel16 incubated with peptide-N-glycosidase-F (PNGaseF); lane B3, purified Δ1-90Cel16 incubated with buffer; and lane B4, purified Δ1-90Cel16 incubated with endoglycosidase-F (EndoF)/PNGaseF.
Enzymatic Deglycosylation of Δ1-90Cel16
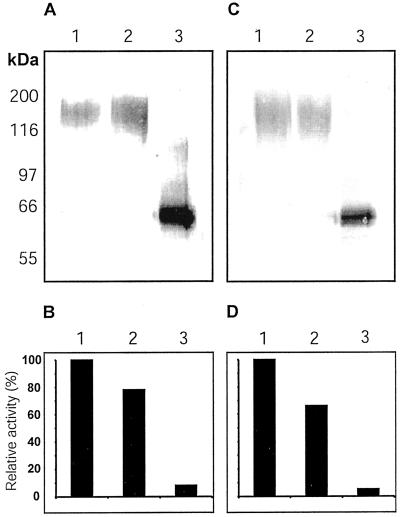
Because the Coomassie-stained Δ1-90Cel16 band was smeared, suggesting that the EGase was glycosylated and because of the large difference between the observed and expected molecular masses of Δ1-90Cel16, we suspected that the recombinant Δ1-90Cel16 was N-glycosylated. Enzymatic removal of N-glycans thus was attempted using either PNGaseF or EndoF/PNGaseF. The reactions were then analyzed in immunoblots with a Cel16 antiserum. Both treatments resulted in a reduction in apparent molecular mass to about 60 kD, matching the expected molecular mass of 59 kD of the non-glycosylated peptide, thus confirming that the recombinant Δ1-90Cel16 expressed in P. pastoris was heavily N-glycosylated (Fig. 2B). Moreover, the use of EndoF and PNGaseF in combination seemed to be more efficient than PNGaseF alone because it gave rise to a sharper band in the immunoblot.
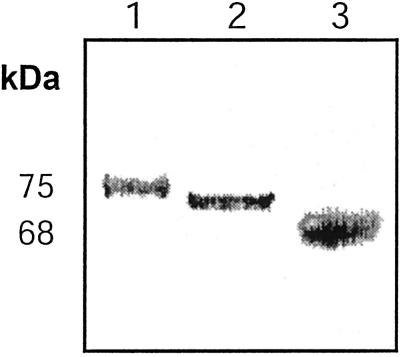
To analyze whether the extensive N-glycosylation affected its enzymatic activity, Δ1-90Cel16 was incubated with EndoF/PNGaseF for 30 min at 37°C followed by western-blot analysis and by the determination of its enzymatic activity using a reducing ends based endoglucanase assay. The deglycosylation was effective, resulting in a reduction in apparent molecular mass to 60 kD, but it also resulted in the loss of more than 95% of the enzymatic activity of Δ1-90Cel16 (Fig. 3, A and B). Upon action of PNGaseF, the original Asn residue comprised within the glycosylation site is changed to an Asp residue, creating a new, additional negative charge. This introduced charge can in turn locally modify the conformation of the peptide. The inactivation of Δ1-90Cel16 following deglycosylation with EndoF/PNGaseF could very well be due to such changes in conformation. Although deglycosylation with EndoF or EndoH is gentler and leaves a GlcNAc residue attached to the peptide, these treatments can also influence the structure of the peptide, although no modification of the local charge is introduced. Likewise, deglycosylation of Δ1-90Cel16 with EndoH resulted in a reduction in apparent molecular mass to 60 kD and also abolished the activity of Δ1-90Cel16 (Fig. 3, C and D). Adding boiled EndoF/PNGaseF or EndoH to Δ1-90Cel16 had no effect on the enzymatic activity of Δ1-90Cel16 (data not shown). Because no proteolytic degradation of Δ1-90Cel16 could be detected following deglycosylation, it seems that the N-glycans have a protective or stabilizing function for Δ1-90Cel16. When analyzing the native B. napus Cel16 protein by western blot, we found the molecular mass to be close to 72 kD (Fig. 4). Furthermore, in western analysis the KOR protein has also been reported to have an apparent molecular mass of 72 kD, in comparison with the theoretical molecular mass 69.2 kD (Zuo et al., 2000). The tomato membrane-anchored EGase, Cel3, was immunodetected as a 93- and 88-kD protein in comparison with the deduced molecular mass of 68.5 kD (Brummell et al., 1997). Plant N-glycans are approximately 1 to 2.2 kD in size (Lerouge et al., 1998; Haruko and Haruko, 1999), and therefore it cannot be excluded that Cel16, KOR, and Cel3 are N-glycosylated in planta. At least Δ1-90Cel16 seems to require the presence of some N-linked oligosaccharides for its activity or its stability; therefore, we used the fully N-glycosylated Δ1-90Cel16 for all further characterizations of the enzymatic properties of Δ1-90Cel16.
Figure 3.
Western analysis (A and C) and determination of enzymatic activity (B and D) of Δ1-90Cel16 upon deglycosylation with EndoF/PNGaseF (A and B) and endoglycosidase-H (EndoH; C and D). Immunoblots were probed with a Cel16 antiserum at a 1:1,500 (w/v) dilution. Δ1-90Cel16 activity was measured with CMC4M (0.1%, w/v) in 50 mM potassium phosphate, pH 6.0, and 250 mM NaCl, using a reducing end assay. For deglycosylation reactions and controls, Δ1-90Cel16 was incubated in buffer only on ice (lanes A1/C1 and bars B1/D1), in buffer only at 37°C (lanes A2/C2 and bars B2/D2), and with glycosidase at 37°C (lanes A3/C3 and bars B3/D3).
