Summary
Systematic reviews require substantial time and effort. This study compared the results of conducting reviews by human reviewers with those conducted with Artificial Intelligence (AI). We identified 11 AI tools that could assist in conducting a systematic review. None of the AI tools could retrieve all articles that were detected with a manual search strategy. We identified tools for deduplication but did not evaluate them. AI screening tools assist the human reviewer in presenting the most relevant article on top, which could reduce the number of articles that need to be screened on title and abstract, and on full text. There was a poor inter-rater reliability to evaluate the risk of bias between AI tools and the human reviewers. A summary table created by AI tools differs substantially from manually constructed summary tables. This study highlights the potential of AI tools to support systematic reviews, particularly during screening phases, but not to replace human reviews.
Subject areas: Medical research, Artificial intelligence
Graphical abstract

Highlights
-
•
Systematic reviews require much time; we evaluated the performance of AI tools
-
•
Eleven AI tools were identified to support conducting systematic reviews
-
•
Screening with AI may reduce workload during screening phases
-
•
Complete replacement of human reviewers by AI tools is not yet possible
Medical research; Artificial intelligence
Introduction
In research, the quality of evidence is partially defined by the hierarchy of study designs, with systematic reviews positioned at the top ranks of that pyramid.1,2 By analyzing the breadth of available studies on a specific research question, systematic reviews can elucidate areas where there is insufficient or conflicting evidence.3,4 This allows researchers to pinpoint gaps that need further investigation.3,5 However, due to the ever-increasing number of new research being published, a huge amount of time, resources and effort is required to properly conduct a systematic review.6 Some researchers have estimated that the duration from protocol registration to publication can range between 6 and 11 months, to as long as 3 years.6,7,8,9 Consequently, it is plausible that many of these reviews may already be considered outdated and necessitate updating prior to publication.10 The recommendations set forth in the Cochrane Handbook for Systematic Reviews of Interventions advocate that the last search should occur within 6 months of publication.2 One possible way to reduce the time and effort required to produce a systematic review of the literature is the application of artificial intelligence (AI) within the review process.11
Countless AI tools have been and are still being developed, which aim to enhance the efficiency of, or to assist in various stages of the systematic review process.12,13 These methods employ various algorithms: natural language processing for interpreting human-generated texts,14 generative AI (GenAI) for crafting new content in a variety of contexts,15 or Machine Learning to predict outcomes and categorize observations based on human feedback or solely based on the learning statistical model.16,17 AI tools can shorten the systematic review process, albeit with the risks that the integrity of the study quality could be compromised.3,18 Given that the modalities of AI are often characterized by a lack of transparency and explainability, these tools could pose ethical risks, bias, and plagiarism dilemmas.19,20,21 The applications of AI are not limited to assisting in literature reviews.22 AI is transforming medicine by enhancing diagnostic accuracy, personalizing decision-making, and optimizing clinical workflows.23,24 Through the integration of machine learning algorithms with medical imaging, electronic health records, and genomic data, AI can assist healthcare providers during daily routine care.25 Early AI models demonstrated subhuman performance, while current AI models match human intelligence and occasionally surpass it.26 It is expected that AI models will outperform humans, whereby human-AI interactions will become more important in a broad variety of fields.26
With the advent of GenAI resulting in an increased interest from academia and a strong potential for the application within the writing process, journals and publishers were forced to update their editorial policies.27 Currently, “Science” remains the sole journal that entirely prohibits the use of GenAI for the generation of texts, images, or graphs, while other journals permit its use, yet request a full disclosure.28,29 Disclosures can be incorporated in the acknowledgments and/or the methods section, and any text generated by AI should be appended to the supplemental information.30 Furthermore, journals explicitly preclude GenAI from being designated as an author, given that AI cannot be held accountable or responsible for the content created.29
Using AI to conduct reviews within abbreviated time frames warrants a thorough investigation. Prior studies have reviewed the implementation of AI tools10,13,31,32,33 across one or multiple stages of the systematic review process, in particular for title and abstract screening34,35 and for the assessment of risk of bias.36 However, to our knowledge, an evaluation of the accuracy and trustworthiness of AI for systematic literature review as compared with reviews undertaken manually has not been undertaken.20 Therefore, the objective of this study is to compare the outcomes of already conducted systematic reviews by human reviewers (Table 1) with those conducted automatically with the assistance of AI, and to evaluate the degree of agreement between both approaches.
Table 1.
Overview of the four systematic reviews that are included in this article
| Review | PROSPERO registrationa | Review domain | PICO question | PubMed string |
|---|---|---|---|---|
| Efficacy review | CRD42022360160 | Treatment efficacy in anesthesiology | To investigate the efficacy of current treatment modalities (Intervention/Control) for patients with Persistent Spinal Pain Syndrome Type II (Population) in terms of pain (Outcome). | (((((((((((FBSS) OR (“failed back surgery syndrome”)) OR (PSPS)) OR (“persistent spinal pain syndrome”)) OR (“failed neck surgery syndrome”)) OR (FNSS)) OR (“failed back surgery”)) OR (“failed neck surgery”)) OR (failed back surgery syndrome[MeSH Terms])) AND ((((((((((((((((((((((((((((((((((((((((((((((((((((((((((((((((treatment) OR (treatments)) OR (surgery)) OR (surgeries)) OR (operation)) OR (operations)) OR (“re-operation”)) OR (“re-operations”)) OR (reoperation)) OR (reoperations)) OR (therapy)) OR (therapies)) OR (procedure)) OR (procedures)) OR (management)) OR (managements)) OR (remedy)) OR (remedies)) OR (SCS)) OR (“spinal cord stimulation”)) OR (“dorsal column stimulation”)) OR (neurostimulation)) OR (BMT)) OR (“best medical treatment”)) OR (“conservative treatment”)) OR (infiltration)) OR (infiltrations)) OR (injection)) OR (injections)) OR (ablation)) OR (ablations)) OR (placebo)) OR (nocebo)) OR (neuromodulation)) OR (physiotherapy)) OR (physical therapy)) OR (education)) OR (“rehab program”)) OR (“rehabilitation program”)) OR (strategy)) OR (“vocational therapy”)) OR (“occupational therapy”)) OR (“online intervention”)) OR (“psychology”)) OR (“ergotherapy”)) OR (“ergonomics”)) OR (“exercise therapy”)) OR (advice)) OR (“internet based intervention”)) OR (“web-based intervention”)) OR (“educational activities”)) OR (“rehabilitation”)) OR (therapeutics)) OR (therapeutics[MeSH Terms])) OR (rehabilitation[MeSH Terms])) OR (physical therapy modalities[MeSH Terms])) OR (exercise therapy[MeSH Terms])) OR (internet based intervention[MeSH Terms])) OR (occupational therapy[MeSH Terms])) OR (rehabilitation, vocational[MeSH Terms])) OR (general surgery[MeSH Terms])) OR (pain management[MeSH Terms])) OR (placebos[MeSH Terms])) OR (spinal cord stimulation[MeSH Terms]))) AND ((pain) OR (pain measurement[MeSH Terms]))) AND (((((RCT) OR (“randomized controlled trial”)) OR (“controlled clinical trial”)) OR (“clinical trial”)) OR (clinical trial[Publication Type])) |
| Gut microbiome review | CRD42023430115 | Gut microbiota | To investigate perturbations in gut microbiota (Outcome) in patients with chronic pain (Population). | (((((((((((((chronic OR persistent OR prolonged OR intractable OR long-lasting OR “long lasting” OR long-term OR “long term” OR musculoskeletal OR widespread OR neuropathic OR cancer)) AND (pain)) OR (“complex regional pain syndrome”)) OR (headache)) OR (neuralgia)) OR (“Chronic Pain”[Mesh])) OR (“Pain, Intractable”[Mesh])) OR (“Complex Regional Pain Syndromes”[Mesh])) OR (“Cancer Pain”[Mesh])) OR (“Headache”[Mesh])) OR (“Neuralgia”[Mesh])) OR (“Musculoskeletal Pain”[Mesh])) AND (((((((((gastrointestinal OR gut OR intestinal OR fecal OR feces OR feces OR bacterial OR microbial)) AND ((microbiota OR microbiome OR flora OR dysbiosis))) OR (“gastrointestinal microbiome”)) OR (“gut-brain axis”)) OR (“gut brain axis”)) OR (“Gastrointestinal Microbiome”[Mesh])) OR (“Microbiota”[Mesh])) OR (“Brain-Gut Axis”[Mesh])) |
| Rehabilitation review | CRD42022346091 | Rehabilitation | To make an inventory of rehabilitation interventions (Intervention) to improve work participation (Outcome) in patients with chronic spinal pain (Population). | (((((“laminectoma” OR “radiculopatha” OR “sciatica” OR “radiculopathy”[MeSH Terms] OR “laminectomy”[MeSH Terms] OR “sciatica”[MeSH Terms])) AND ((((“chronic” OR “persistent” OR “long-term” OR “intractable” OR “long lasting”)) AND ((“pain”))) OR ((“chronic pain”[MeSH Terms] OR “pain, intractable”[MeSH Terms])))) OR ((“failed back surgery syndrome”) OR (“FBSS”) OR (“persistent spinal pain syndrome”) OR (“persistent spinal pain syndrome type 2”) OR (“PSPS-T2”) OR (“failed back surgery”) OR (“failed neck surgery”) OR (“failed surgery”) OR (“failed neck surgery syndrome”) OR (“FNSS”) OR (“post laminectomy syndrome”) OR (“post surgery syndrome”) OR (“post lumbar surgery syndrome”) OR (“postoperative back pain”) OR (“postoperative neck pain”) OR (“postoperative neuropathic pain”) OR (“failed back surgery syndrome”[MeSH Terms]))) AND ((“rehab programa”) OR (“rehabilitation programa”) OR (“support”) OR (“strategy”) OR (“recovery of function”) OR (“neurophysiotherapy”) OR (“physical therapy”) OR (“vocational therapy”) OR (“vocational education”) OR (“physiotherapy”) OR (“occupational therapy”) OR (“online intervention”) OR (“psychology”) OR (“ergotherapy”) OR (“ergonomics”) OR (“exercise therapy”) OR (“rehabilitation”[MeSH Terms]) OR (“rehabilitation, vocational”[MeSH Terms]) OR (“physical therapy modalities”[MeSH Terms]) OR (“occupational therapy”[MeSH Terms]) OR (“exercise therapy”[MeSH Terms]) OR (“ergonomics”[MeSH Terms]) OR (“advice”) OR (“education”) OR (“Internet-Based Intervention"[Mesh]) OR (“internet-based intervention”) OR (“web-based intervention”) OR (“educational activities”))) AND ((“return to work”[MeSH Terms]) OR (“unemployment”[MeSH Terms]) OR (“employment”[MeSH Terms]) OR (“sick leave”[MeSH Terms]) OR (“absenteeism"[MeSH Terms]) OR (“presenteeism”[MeSH Terms]) OR (“work performance"[MeSH Terms]) OR (“return to work”) OR (“RTW”) OR (“returning to work”) OR (“back to work”) OR (“work resumption”) OR (“stay at work”) OR (“re-employment”) OR (“employment”) OR (“unemployment”) OR (“sick leave”) OR (“disability leave”) OR (“work absence”) OR (“absenteeism”) OR (“presenteeism”) OR (“job performance”) OR (“work performance”) OR (“work disability”) OR (“occupational disability”) OR (“employment status”) OR (“work status”) OR (“work”)) |
| Return to work review | CRD42024501152 | Return to work | To explore employment or return to work (Outcome) after implantable electrical neurostimulation (Intervention) in patients with chronic pain (Population). | (((chronic OR persistent OR (long-term) OR intractable OR (long lasting)) AND (pain)) OR (“chronic pain”[MeSH Terms] OR “pain, intractable”[MeSH Terms]) ) OR (causalgia) OR (reflex sympathetic dystrophy) OR (failed back surgery syndrome) OR (postlaminectomy syndrome) OR (CRPS) OR (complex regional pain syndromea) OR (FBSS) OR (failed neck surgery syndrome) OR (FNSS) OR (peripheral neuropathy) OR (neuropathic pain) OR (angina pectoris) OR (peripheral vascular diseasea) OR (peripheral nervous system diseasea) OR (“causalgia”[MeSH Terms]) OR (“reflex sympathetic dystrophy”[MeSH Terms]) OR (“failed back surgery syndrome”[MeSH Terms]) OR (“complex regional pain syndromes”[MeSH Terms]) OR (“neuralgia”[MeSH Terms]) OR (“angina pectoris”[MeSH Terms]) OR (“peripheral vascular diseases”[MeSH Terms]) AND ((neurostimulata) OR (spinal cord stimulata) OR (dorsal column stimulata) OR (dorsal root ganglion stimulata) OR (deep brain stimulata) OR (peripheral nerve stimulata) OR (sacral nerve stimulata) OR (motor cortex stimulata) OR (occipital nerve stimulata) OR (vagus nerve stimulata) OR (epidural motor cortex stimulata) OR (pain stimulata) OR (closed loop stimulata) OR (“spinal cord stimulation”[MeSH Terms]) OR (“deep brain stimulation”[MeSH Terms]) OR (“vagus nerve stimulation”[MeSH Terms])) AND ((returna to work) OR (RTW) OR (employment) OR (absenteeism) OR (presenteeism) OR (work performance) OR (work disability) OR (work resumption) OR (sick leave) OR (work status) OR (occupational disability) OR (work disability) OR (employment status) OR (work absence) OR (unemployment) OR (back to work) OR (work resumption) OR (work participation) OR (vocational rehabilitation) OR (vocational outcome) OR (“return to work”[MeSH Terms]) OR (“employment”[MeSH Terms]) OR (“work performance”[MeSH terms]) OR (“sick leave”[MeSH Terms]) OR (work limitations) OR (work impairment) OR (occupation) OR (reintegration)) |
All protocols have been registered in the International Prospective Register of Systematic Reviews (PROSPERO).