Figure 4.
Western analysis of B. napus Cel16 protein in crude leaf homogenate. Lane 1, Prestained 75-kD molecular mass marker protein; lane 2, the Cel16 antiserum (dilution 1:1,500, w/v) recognizes a 72-kD protein in the crude B. napus leaf homogenate (total protein loaded was 1.0 μg); lane 3, prestained 68-kD molecular mass marker protein.
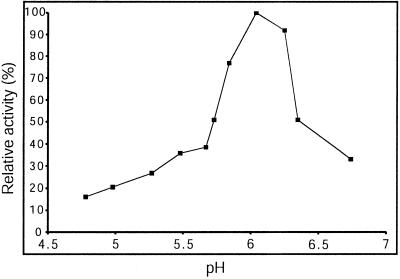
pH Profile
Before studying the substrate specificity of Δ1-90Cel16, the influence of pH on its enzymatic activity was investigated. Using CMC4M as substrate, the EGase activity of Δ1-90Cel16 was measured at various pH values between 4.8 and 6.7 at a constant concentration of NaCl of 250 mm. The pH optimum was 6.0 (Fig. 5). As a consequence, further investigations of the enzymatic properties and substrate specificity of Δ1-90Cel16 were carried out at pH 6.0 and 250 mm NaCl.
Figure 5.
Effect of pH on the enzymatic activity of Δ1-90Cel16. Assays were performed in 50 mm potassium phosphate and 250 mm NaCl, using CMC4M as substrate.
Enzymatic Properties of Δ1-90Cel16
The substrate specificity of Δ1-90Cel16 was determined using various cellulose derivatives as well as other cell wall carbohydrate polymers and oligosaccharides. Hydrolysis of polymers was monitored using a reducing end assay, whereas the composition of the reaction products from oligosaccharides was analyzed by high-performance anion-exchange chromatography on a Carbo-Pac PA-1 column. Because of the homology between membrane-anchored EGases and secreted EGases, various β-1,4-glucans that are known to be substrates for the plant-secreted EGases were tested as well as other, distinct polymers.
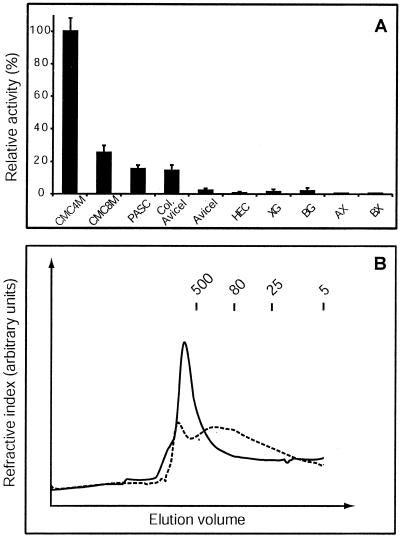
Of all the cellulose derivatives, CMC4M, CMC8M, phosphoric acid swollen cellulose (PASC), and colloidal Avicel could be hydrolyzed by Δ1-90Cel16, whereas Avicel and hydroxyethylcellulose (HEC) could not. Furthermore, Δ1-90Cel16 was not active on tamarind xyloglucan, barley (1→3),(1→4)-β-d-glucan, wheat (Triticum aestivum) arabinoxylan, or birchwood xylan (Fig. 6A). CMC4M proved to be the best substrate, about 3.5 times better than CMC8M. This is most likely due to the fact that CMC4M has a two times lower degree of substitution with carboxymethyl groups than CMC8M. In HEC, hydroxyl groups are substituted with hydroxyethyl groups with a degree of substitution of about 25 hydroxyethyl groups per 10 anhydro-Glc units, preventing the polymer from crystallizing. Substitution with carboxymethyl and hydroxyethyl groups seems to interfere with the action of Δ1-90Cel16 on the modified cellulose, showing that Δ1-90Cel16 exhibits a marked preference for low- and un-substituted cellulose chains. Although often described as amorphous, PASC has a high surface to volume ratio (Ong et al., 1993) and is probably a low-crystallinity form of cellulose (Atalla, 1993). In PASC, more glucan chains are thought to be accessible to the solvent as a result of swelling and disruption of crystallinity. Avicel is a heterogeneous microcrystalline cellulose preparation, in which microfibrils are aggregated, whereas colloidal Avicel is a homogenous suspension in which the microfibrils are mostly disaggregated and the number of available binding regions for an EGase is increased. The fact that PASC is a better substrate than colloidal Avicel (Fig. 6A), therefore, is due to the swelling and lower crystallinity of PASC giving more accessible sites for Δ1-90Cel16. The reason for Avicel not to be degraded by Δ1-90Cel16 (Fig. 6A) is probably due to the highly crystalline, aggregated structure, making it sterically impossible for Δ1-90Cel16 to bind to the backbone of the cellulose chains.
Figure 6.
Substrate specificity of Δ1-90Cel16 (A) and analysis of the reaction products using size exclusion chromatography (B). Substrates used were: CMC4M-4M (CMC4M), CMC4M-8M (CMC8M), PASC, colloidal Avicel (Col. Avicel), Avicel, HEC, tamarind xyloglucan (XG), barley β-1,3-β1.4-glucan (BG), wheat arabinoxylan (AX), and birchwood xylan (BX). Fifty-five nanograms microliters−1 of Δ1-90Cel16 was incubated with each substrate (0.1%, w/v). Error bars represent se from eight independent assays (A). Size-exclusion chromatography of CMC4M untreated (t = 0, solid line) and after hydrolysis overnight with 55 ng μL−1 of Δ1-90Cel16 (dotted line; B). The molecular mass of the eluted components was estimated by comparing their retention times with those of dextran standards (kilodaltons).