Results
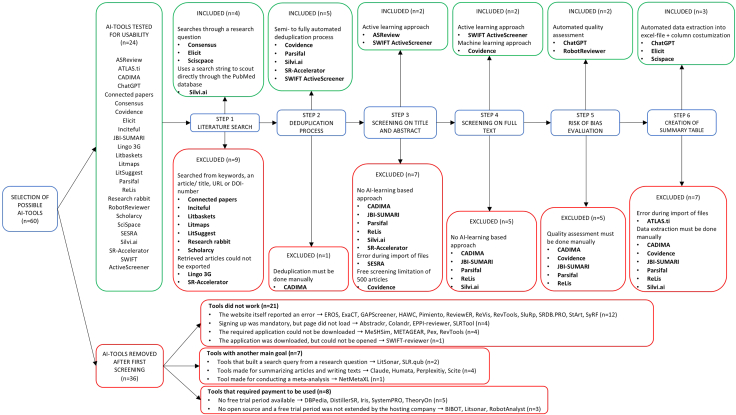
In total, 60 different AI-tools were identified, of which 36 tools were excluded after first screening (Figure 1). Consecutively, the usability of the remaining 24 tools was tested, whereafter a total of 11 different AI-tools were eligible for at least one stage of the systematic review process. There were 4 tools selected for the literature search, 5 for the deduplication process, 2 for screening on title and abstract, 2 for full text screening, 2 for the risk of bias evaluation, and 3 tools for the creation of the summary table (Table S1). Considering the rapid evolution within the field of AI, the present compilation of tools serves merely as a current overview rather than a definitive guide. The findings of the manual approach and the results of the AI tools for the different stages of the systematic review process are presented in Tables 2, 3, and S2.
Figure 1.
In- and excluded AI tools on all stages of the review process with rationale
Abbreviations. AI: artificial intelligence; n: number of tools.
Table 2.
Manual screening versus screening with AI tools for search strategy, screening on title and abstract, screening on full text, and risk of bias evaluation
| Efficacy review | Gut microbiome review | Rehabilitation review | Return to work review | All reviews combined | |
|---|---|---|---|---|---|
| Results for Search strategy | |||||
| MANUALLY |
n = 1140 articles retrieved ➣NNR 22.79 ➣precision 4.39% |
n = 6285 articles retrieved ➣NNR 299.29 ➣precision 0.33% |
n = 1289 articles retrieved ➣NNR 214.83 ➣precision 0.47% |
n = 4135 articles retrieved ➣NNR 63.62 ➣precision 1.57% |
n = 12849 articles retrieved ➣NNR 90.47 ➣precision 1.11% |
| CONSENSUS |
n = 238 articles retrieved ➣ recall 18% ➣ NNR 26.46 ➣ precision 3.78% |
n = 240 articles retrieved ➣ recall 14.29% ➣ NNR 80 ➣ precision 1.25% |
n = 242 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 233 articles retrieved ➣ recall 7.69% ➣ NNR 46.6 ➣ precision 2.15% |
n = 953 articles retrieved ➣ recall 11.97% ➣ NNR 56.09 ➣ precision 1.78% |
| ELICIT |
n = 394 articles retrieved ➣ recall 2% ➣ NNR 394 ➣ precision 0.25% |
n = 395 articles retrieved ➣ recall 19.05% ➣ NNR 98.75 ➣ precision 1.01% |
n = 304 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 200 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 1293 articles retrieved ➣ recall 3.52% ➣ NNR 258.6 ➣ precision 0.39% |
| SCISCPACE |
n = 90 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 90 articles retrieved ➣ recall 4.76% ➣ NNR 90 ➣ precision 1.11% |
n = 90 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 90 articles retrieved ➣ recall 0% ➣ NNR/➣ precision 0% |
n = 360 articles retrieved ➣ recall 0.7% ➣ NNR 360 ➣ precision 0.28% |
| Results for Screening on title and abstract when reaching the stopping criterion | |||||
| MANUALLY |
n = 708 screened, n = 105 included ➣NNR 6.74 ➣precision 14.83% |
n = 3544 screened, n = 37 included ➣NNR 95.78 ➣precision 1.04% |
n = 854 screened, n = 38 included ➣NNR 22.47 ➣precision 4.45% |
n = 2537 screened, n = 104 included ➣NNR 24.4 ➣precision 4.10% |
n = 7643 screened, n = 284 included ➣NNR 26.88 ➣precision 3.72% |
| ASREVIEW | Provided input: ➣ 5 relevant (RCT) ➣10 irrelevant (5 protocol and 5 SR) |
Provided input: ➣ 5 relevant (gut microbiota composition) ➣10 irrelevant (5 HPA-axis measures and 5 SR) |
Provided input: ➣ 5 relevant (return to work) ➣10 irrelevant (5 medical/pharmacological interventions and 5 SR) |
Provided input: ➣ 5 relevant (return to work) ➣10 irrelevant (5 acute pain and 5 SR) |
|
|
n = 535 screened, n = 104 included ➣ recall 99.05% ➣ NNR 5.14 ➣ precision 19.44% |
n = 557 screened, n = 31 included ➣ recall 83.78% ➣ NNR 17.96 ➣ precision 5.57% |
n = 467 screened, n = 38 included ➣ recall 100% ➣ NNR 12.30 ➣ precision 8.13% |
n = 595 screened, n = 101 included ➣ recall 97.12% ➣ NNR 5.89 ➣ precision 16.97% |
n = 2154 screened, n = 274 included ➣ recall 96.48% ➣ NNR 7.86 ➣ precision 12.72% |
|
| SWIFT ACTIVE SCREENER |
n = 429 screened, n = 105 included ➣ recall 100% ➣ NNR 4.08 ➣ precision 24.48% |
n = 810 screened, n = 33 included ➣ recall 89.19% ➣ NNR 24.57 ➣ precision 4.07% |
n = 543 screened, n = 36 included ➣ recall 94.74% ➣ NNR 15.08 ➣ precision 6.63% |
n = 539 screened, n = 104 included ➣ recall 100% ➣ NNR 5.18 ➣ precision 19.3% |