Δ1-90Cel16 was unable to hydrolyze the cello-oligosaccharides cellotriose, cellotetraose, cellopentaose, and p-nitrophenyl-β-cellotriose, indicating that these were too small to allow an efficient binding of the enzyme. The Xyl oligosaccharide 1,4-β-xylopentaose could not be hydrolyzed by Δ1-90Cel16 either. Cel16 is predicited to exhibit an inverting hydrolysis mechanism (Henrissat, 1991). Inverting enzymes cannot perform transglycosyltaion reactions. In accordance, no transglycosylation activity could be detected for Δ1-90Cel16 upon incubation with any of the oligosaccharides mentioned above and subsequent analysis using high-performance anion-exchange chromatography. In conclusion, in contrast to many secreted EGases, Δ1-90Cel16 did not hydrolyze either the commonly used EGase substrate HEC nor XG. Δ1-90Cel16 only hydrolyzed low- or unsubstituted cellulose derivatives. However, we cannot exclude that the specific activities of the recombinant Δ1-90Cel16 may not necessarily be identical to those of the protein in its native environment, particularly because the glycosylation patterns are different.
Size-Exclusion Chromatography
The activity of Δ1-90Cel16 could be measured viscometrically, suggesting that Δ1-90Cel16 is an endo-acting enzyme. However, to further investigate whether Δ1-90Cel16 is a true endo-acting enzyme or a processive enzyme (both endo- and exo-acting), we analyzed Δ1-90Cel16-hydrolyzed CMC4M by size exclusion chromatography (Fig. 6B). Untreated CMC4M eluted as a major peak bigger than 500 kD. Δ1-90Cel16 hydrolysis of CMC4M resulted in accumulation of lower molecular mass CMC4M fragments with a broad size range of 5 to 500 kD. No cello-oligosaccharides or Glc could be detected. This proved that Δ1-90Cel16 is a true non-processive endo-hydrolase.
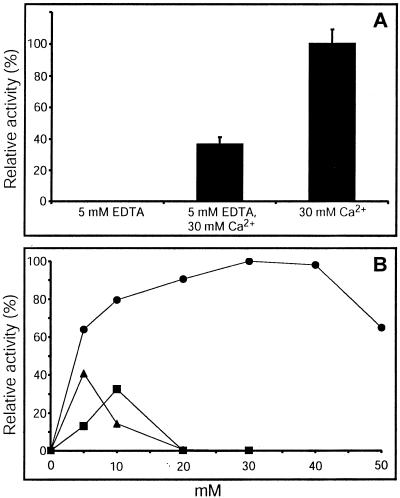
Effect of EDTA and Divalent Cations
Using CMC4M as substrate, we found that the presence of 5 mm EDTA totally abolished the enzymatic activity of Δ1-90Cel16 (Fig. 7A). This effect could be compensated by the addition of CaCl2 (Fig. 7A). CaCl2 strongly enhanced activity of Δ1-90Cel16 in a concentration-dependent manner up to about 40 mm (Fig. 7B). At higher CaCl2 concentrations, the activity of Δ1-90Cel16 declined. MgCl2 and ZnCl2 had much less effect on enzyme activity at concentrations up to 10 mm but were detrimental at higher concentrations (Fig. 7B). Thus, the activity of Δ1-90Cel16 appears to be strongly dependent on calcium. Calcium has been shown to increase stability of many proteins including EGases but some EGases have been reported to require the presence of calcium for their enzymatic activity. Such a strong dependence on calcium has previously been reported for some bacterial EGases, where calcium has been shown to either have a direct effect on the enzymatic properties of the enzyme (Chauvaux et al., 1990) or a general tertiary structure stabilizing effect (Welfle et al., 1995).
Figure 7.
Inhibition of Δ1-90Cel16 by 5 mm EDTA (A) and influence of Ca2+ (A and B, ●), Zn2+ (B, ▴), and Mg2+ (B, ▪) on the activity of Δ1-90Cel16. Addition of 30 mM Ca2+ corresponds to 100% activity. Error bars represent se from six independent assays. Fifty-five nanograms microliters−1 of Δ1-90Cel16 was incubated overnight with CMC4M (0.1%, w/v) in 50 mm potassium phosphate buffer and 250 mm NaCl, pH 6.0, in the presence of EDTA and/or divalent cations. Δ1-90Cel16 activity consequently was determined using a reducing assay.
DISCUSSION
The new class of membrane-anchored EGases, of which three genes (KOR, KOR2, and KOR3) have been identified in Arabidopsis, has gained much attention due to a potential function in cellulose biosynthesis (Brummell et al., 1997; Nicol et al., 1998; Zuo et al., 2000; Mølhøj et al., 2001a, 2001b). To our knowledge, these membrane-anchored EGases and a maize membrane-associated exo-β-d-glucanase (Kim et al., 2000) are the only known classes of plant plasma membrane-associated glycohydrolases.
Here, we report the enzymatic properties of the catalytic domain of a membrane-anchored EGase from B. napus, Cel16, expressed as a soluble and functional enzyme, Δ1-90Cel16, leaving out the predicted intracellular N-terminal and transmembrane domains, using the P. pastoris expression system. This expression system has recently been used for the recombinant expression of secreted plant EGases (Ferrarese et al., 1998). We initially tried to express Δ1-90Cel16 in Escherichia coli as a fusion protein to thioredoxin but could not detect any enzymatic activity using CMC4M as substrate.