n = 2321 screened, n = 278 included ➣ recall 97.89% ➣ NNR 8.35 ➣ precision 11.97% |
| Results for Screening on full text | |||||
| MANUALLY |
n = 110 screened, n = 50 included ➣NNR 2.2 ➣precision 45.45% |
n = 42 screened, n = 21 included ➣NNR 2 ➣precision 50% |
n = 48 screened, n = 6 included ➣NNR 8 ➣precision 12.5% |
n = 129 screened, n = 65 included ➣NNR 1.98 ➣precision 50.39% |
n = 329 screened, n = 142 included ➣NNR 2.32 ➣precision 43.16% |
| COVIDENCE |
n = 103 screened, n = 50 included ➣ recall 100% ➣ NNR 2.06 ➣ precision 48.54% |
n = 42 screened, n = 21 included ➣ recall 100% ➣ NNR 2 ➣ precision 50% |
n = 46 screened, n = 6 included ➣ recall 100% ➣ NNR 7.67 ➣ precision 13.04% |
n = 129 screened, n = 65 included ➣ recall 100% ➣ NNR 1.98 ➣ precision 50.39% |
n = 320 screened, n = 142 included ➣ recall 100% ➣ NNR 2.25 ➣ precision 44.38% |
| SWIFT ACTIVE SCREENER |
n = 92 screened, n = 50 included ➣ recall 100% ➣ NNR 1.84 ➣ precision 54.35% |
n = 37 screened, n = 21 included ➣ recall 100% ➣ NNR 1.76 ➣ precision 56.76% |
n = 40 screened, n = 6 included ➣ recall 100% ➣ NNR 6.67 ➣ precision 15% |
n = 110 screened, n = 65 included ➣ recall 100% ➣ NNR 1.69 ➣ precision 59.09% |
n = 279 screened, n = 142 included ➣ recall 100% ➣ NNR 1.96 ➣ precision 50.9% |
| Results for Risk of bias evaluation | |||||
| MANUALLY | Cochrane Risk of Bias Tool | Newcastle-Ottawa Scale | Modified Downs and Black checklist | Modified Downs and Black checklist | |
| CHATGPT (GPT-4-turbo) | → 3 articles not rated ➣ Kappa = −0.19 (95% CI from −0.32 to-0.05) |
➣ Kappa = −0.03 (95%CI from −0.20 to 0.14) | ➣ Kappa = 0 (95% CI from 0 to 0) | → 1 article not rated ➣ Kappa = −0.02 (95% CI from −0.17 to 0.12) |
|
| ROBOTREVIEWER | → 4 RCTs not rated ➣Output with two level categorization: low versus high/unclear for random sequence generation, allocation concealment, blinding of participants and personnel and blinding of outcome assessment |
Wrong risk of bias tool | Wrong risk of bias tool | Wrong risk of bias tool | |
Abbreviations. CI: confidence interval; n: number of articles; NNR: number needed to read; RCT: randomized controlled trial.
Table 3.
Observed agreement between manual extraction and data extraction with ChatGPT, Elicit, and SciSpace for creating the summary table
| Metric | CHATGPT | ELICIT | SCISPACE | Number of applicable reviews | Kappa [95% CI] between tools |
|---|---|---|---|---|---|
| First author | 97.22% | 96.88% | 84.85% | 4 | κ = −0.16 [−0.265 to 0.048] |
| Year of publication | 100% | 62.5% | 63.64% | 4 | κ = −0.11 [−0.288 to 0.067] |
| Study design | 37.5% | 41.67% | 30.77% | 2 | κ = 0.73 [0.435 to 1] |
| Country of publication | 90% | 100% | 75% | 1 | κ = −0.09 [−0.24 to 0.06] |
| Sample size | 69.44% | 81.25% | 69.7% | 4 | κ = 0.44 [0.24 to 0.66] |
| Age | 55.56% | 31.25% | 42.42% | 4 | κ = 0.42 [0.18 to 0.67] |
| Sex | 55.56% | 46.88% | 42.42% | 4 | κ = 0.21 [−0.005 to 0.42] |
| BMI | 70% | 20% | 30% | 1 | κ = 0.23 [−0.22 to 0.68] |
| Population | 15% | 83.33% | 39.89% | 2 | κ = 0.13 [−0.14 to 0.40] |
| Treatment modality | 90% | 70% | 50% | 1 | κ = 0.13 [−0.23 to 0.49] |
| Interpretation results | 70% | 70% | 40% | 1 | κ = 0.45 [−0.01 to 0.91] |
| Microbiome data | 30% Alpha-diversity 50% Beta-diversity 10% Relative abundance |
80% Alpha-diversity 70% Beta-diversity 50% Relative abundance |
40% Alpha-diversity 60% Beta-diversity 10% Relative abundance |
1 | κ = 0.20 [−0.23 to 0.62] κ = 0.38 [0.02 to 0.75] κ = −0.08 [−0.24 to 0.07] |
| Type of rehabilitation program | 0% | 100% | 20% | 1 | κ = −0.37 [−0.62 to −0.12] |
| Comparator intervention | 83.33% | 100% | 100% | 1 | |
| Work participation | 0% | 0% | 0% | 1 | κ = 0.11 [−0.64 to 0.86] |
| Type of neuromodulation | 70% | 100% | 87.5% | 1 | κ = 0.45 [0.34 to 0.57] |
| Work-related outcomes Follow-up intervals |
60% work 70% intervals |
75% work 75% intervals |
62.5% work intervals 87.5% |
1 | κ = 0.81 [0.39 to 1] κ = 0.7 [−0.06 to 1] |
| Work status at baseline and after neuromodulation | 30% baseline 20% neuromodulation |
62.5% baseline 50% neuromodulation |
75% baseline 50% neuromodulation |
1 | κ = 0.55 [−0.02 to 1] κ = 0.49 [0.05 to 0.94] |
| Percentage return to work | 10% | 25% | 12.5% | 1 | κ = 0.51 [−0.28 to 1] |
Abbreviations. BMI: body mass index, CI: confidence interval.
Findings from stage 1: Search strategy
Four different AI tools were retrieved for conducting the search strategy, each sourcing different databases. Consensus and Elicit both searched Semantic Scholar. SciSpace sourced research articles and related data from various reputable platforms, including Semantic Scholar, OpenAlex, Google Scholar, and other trusted repositories. Silvi.ai directly sourced data from the databases PubMed and ClinicalTrials. The latter was not further considered since a pure comparison of the retrieved articles by humans and AI only for PubMed would not provide correct metrics compared to the other AI tools. Recall (i.e., how many of the relevant studies were found by the search strategy) was highest for Consensus, yet the highest recall was 18% for the efficacy review. For precision, meaning how many retrieved studies were relevant, Consensus scored the best. To find one relevant article, reviewers needed to screen up to 394 articles with Elicit, depending on the research topic.
Findings from stage 2: Deduplication process
For deduplication, Covidence, Parsifal, Silvi.ai, SWIFT ActiveScreener, and SR-accelerator were identified as potential AI tools. A comprehensive quantitative tool-by-tool comparison to the human approach of deduplication through EndNote was not included. Therefore, we do not report on the performance of these tools in this study.
Findings from stage 3: Screening on title and abstract
Two different AI tools, ASReview and SWIFT ActiveScreener, were compared to manual screening. For both tools, high values for recall were revealed, namely 96.48% for ASReview and 97.89% for SWIFT ActiveScreener. Overall, SWIFT ActiveScreener had slightly better recall, while ASReview had the lowest NNR (NNR of 7.86 compared to 8.35 for SWIFT ActiveScreener) and a slightly better precision.
Findings from stage 4: Screening on full text
Both Covidence and SWIFT ActiveScreener were able to help with screening on full text. SWIFT ActiveScreener had the highest precision (50.9% compared to 44.38%) and lowest NNR, making it the most efficient AI tool for this stage.
Findings from stage 5: Risk of bias evaluation
ChatGPT (GPT-4-turbo) and RobotReviewer were selected as potential tools to evaluate risk of bias. For ChatGPT, only poor inter-rater reliability values were revealed. Only the efficacy review could be evaluated with RobotReviewer, since this tool only includes RCTs. RobotReviewer provided an output with two levels (i.e., low versus high/unclear risk), which was not compatible with the 3-level scoring of the manual scoring tool.
Findings from stage 6: Creation of summary table
Firstly, when comparing the AI-customized columns with those manually made, both ChatGPT, Elicit and SciSpace were able to create the columns as preferred (Table S2). Elicit was not able to retrieve data from 2 articles from the rehabilitation review and from 2 articles from the return to work review. SciSpace could not retrieve data from 1 article from the rehabilitation review and from 2 articles from the return to work review. Secondly, the extracted data was compared with manually extracted data. For ChatGPT, extractions matched the manually extracted data perfectly for year of publication. Elicit could extract the country of publication perfectly, as well as the type of rehabilitation program and comparator intervention in the rehabilitation review, and the type of neuromodulation in the return to work review. SciSpace also extracted the comparator intervention in the rehabilitation review perfectly. Overall, performance for the three AI tools ranged between 0% and 100%. Inter-rater reliability between the different AI tools was also evaluated. Author extraction and year of publication had very poor kappa values. Sample size and age had moderate reliabilities of 0.44 and 0.42, respectively, while sex distribution had fair reliability. For study interpretation, a moderate reliability was found between AI tools.
Discussion
Conducting a systematic review is a time-consuming task, whereby it has been proposed that AI could support this process, specifically during the screening phases.35,37 A comparison between manually conducted systematic reviews and reviews assisted by AI is crucial to determine the added value of AI in conducting systematic reviews. In the current study, AI was deemed suitable for 2 stages in the pipeline of conducting a systematic review, namely for screening on title and abstract, and screening on full text based on high recall values. AI seemed to be inadequate to conduct a search strategy due to the low recall values, not appropriate to perform a risk of bias evaluation due to low inter-rater reliability and not capable of creating a summary table with results comparable to the manual approach.
This study revealed a broad variety of AI tools that could assist in conducting specific phases of a systematic review, demonstrating the drastic investments in AI for academic research during the past years.33,34 Some tools are developed to automate specific tasks, while others are more ambitious, by advancing the use of AI in the interpretation of data. The primary constraint of the utilization of multiple AI tools is that all tools operate on standalone platforms, and because they do not integrate with other tools, data has to be exported constantly.32 Should a singular comprehensive, freely available tool emerge in the future, the challenges associated with converting data files to appropriate formats and the necessity of managing multiple accounts and passwords would be significantly eliminated. Perhaps the problem is with the interface of the tools, which are aimed at human-machine interaction instead of machine-machine interaction, rather than with the backend capabilities of the AI tools. As an alternative, batch scripting options for every AI tool would enable the creation of an optimal pipeline with interchangeable components in which an AI tool performs one of the stages, the file is saved, formatting of the file is conducted, and through a command line, the adapted file is imported into the next AI tool, after which the same process is conducted.