We found that the P. pastoris-expressed truncated membrane-anchored EGase, Δ1-90Cel16, was highly N-glycosylated and although the native B. napus Cel16 seemed not to be N-glycosylated to the same degree as the recombinant Δ1-90Cel16, one cannot exclude that it also contains some N-glycans. Δ1-90Cel16 was inactivated when deglycosylated with EndoF/PNGaseF or EndoH, probably due to a change in conformation or loss of stability. The N-glycans may very well help keep the correct folding of the truncated protein.
Secreted EGases have been proposed to act primarily on xyloglucan (Hayashi, 1989; Brummell et al., 1994) but a secreted EGase has recently been shown to release cello-oligosaccharides from the cell wall of poplar suspension cell cultures (Ohmiya et al., 2000). Thus, potential in vivo substrates for membrane-anchored EGases could be xyloglucan and/or amorphous cellulose in the cell wall. We found that Δ1-90Cel16 hydrolyzed CMC4M, CMC8M, PASC, and colloidal Avicel but not xyloglucan, β-glucan, or xylan. Δ1-90Cel16 did not hydrolyze cellopentaose, suggesting that it may have more than five Glc-binding subsites. This fits well with the observation that EGase activity was detected using CMC as substrate in Suc density/gradient fractions enriched for Cel3, a tomato homolog of Cel16, and KOR (Brummell et al., 1997). Because xyloglucan is not a substrate for Δ1-90Cel16, it is unlikely that plant membrane-anchored EGases are involved in xyloglucan integration or metabolism. This observation fits well with the fact that genes encoding plant membrane-anchored EGases have also been identified in several grasses like barley (accession no. AAB040769), maize (accession no. AW065538), and rice (accession no. C25232), where xyloglucan is only a minor component of the primary cell wall (Carpita, 1996). Altogether, this leads us to propose that the natural substrate of Cel16 is a component of cellulose. However, although the recombinant Δ1-90Cel16 does not degrade xyloglucans, it cannot be excluded that Cel16 could act in concert with xylosidases to degrade xyloglucans in planta.
The Arabidopsis mutant, rsw2, which is an allelic mutant of KORRIGAN (H. Höfte, personal communication; Lane et al., 2001), shows a similar temperature-sensitive, radial swelling phenotype as the cellulose synthase rsw1 mutant (Baskin et al., 1992; Arioli et al., 1998), and produces about 50% less cellulose in the roots than wild-type-grown plants at the restrictive temperature (Peng et al., 2000). Furthermore, rsw2 shows little changes in the production of matrix polysaccharides. The reason for the KORRIGAN mutant to have thicker cell walls, increased cell diameter, holes in cell walls, and some collapsed cells, (Nicol et al., 1998) is probably due to the lower levels of cellulose as found in the rsw2 mutant (Peng et al., 2000), giving less hydrogen bindings and less dense cell walls. The substrate preference of Δ1-90Cel16 for low-substituted amorphous cellulose, the polymeric nature of the end reaction products of Δ1-90Cel16 hydrolysis, and the lower level of cellulose found in the rsw2 mutant, point to a role of Cel16 and KOR in cellulose biosynthesis.
One can think of three possible functions for plant membrane-anchored EGases in cellulose synthesis: (a) a proofreading activity involved in trimming off disordered amorphous cellulose chains, (b) determination of the length of individual cellulose chains during or subsequent to microfibril assembly, or (c) termination of the whole cellulose synthesis releasing the cellulose microfibril from the synthase complex. Chapple and Carpita (1998) suggested that membrane-anchored EGases may proofread the glucan chains and excise disordered amorphous cellulose. The second possibility regards determination of chain length. Microfibrils are often much longer than individual cellulose chains and at many points along the length of a microfibril cellulose chains end and new ones start (Delmer, 1999). These breaks have been proposed to help the arrangement of microfibrils in the apoplastic spaces, eventually mediating a flexibility to the microfibrils and helping the cellulose molecules to associate by Van der Waals and hydrogen bonds to form microfibrils. These breaks could be due to the activity of membrane-anchored EGases. The third possibility is a function in cellulose chain termination (Delmer, 1999). This would mean that membrane-anchored EGases control the molecular mass of individual cellulose microfibrils. The fact that an Acetobacter xylinum-secreted EGase seems to be involved in releasing the cellulose from the cell (Oikawa et al., 1997) suggests that plant membrane-anchored EGases very well could have the same function. The substrate specificity of Δ1-90Cel16 reported here implies that membrane-anchored EGases can perform each of the three proposed functions.
A novel mechanism for cellulose synthesis has been proposed for Agrobacterium tumefaciens (Matthysse et al., 1995a, 1995b). This model involves transfer of Glc from UDP-Glc to form a lipid-linked cello-oligosaccharide. Subsequent polymerization is catalyzed by transglycosylation of cellobiose or cello-oligosaccharides from the lipid through the action of the membrane-bound EGase, CelC. In a similar manner, plant membrane-anchored EGases could be involved in an early step of cellulose synthesis in providing short primers of the required length for chain elongation. However, it is not known whether cellulose biosynthesis in plants requires a primer.