AI tools that run a search strategy (stage 1) miss a large number of articles that were identified in the manually conducted reviews, as indicated by low recall values. Both Consensus and Elicit only search through Semantic Scholar, thereby overlooking licensed journals and articles behind paywall, explaining the low rate of matches with manually conducted search strategies.38 These tools are also inadequately equipped due to their non-reproducible and frequently incomplete search capabilities.39 Further developments to the search methodologies of these AI tools are needed to facilitate the literature search, which would consequently make the formulation of a specific search string for every database unnecessary. This study explored the potential of AI for the automation of the deduplication process and identified 5 potential AI tools. Previously, SR-accelerator presented high sensitivity (0.90 [95% CI = 0.84–0.95]) and was thereby identified as one of the more accurate tools for deduplication.40,41 These AI tools are created to automate the deduplication process by identifying and removing the same study from a group of uploaded records, while Endnote is considered a semi-automatic tool since it combines software with human checking.42
For screening on title and abstract, two different AI tools were evaluated, whereby both tools were developed to support the screening process instead of automating it. This means that screening on title and abstract still needs to be conducted by human reviewers, and that AI tools aid the reviewer by altering the order in which articles are presented to the human reviewer. An AI-tool that could screen independently and replace the human reviewer was not found. A key element in screening on title and abstract is the implementation of a stopping criterion, namely the moment that screening additional articles will no longer lead to the identification of relevant articles.43 This point presents a perfectly balanced trade-off between the time spent to keep screening articles and the probability of identifying a new relevant article.44 There is currently a lack of standardized stopping rules whereby some reviewers stop when they run out of time (pragmatic approach), when a specified number of irrelevant articles are seen in a row (heuristic approach), or when a pre-specified number of relevant documents has been identified (sampling approach).45 Novel, more complicated ways of determining a stopping rule are currently being proposed with flexible statistical stopping criteria based on error stability or rejecting a hypothesis of missing a given recall target.44,45 The current study merged stopping roles from two other studies to incorporate both a heuristic and sampling approach.17,35 This stopping criterion implied that a substantial proportion of articles remained unscreened, while almost all articles that were included when manually screening were retrieved. A similar finding was revealed by Muthu (2023), who tested ASReview on three different reviews with increased complexity in the structure of the research question.46 It was concluded that after screening 30% of the articles, the tool identified 97%, 86%, and 64% of relevant articles for easy, intermediate, and advanced complexity of the research question.46 Besides ASReview, SWIFT ActiveScreener was also identified as a useful AI tool in the current work. An accuracy of 97.9% was previously found for SWIFT ActiveScreener.47 Despite the efficiency gains, both tools lack the functionality to specify the rationale for article exclusion, which could potentially augment the learning capabilities of the tools. For full text screening, Covidence and SWIFT ActiveScreener could be used to support screening due to their perfect recall. However, the benefits are rather limited since all articles still need to be screened to retrieve all articles that were deemed relevant with the manual screening approach. This is confirmed by Liu et al.,48 mentioning that the potential time and resource savings associated with the AI-tool SWIFT ActiveScreener are limited to its application in title and abstract screening. This leads to the suggestion that for full text screening, AI tools could support earlier identification of relevant articles and thus enable future workload reduction if tool-specific stopping criteria are applied, however, a full screening of all articles was conducted to reach this conclusion.
For risk of bias evaluation, ChatGPT created an additional category when a specific criterion was unclear, thereby forming a classification system with an additional category compared to the revised Cochrane Risk of Bias Tool. Additionally, inter-rater reliability was very low, thereby making this tool inefficient to evaluate the risk of bias. Moreover, concerns regarding the AI tool RobotReviewer pertain to the self-constructed domains, which hinder a comprehensive comparison with the manual methodology. Thereby, the tool also overlooks some core domains (e.g., missing data) along with an overall risk of bias, making the tool currently inadvisable to use.36,49,50
The final stage that was evaluated in this study was the creation of the summary table. All three tools were capable of customization of columns of the summary table and clearly presented the extracted information in separate columns. Extraction difficulties did arise when information was presented in visual formats or tables rather than in textual form, or when the data was not clearly presented. This points out that human interventions are necessary to conduct the data extraction successfully. A solution to this may be to first provide the figures to tools that can derive data from the figures. This probably caused the limited inter-rater reliability between the different AI tools, which is problematic when only AI tools are used for this task.
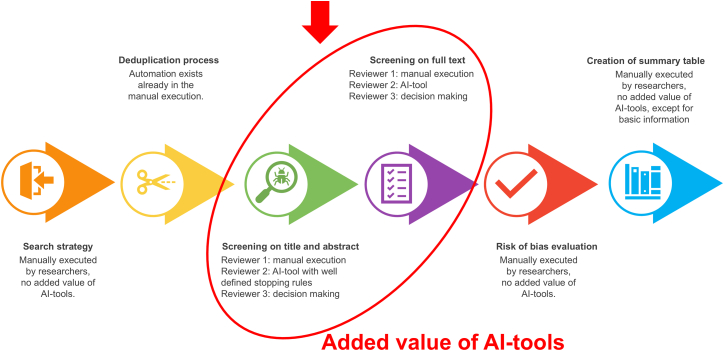
Combining the results of the different stages of a systematic review, current implementations of AI tools showed their ability to serve a supportive role, yet AI is not capable of complete substitution. Human judgment remains critical for tasks requiring contextual understanding, as AI might miss nuanced interpretations or domain-specific insights, and for critical appraisal, as determining the quality of studies often requires subjective judgment and expertise. Screening on title and abstract and screening on full text could receive support from AI tools (Figure 2). The AI tools could provide a benefit to the human reviewer by constantly presenting the most relevant article on top during the screening process. The lack of standardization about the stopping rule and the (although limited) number of articles that were not withheld by implementing a stopping rule, led to the suggestion to only embed the use of AI tools in the title and abstract screening phase for the second reviewer. We propose that the first and third reviewers keep screening manually without a stopping rule, and that the time burden of the second reviewer can be reduced by using ASReview or SWIFT ActiveScreener for screening on title and abstract, and by Covidence or SWIFT ActiveScreener for full text screening. With this approach, a new category of conflicts will arise, namely, included/excluded versus not screened. Those could then, along with the other discrepancies, be resolved by the third reviewer. Finally, the training of large language models is important, and the performance of these models may vary significantly depending on the domain-specific data.51
Figure 2.
Overview of where AI tools can assist in the approach of conducting a systematic review in medical sciences
Limitations of the study
Presumably, the main limitation of this study is the lack of evaluation of the time efficiency, meaning that a concrete estimate for the time gain for the second reviewer cannot be provided. If a specific AI tool reduces workload, with an equivalent accuracy, then the use of the tool will be justifiable. While exact time savings were not recorded, the NNR provides an indirect indication for relative time efficiency. Second, during the screening on title and abstract, one stopping rule was used across all AI tools, instead of relying on tool specific rules. Thirdly, only AI tools without payment requirements were selected to ensure that the tools that are proposed in this work are accessible for every researcher. This selection criterion has excluded several tools, some of which may be suitable for conducting specific stages of a systematic review. This also led to the retrieval of a lot of AI software applications and fewer GPT-based models. In case GPT-based models are of main interest, GPTscreenR52 and Bio-SIEVE53 seem to be promising for screening processes, PROMPTHEUS54 for data extraction and creating a summary table, and CRUISE-Screening55 for conducting living literature reviews. Finally, the training of large language models is important and performance of these models may vary significantly depending on the domain-specific data.51 Only when, as brought up by Van Dijk et al.,35 AI tools are recognized as an official component of the review process, can academic sciences really start to benefit from their potential.