In conclusion, the characterization of the substrate specificity of Δ1-90Cel16 is an important step toward an understanding of the function of plant membrane-anchored EGases because it provides a direct evidence that cellulose is the substrate of these enzymes. The N-terminal, predicted intracellular domain of membrane-anchored EGases may be involved in localizing the enzyme in the vicinity of the cellulose-synthesizing terminal complex by specific interactions and it will be interesting to elucidate whether membrane-anchored EGases interact with or are integrated parts of the terminal complex as the catalytic subunit of a cellulose synthase has been shown to be (Kimura et al., 1999).
MATERIALS AND METHODS
Δ1-90Cel16 Gene Construct
A full-length cDNA encoding Cel16 was previously isolated and cloned (Mølhøj et al., 2001a). The nt271-1863 region of the cDNA clone Cel16 (Mølhøj et al., 2001a) corresponding to the presumed catalytic domain (Leu91-Pro621) was amplified using PCR and proofreading polymerase. The cDNA fragment corresponding to the truncated protein was obtained using synthetic oligonucleotides: 5′-CAGGAATTCTTGATCGTCAAAACTGTGCC-G-3′ and 5′-TAATAGCGGCCGCTCAAGGTTTCCATGGTGCTGGTG3′. EcoRI (in italic in the sequence) and NotI (underlined in the sequence) sites were introduced for cloning as well as one stop codon at the 3′ end (in bold in the sequence). After digestion, the PCR fragment was cloned in the EcoRI and NotI sites of the pPICZαA Pichia pastoris expression vector (Invitrogen, Copenhagen), yielding a construct named pPICZαA-Δ1-90Cel16. The truncated Cel16 cDNA fragment thus was inserted in frame at the 3′ end of the Saccharomyces cerevisiae α-factor secretion signal to ensure secretion of the recombinant proteins and a stop codon had been included before the myc epitope-(His)6-tag. Following sequencing, pPICZαA-Δ1-90Cel16 was linearized with PmeI and used for the transformation of P. pastoris wild-type X-33 strain. Transformation was performed by electroporation according to the manufacturer (Invitrogen). As a negative control, the pPICZαA vector linearized with PmeI was used for the transformation. Transformants were selected on plates with yeast extract peptone dextrose medium containing 100 μg mL−1 zeocin.
Production of a Polyclonal Antibody toward the C Terminus of Cel16
A region located at the C terminus of Cel16 was expressed recombinantly in Escherichia coli as a fusion protein with 6× His-dihydrofolate reductase (N terminally) to be used as an antigen to produce an antiserum to Cel16. For this, the nucleotide 1,456 through 1,863 region corresponding to Gln486-Pro621 and belonging to the presumed periplasmatic enzyme domain of Cel16 was amplified from the full-length cDNA clone (Mølhøj et al., 2001a) using the PCR technique. The PCR primers (5′-GGGGAAGATCTCA-GATTGATTACATACTAGGTAAAAAC-3′ and 5′-TTCAC-CTGCAGAGGTTTCCATGGTGCTGGTGGG-3′) had been designed to introduce a BglII and a PstI restriction sites (underlined in the sequences) at the 5′- and 3′-ends, respectively. The PCR product was then cloned into the pQE-40 vector (Qiagen, Albertslund, Denmark) that had been digested with BglII and PstI. The resulting plasmid was checked by sequencing and transformed into E. coli SG13009[pREP4]. The transformed cells were cultured in LB medium until optical density (OD)600 reached 0.6. Expression of the recombinant protein was then induced with 2 mm isopropyl-β-thiogalactopyranoside and growth was continued for another 4 h. Cells were harvested by centrifugation. Cell lysis and purification of the recombinant protein under denaturing conditions using an Ni-NTA Agarose column were performed according to the manufacturer (Qiagen). Rabbits were immunized both intramuscularly in the hind leg and subcutaneously in the neck region at 4-week intervals with 70 μg of antigen emulsified with RIBI adjuvant R-730 (RIBI ImmunoChem Research Inc., Hamilton, MT). Pre-immune serum was collected before the first injection and antisera were collected 12 d after each of four injections.
Screening of P. pastoris Clones and Recombinant Expression of Δ1-90Cel16
About 50 transformants were screened for high secretion of the recombinant protein as follows: 10 mL of buffered complex glycerol media (BMGY) supplemented with 100 μg mL−1 zeocin in a 100-mL flask was inoculated with a single colony and grown in a shaking incubator (28°C, 240 rpm) for 24 h until OD600 reached 2 to 6. Cells were harvested by centrifugation and resuspended in 10 mL of buffered complex methanol media at OD600 = 1. The cultures were incubated another 96 h. One-milliliter culture medium aliquots were taken out and methanol (final concentration 0.75%–1%, v/v) was added every 24 h. The medium aliquot samples (200 μL) were analyzed by immuno dot blot using ProBlott membrane (Applied Biosystems, Foster City, CA), a Bio-Dot Apparatus (Bio-Rad, Herlev, Denmark), and the polyclonal antibody raised against the C terminus of Cel16. The transformant with the highest level of secreted recombinant protein was chosen for large-scale expression. Due to highest expression levels after 24 h, the best expressing clone, T4, was further analyzed at 4, 8, 12, and 24 h of expression and used for large-scale (2 L) expression in flasks using the same conditions described above. Time for optimal expression was determined to about 24 h.
Purification of Δ1-90Cel16 Recombinantly Expressed in P. pastoris
A colony of the transformant strain T4 was used to inoculate 5 mL of BMGY (100 μg mL−1 zeocin). After 12 h growth in a shaking incubator (28°C, 230 rpm), the whole culture was used to inoculate 200 mL of BMGY (100 μg mL−1 zeocin) in a 600-mL flask. This culture was grown for 12 h under the same conditions until OD600 reached 2 to 6. Cells were harvested by centrifugation and resuspended into 2 × 1 L of buffered complex methanol media in two 5-L flasks at OD600 = 1. These cultures were grown for 24 h. The cells (OD600 15–23) were harvested by centrifugation (13,000g for 10 min) and discarded. The supernatant (2 L) was further filtrated through folded filters (Whatman, Albertslund, Denmark) and supplemented with protease inhibitors (0.5 mm phenylmethylsulfonylfluoride, 2 mm EDTA, 1 μm pepstatin, and 10 μm E-64 {N-[N-(l-3-transcarboxyoxirane-2-carbonyl)-l-leucyl]-agmatine, Boehringer Mannheim, Kvistgaard, Denmark}) to minimize the extracellular protease activity present in the P. pastoris culture. All of the following steps were performed at 4°C.
The supernatant was filtered to a final volume of 100 mL using a Minitan II unit equipped with a high-flux biomax polysulfone membrane (both from Millipore, Glostrup, Denmark) and supplemented with Complete protease inhibitor cocktail (Boehringer Mannheim). Before the crude extract was applied to an SP-Sepharose Fast Flow XK25 column (Amersham Pharmacia Biotech, Birkeroed, Denmark), pH and conductivity were adjusted with 1 m acetic acid and water, respectively. The SP-Sepharose column was equilibrated with 100 mm Na-acetate, pH 5.5. After a washing step using the same buffer, the column was eluted stepwise with 100 mm Na-Acetate, pH 5.5, containing 0.3, 0.5, and 1 m NaCl. Δ1-90Cel16 eluted with 100 mm Na-Acetate, pH 5.5, and 0.5 m NaCl. Fractions containing Δ1-90Cel16 were pooled and concentrated about 6-fold using Centricon Plus-20 (Biomax-100; Amicon, Roskilde, Denmark). All purification steps were followed using SDS-PAGE stained with Coomassie Brilliant Blue, and the identity of the purified protein was checked by western analysis. The concentration of solutions of pure Δ1-90Cel16 was estimated spectrophotometrically at 280 nm using a calculated extinction coefficient, E0.1% = 2.01 (Gill and von Hippel, 1989).
SDS-PAGE and Western Blotting
SDS-PAGE was performed in 10% (w/v) acrylamide, and proteins were visualized with Coomassie Brilliant Blue R-250. For western blots, proteins were transferred onto ProBlott polyvinylidene difluoride membranes (Applied Biosystems) by semidry blotting. For immunoreaction, incubation with the primary antibody (O.N. at 4°C), diluted 1:1,500 (v/v), and subsequently with alkaline phosphatase-conjugated swine anti-rabbit antibody (3 h at room temperature; Dako, Glostrup, Denmark), diluted 1:2,000 (v/v), was carried out in Tris-buffered saline/5% (v/v) horse serum/2% (v/v) Tween 20 (Blake et al., 1984). Proteins in Brassica napus cv Bridger leaves were extracted using ice-cold 0.2 m NaOH, 2% (v/v) mercaptoethanol, centrifuged at 12.000 rpm for 10 min at 4°C, and precipitated by addition of ice-cold acetone (Dal Degan et al., 1994).
Polymeric Substrates Used
CMC4M-8M, high viscosity (CMC8M, Sigma, Copenhagen), CMC4M-4M (CMC4M, Megazyme, Bray, Ireland), Avicel PH101 (Sigma-Aldrich, Copenhagen), PASC, HEC (Dir. Prof. Dr. J. Demeester, Centre for Standards, Gent, Belgium), barley (1→3),(1→4)-β-d-glucan medium viscosity (Megazyme), wheat arabinoxylan medium viscosity (Ara:Xyl = 41:59; Glu, Gal, and Man <1%; Megazyme), birchwood xylan (>90% [w/v] Xyl residues; Sigma), colloidal Avicel PH101, and tamarind xyloglucan (Megazyme) were prepared as 1% (w/v) solutions in 50 mm potassium phosphate, pH 6.0. Cellotriose, cellotetraose, p-Nitrophenyl-β-cellotriose, cellopentaose (Calbiochem, Darmstadt, Germany), and 1,4-β-xylopentaose (Megazyme) were prepared as 20 mm solutions in 50 mm potassium phosphate, pH 6.0. CMC8M contains 6.5 to nine carboxymethyl groups per 10 anhydro-Glc units, whereas CMC4M contains approximately four carboxymethyl groups per 10 anhydro-Glc units. HEC contains about 25 hydroxyethyl groups per 10 anhydro-Glc units. PASC was prepared from Avicel PH101 (microcrystalline cellulose) by using 85% (w/v) phosphoric acid (Wood, 1988), and stored in 5 mm NaN3 aqueous suspension. Colloidal Avicel was obtained by milling Avicel PH101 (10 g L−1) for 5 h at 4°C in distilled water with a shaker (Pagés et al., 1997). After sedimentation (for 12 h at 4°C), two distinct phases were obtained: the pellet, containing the non-disaggregated cellulose particles, and the supernatant, corresponding to a homogenous suspension in which the cellulose particles were mostly disaggregated. The supernatant consisting of a colloidal suspension was concentrated by ultrafiltration with an Amicon concentrator and a 10K Amicon Diaflo membrane (Amicon). The cellulose concentration was calculated from dry weight of an aliquot fraction.