AI showed its ability to serve a supportive role in the systematic review process, specifically for screening, yet it is not capable of complete substitution. Overall, AI tools are not ready to serve as a stand-alone reviewer. Human involvement remains required for contextual interpretation, nuanced judgment, and comprehensive analysis.
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact, Lisa Goudman (lisa.goudman@vub.be).
Materials availability
This study did not generate new reagents, plasmids, or mouse lines.
Data and code availability
-
•
The datasets used in this article were previously collected and described, with sources described in the key resources table.
-
•
The software and code used in this study are available in https://www.r-project.org/. Autonomous generated codes used in the STAR Methods section of the text are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
Acknowledgments
The authors like to thank Laure Booghmans for her help with this article.
Author contributions
MM: conception, interpretation, visualizations, and writing; GN: interpretation and writing; NW: interpretation and writing; LG: conception, analysis, interpretation, visualizations, and writing. All authors approved the final version of the article.
Declaration of interests
Lisa Goudman is a postdoctoral research fellow funded by the Research Foundation Flanders (FWO), Belgium (project number 1211425N). Maarten Moens received a research mandate by Research Foundation Flanders (FWO), Belgium (project number 1801125N), and has received speaker fees from Medtronic. STIMULUS received independent research grants from Medtronic. There are no other conflicts of interest to declare.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Efficacy review (PROSPERO CRD42022360160) | Goudman et al.56 | https://doi.org/10.1038/s43856-025-00778-x |
| Gut microbiome review (PROSPERO CRD42023430115) | Goudman et al.57 | https://doi.org/10.3389/fimmu.2024.1342833 |
| Rehabilitation review (PROSPERO CRD42022346091) | Callens et al.58 | https://doi.org/10.2340/jrm.v56.25156 |
| Return to work review (PROSPERO CRD42024501152) | Under review | PROSPERO: CRD42024501152 |
Experimental model and study participant details
Experimental model
This study is a comparison between human reviewers versus AI-tools to conduct a systematic review. Four systematic reviews are used as reference works, wherefore the study utilizes existing literature rather than employing the experimental models prevalent in the life sciences.
Ethics approval and consent to participate
Ethical approval was not sought for this study because the study does not contain any animal or direct human participants for experiments.
Method details
Study design
In the present study, a comparison is made between a manual screening approach and screening with AI-tools for different stages of the systematic review process. The following six stages of a systematic review were considered: search strategy, deduplication, screening on title and abstract, screening on full text, risk of bias evaluation and creation of the summary table. For this comparison, 4 recent systematic reviews were included, all manually conducted by STIMULUS research group in Belgium. An overview of these 4 reviews is provided in Table 1, which are denoted as the efficacy review,56 gut microbiome review,57 rehabilitation review,58 and return to work review.59
The artificial intelligence (AI) tools
To identify all possible AI-tools that could assist when conducting a systematic review, a thorough literature search was conducted in March 2024. Inclusion criteria were: the tool was available in March 2024 (i.e., the website of this tool still existed and the tool was available to use online or could be downloaded), did not require payment (i.e., open source, had a free trial period or in case this was not automatically available, the trial was extended by the company hosting the tool), and could carry out at least one of the six stages. AI-Tools were included when they could be executed (at least partially) automated, and under the condition that no difficulties occurred during the execution (e.g., importing errors). Where exporting-related issues could be resolved through payment, this was performed. AI-tools were included when they could perform a specific stage, meaning that the literature search occurred through a research question or a search string, the screening stages were carried out using an AI-learning based approach, or columns for the summary table could be customized prior to data extraction.
The AI-tools could be open-ended versus task-specific and generative versus non-generative. Tools were compared based on their specific performance during each stage separately, whereby the same output was compared between the different tools.
Stages of the systematic review: Manual versus AI approach
In the four systematic reviews on which the manual approach is compared with the AI approach, the 6 different stages of the systematic review were undertaken identically. These crucial execution stages were executed without strategic decisions from the research team and were derived from the 24-step guide proposed by Muka et al.60 In the first stage, defining a research question, defining a search strategy, and running the strategy in multiple databases were merged into search strategy. Deduplication involved collecting references and abstracts into a single file, followed by the elimination of duplicates. Screening on title and abstract entailed screening, comparing and selecting on title and abstract by at least two reviewers. The fourth stage combined full text screening based on applying selection criteria and searching for additional references. The fifth stage corresponds to evaluation of study quality and risk of bias. The last stage is a combination of preparing the database for analysis and conducting descriptive synthesis. For the four reviews, all stages of the systematic review process were executed in March 2024.
Stage 1: Search strategy
For each review, a research question was created according to the PICO (evidence-based search strategy focusing on patient/population, intervention, comparison and outcome) framework. A final search string was built and then entered in PubMed, Web of Science, Embase and Scopus, to collect all eligible articles. No limitations were applied to the search strategy. The literature search with the AI-tools was undertaken by either using the original research question or the PubMed search string, depending on the requirements of the AI-tool.
Stage 2: Deduplication
In the manual approach, the removal of duplicates was conducted with the assistance of Endnote or Rayyan, or a combination of both. Once detected, the duplicates were removed automatically in Endnote and manually in Rayyan. With the AI-tools this process was semi- to fully automated. Dependent on the possibilities of each tool, deduplication was conducted by using one file containing records from all databases, or the files from the different databases were uploaded separately.
Stage 3: Screening on title and abstract
Following deduplication, manual title and abstract screening was performed with the assistance of Rayyan. Decisions to select articles for further assessment were based on predetermined eligibility criteria, to minimize the chance of including non-relevant articles. The AI-tools were also provided with these eligibility criteria. In the AI screening, the AI-tools automatically presented the most relevant articles first, based on the ongoing decisions with respect to inclusion and exclusion (i.e., active learning). Human reviewers were still responsible for screening on title and abstract, however the order in which articles were presented was different. To assure that AI-screening was not continued after all relevant articles were found, we implemented a threefold ‘stopping criterion’: (1) a predetermined selection of key articles is detected, (2) minimally 10% of the total dataset was screened, and (3) a sequence of 100 consecutive irrelevant articles was reached.17,35 Screening on title and abstract was terminated when all articles were retrieved or when the stopping criterion was reached.