Determination of Enzyme Activity
During purification, fractions were assayed viscometrically for their EGase activity in an Ostwald microviscometer (Schott-Geräte, Hofheim, Germany) at 35°C in 2 mL. The reaction was initiated by the addition of 1 mL of enzyme to 1 mL of a 1% (w/v) CMC4M solution in 50 mm potassium phosphate, pH 6.0. Viscosity was measured at time intervals, as described by Truelsen and Wyndaele (1991).
The purified Δ1-90Cel16 was assayed at room temperature for its EGase activity. Δ1-90Cel16 was incubated with 0.1% (w/v) polymer or 2 mm oligosaccharide and 50 mm potassium phosphate, pH 6.0, overnight at room temperature before measuring reducing ends using the p-hydroxybenzoic acid hydrazine assay for reducing sugars (Lever, 1972). Activity was expressed as the newly released reducing ends using Glc as standard. For Avicel and colloidal Avicel, the reaction mixtures were incubated with gentle agitation to inhibit sedimentation of the polymers.
Enzymatic Deglycosylation
For enzymatic deglycosylation of N-linked glycans, 1.5 μg purified Δ1-90Cel16 was incubated 30 min or overnight at 37°C with PNGaseF or the combined EndoF/PNGaseF from Flavobacterium meningosepticum (Glyko, Oxfordshire, UK) or EndoH (Sigma) in 50 mm potassium phosphate, pH 6.0, before being assayed and analyzed by western blotting.
Enzymatic Properties
pH Profile
The pH profile of the activity of Δ1-90Cel16 was determined using CMC4M (0.1%, w/v) as substrate in 50 mm potassium phosphate, pH 4.7 to 6.8, containing 250 mm NaCl. The enzymatic reaction was allowed to proceed for 12 h at room temperature and subsequently stopped by the addition of cold NaOH to 50 mm. An aliquot of the reaction mixture had been taken out just after addition of Δ1-90Cel16 and the reaction stopped by the addition of cold NaOH for the determination of reducing ends before the action of Δ1-90Cel16. Reducing ends at reaction start and after 12 h were determined as described above.
Substrate Specificity of Δ1-90Cel16
The substrate specificity of Δ1-90Cel16 was established by the determination of the hydrolysis rates for the hydrolysis of a series of cellulose derivatives and other cell wall carbohydrate polymers (see detailed list above) as well as cello-oligosaccharides. Solutions of polymers (0.1%, w/v) or oligosaccharide (2 mm) were incubated with 55 ng μL−1 of Δ1-90Cel16 O.N. at room temperature. Δ1-90Cel16-catalyzed hydrolysis was measured using the reducing end assay described above. For the reactions with oligosaccharides, the compostion of the reaction products was furthermore analyzed by high-performance anion-exchange chromatography on a Carbo-Pac PA-1 column (Dionex, Roedovre, Denmark) and was eluted using a gradient of sodium acetate (from 0–0.5 m in 50 mL) in 0.1 m NaOH and monitored by pulse amperometric detection.
Effect of Divalent Metal Cations
To investigate the effect of divalent metal cations, 5 mm EDTA, or 5 to 50 mm CaCl2, or 5 to 20 mm MgCl2, or 5 to 20 mm ZnCl2 was added to the standard assay (0.1%, w/v) CMC4M in 50 mm potassium phosphate buffer, pH 6.0, containing 250 mm NaCl and 55 ng μL−1 of Δ1-90Cel16). All other conditions were as for the pH profile study.
Size-Exclusion Chromatography
A 0.1% (w/v) solution of CMC4M in 50 mm potassium phosphate, pH 6.0, was incubated with 55 ng μL−1 of Δ1-90Cel16 for 12 h at room temperature. Size exclusion chromatography of the reaction mixture was performed on a Superose 12 column (Amersham Pharmacia Biotech) equilibrated in 50 mm ammonium formate, pH 5.0. Samples containing an equivalent of 170 μg CMC4M were applied to the column and eluted isocratically in the same buffer at a flow rate of 24 mL h−1. The eluent was monitored by refractive index detection. The molecular mass of the eluted components was estimated by comparing their retention times with those of blue dextran standards.
ACKNOWLEDGMENTS
We are grateful to Helle Munck Petersen for technical assistance. Dr. Kylie Joy Nunan is thanked for assistance in the gel-filtration chromatography.
Footnotes
This work was supported by a grant from the Danish National Research Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010269.
LITERATURE CITED
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Atalla RH. The structures of native celluloses. In: Suominen P, Reinikainen T, editors. Trichoderma reesei Cellulases and Other Hydrolases, Foundation for Biotechnical and Industrial Fermentation. Helsinki. 1993. pp. 25–39. [Google Scholar]
- Baskin TI, Betzner AS, Hoggart R, Cork A, Williamson RE. Root morphology mutants in Arabidopsis thaliana. Aust J Plant Physiol. 1992;19:427–437. [Google Scholar]
- Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Catala C, Lashbrook CC, Bennett AB. A membrane-anchored E-type endo-1,4-β-glucanase is localized on golgi and plasma membranes of higher plants. Proc Natl Acad Sci USA. 1997;94:4794–4799. doi: 10.1073/pnas.94.9.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Lashbrook CC, Bennett AB. Plant endo-β-1,4-glucanases: structure, properties and physiological function. Am Chem Soc Symp Ser. 1994;566:100–129. [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, Baulcombe DC, Fincher GB. Virus-induced silencing of a plant cellulose synthase gene. Plant Cell. 2000;12:691–706. doi: 10.1105/tpc.12.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M, Griffing LR. The plant extracellular matrix: news from the cell's frontier. Plant Cell. 1996;8:1451–1463. doi: 10.1105/tpc.8.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala C, Rose JK, Bennett AB. Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol. 2000;122:527–534. doi: 10.1104/pp.122.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C, Carpita N. Plant cell walls as targets for biotechnology. Curr Opin Plant Biol. 1998;1:179–185. doi: 10.1016/s1369-5266(98)80022-8. [DOI] [PubMed] [Google Scholar]
- Chauvaux S, Beguin P, Aubert JP, Bhat KM, Gow LA, Wood TM, Bairoch A. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem J. 1990;265:261–655. doi: 10.1042/bj2650261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Dal Degan F, Rocher A, Cameron-Mills V, von Wettstein D. The expression of serine carboxypeptidases during maturation and germination of the barley grain. Proc Natl Acad Sci USA. 1994;91:8209–8213. doi: 10.1073/pnas.91.17.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E. Multiple endo-1,4-β-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol. 1999;46:39–61. doi: 10.1016/s0070-2153(08)60325-7. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarese L, Trainotti L, Gattolin S, Casadoro G. Secretion, purification and activity of two recombinant pepper endo-β-1,4-glucanases expressed in the yeast Pichia pastoris. FEBS Lett. 1998;422:23–26. doi: 10.1016/s0014-5793(97)01592-5. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Haruko U, Haruko O. Glycobiology of plant glycoprotein epitope: structure, immunogenicity and allergeneicty of plant glucotopes. Trends Glycosci Glycotechnol. 1999;11:413–428. [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- Hayashi T, Ohsumi C. Endo-1,4-β-glucanase in the cell wall of stems of auxin-treated pea seedlings. Plant Cell Physiol. 1994;35:419–424. [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Olek AT, Carpita NC. Cell wall and membrane-associated exo-β-D-glucanases from developing maize seedlings. Plant Physiol. 2000;123:471–486. doi: 10.1104/pp.123.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2086. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE. Temperature sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis thaliana. Plant Physiol. 2001;126:278–288. doi: 10.1104/pp.126.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Laine AC, Gomord V, Faye L. N-Glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol. 1998;38:31–48. [PubMed] [Google Scholar]
- Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Thomas DL, White AR. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995b;177:1076–1081. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995a;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Fry SC, Durachko DM, Cosgrove DJ. The relationship between xyloglucan endotransglycosylase and in-vitro cell wall extension in cucumber hypocotyls. Planta. 1993;190:327–331. doi: 10.1007/BF00196961. [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Johansen B, Ulvskov P, Borkhardt B. The expression of a membrane-anchored endo-1,4-β-glucanase from Brassica napus, orthologous to KOR from Arabidopsis thaliana, is inversely correlated to elongation in light-grown plants. Plant Mol Biol. 2001a;45:93–105. doi: 10.1023/a:1006475908000. [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Jørgensen B, Ulvskov P, Borkhardt B. Two Arabidopsis thaliana genes, KOR2 and KOR3, which encode membrane-anchored endo-1,4-β-D-glucanases are differentially expressed in developing leaf trichomes and their support cells. Plant Mol Biol. 2001b;46:263–275. doi: 10.1023/a:1010688726755. [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya Y, Samejima M, Shiroishi M, Amano Y, Kanda T, Sakai F, Hayashi T. Evidence that endo-1,4-β-glucanases act on cellulose in suspension-cultured poplar cells. Plant J. 2000;24:147–158. doi: 10.1046/j.1365-313x.2000.00860.x. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Kamatani T, Kaimura T, Ameyama M, Soda K. Endo-β-glucanase from Acetobacter xylinum: purification and characterization. Curr Microbiol. 1997;34:309–313. doi: 10.1007/s002849900187. [DOI] [PubMed] [Google Scholar]
- Ong E, Gilkes NR, Miller RC, Jr, Warren RAJ, Kilburn DG. The cellulose-binding (CBDCex) domain of an exoglucanase from Cellulomonas fimi: production in Escherichia coli and characterization of the polypeptide. Biotechnol Bioeng. 1993;42:401–409. doi: 10.1002/bit.260420402. [DOI] [PubMed] [Google Scholar]
- Pagés S, Gal L, Bélaich A, Gaudin C, Tardif C, Bélaich JP. Role of Scaffolding protein CipC of Clostridium cellulyticum in cellulose degradation. J Bacteriol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Hocart CH, Redmond JW, Williamson RE. Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta. 2000;211:406–414. doi: 10.1007/s004250000301. [DOI] [PubMed] [Google Scholar]
- Reiter WD. The molecular analysis of cell wall components. Trends Plant Sci. 1998;3:27–32. [Google Scholar]
- Truelsen TA, Wyndaele R. Cellulase in tobacco callus: regulation and purification. J Plant Physiol. 1991;139:129–134. [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welfle K, Misselwitz R, Welfle H, Politz O, Borriss R. Influence of Ca2+ on conformation and stability of three bacterial hybrid glucanases. Eur J Biochem. 1995;229:726–735. doi: 10.1111/j.1432-1033.1995.tb20520.x. [DOI] [PubMed] [Google Scholar]
- Wood TM. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 1988;160:19–25. [Google Scholar]
- Wu SC, Blumer JM, Darvill AG, Albersheim P. Characterization of an endo-β-1,4-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiol. 1996;110:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Nishizawa N, Wu Y, Kost B, Chua N-H. KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell. 2000;12:1137–1152. doi: 10.1105/tpc.12.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]