Stage 4: Screening on full text
In the manually conducted approach, full text screening was conducted independently by two reviewers. In the AI approach, SWIFT ActiveScreener also applied the active learning approach, whereas the algorithm behind Covidence determined its order of articles in advance, based on the eligibility criteria. For full text screening, no stopping criterion was used.
Stage 5: Risk of bias evaluation
Depending on the study design(s) in the reviews, the revised Cochrane Risk of Bias Tool (RoB 2),61 the Newcastle-Ottawa Scale62,63 or the modified Downs and Black checklist64 were used to evaluate risk of bias. The in-depth assessment for the risk of various types of bias was performed automatically with AI. Comparisons were made on the total scores on the risk of bias tools.
Stage 6: Creation of summary table
Collation of data in a summary table was done manually after reading all included articles thoroughly. The table content was a priori determined and contained author, year, country, study design, population characteristics and main findings. Besides comparing the possibilities of AI for column customization, the extracted data was checked for accuracy. For each review, a selection of 10 articles, based on a variety of publication years, were compared, with exception of the rehabilitation review where all 6 included articles were compared.
Quantification and statistical analysis
Data analysis
To compare the performance of human reviewers versus the AI-tools, sensitivity/recall, precision and number needed to read (NNR) were calculated. Recall is the proportion of all relevant articles identified by the search relative to the total number of relevant articles retrieved. NNR refers to the number of articles to screen/read to identify a relevant article. Precision is the total number of relevant articles included to the total number of citations screened.
Inter-rater reliability was determined between the human reviewers and the AI-tool for risk of bias evaluation using Cohen's weighted kappa coefficient (κ) for ordinal data with equal-spacing weighting, and for comparing AI-tools for the creation of the summary table using Fleiss’ kappa coefficient (κ). For the latter, articles with incomplete data were ignored. Kappa values were interpreted as follows: <0.00 = poor; 0.00–0.20 = slight; 0.21–0.40 = fair; 0.41–0.60 = moderate; 0.61–0.80 = substantial; 0.81–1.00 = almost perfect reliability.65 Observed agreement was selected as metric to compare data extraction for the summary table. All analyses were performed in R Studio version 4.2.3.
Published: September 12, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2025.113559.
Supplemental information
References
- 1.Wallace S.S., Barak G., Truong G., Parker M.W. Hierarchy of Evidence Within the Medical Literature. Hosp. Pediatr. 2022;12:745–750. doi: 10.1542/hpeds.2022-006690. [DOI] [PubMed] [Google Scholar]
- 2.Julian P.T., Higgins J.T., Chandler J., Cumpston M., Li T., Page M.J., Welch V.A. © 2019 The Cochrane Collaboration; 2019. Cochrane Handbook for Systematic Reviews of Interventions. [DOI] [Google Scholar]
- 3.Fabiano N., Gupta A., Bhambra N., Luu B., Wong S., Maaz M., Fiedorowicz J.G., Smith A.L., Solmi M. How to optimize the systematic review process using AI tools. JCPP Adv. 2024;4 doi: 10.1002/jcv2.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bangdiwala S.I. The importance of systematic reviews. Int. J. Inj. Contr. Saf. Promot. 2024;31:347–349. doi: 10.1080/17457300.2024.2388484. [DOI] [PubMed] [Google Scholar]
- 5.Iddagoda M.T., Flicker L. Clinical systematic reviews – a brief overview. BMC Med. Res. Methodol. 2023;23:226. doi: 10.1186/s12874-023-02047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borah R., Brown A.W., Capers P.L., Kaiser K.A. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open. 2017;7 doi: 10.1136/bmjopen-2016-012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark J., McFarlane C., Cleo G., Ishikawa Ramos C., Marshall S. The Impact of Systematic Review Automation Tools on Methodological Quality and Time Taken to Complete Systematic Review Tasks: Case Study. JMIR Med. Educ. 2021;7 doi: 10.2196/24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dicks L.V., Walsh J.C., Sutherland W.J. Organising evidence for environmental management decisions: a '4S' hierarchy. Trends Ecol. Evol. 2014;29:607–613. doi: 10.1016/j.tree.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Beller E.M., Chen J.K.-H., Wang U.L.-H., Glasziou P.P. Are systematic reviews up-to-date at the time of publication? Syst. Rev. 2013;2:36. doi: 10.1186/2046-4053-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beller E., Clark J., Tsafnat G., Adams C., Diehl H., Lund H., Ouzzani M., Thayer K., Thomas J., Turner T., et al. Making progress with the automation of systematic reviews: principles of the International Collaboration for the Automation of Systematic Reviews (ICASR) Syst. Rev. 2018;7:77. doi: 10.1186/s13643-018-0740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos Á.O.D., da Silva E.S., Couto L.M., Reis G.V.L., Belo V.S. The use of artificial intelligence for automating or semi-automating biomedical literature analyses: A scoping review. J. Biomed. Inform. 2023;142 doi: 10.1016/j.jbi.2023.104389. [DOI] [PubMed] [Google Scholar]
- 12.Golan R., Reddy R., Muthigi A., Ramasamy R. Artificial intelligence in academic writing: a paradigm-shifting technological advance. Nat. Rev. Urol. 2023;20:327–328. doi: 10.1038/s41585-023-00746-x. [DOI] [PubMed] [Google Scholar]
- 13.Blaizot A., Veettil S.K., Saidoung P., Moreno-Garcia C.F., Wiratunga N., Aceves-Martins M., Lai N.M., Chaiyakunapruk N. Using artificial intelligence methods for systematic review in health sciences: A systematic review. Res. Synth. Methods. 2022;13:353–362. doi: 10.1002/jrsm.1553. [DOI] [PubMed] [Google Scholar]
- 14.Wyatt J.M., Booth G.J., Goldman A.H. Natural Language Processing and Its Use in Orthopaedic Research. Curr. Rev. Musculoskelet. Med. 2021;14:392–396. doi: 10.1007/s12178-021-09734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preiksaitis C., Rose C. Opportunities, Challenges, and Future Directions of Generative Artificial Intelligence in Medical Education: Scoping Review. JMIR Med. Educ. 2023;9 doi: 10.2196/48785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinger L., Gazendam A., Ekhtiari S., Bhandari M. Machine learning and artificial intelligence in research and healthcare. Injury. 2023;54 doi: 10.1016/j.injury.2022.01.046. S69–s73. [DOI] [PubMed] [Google Scholar]
- 17.Boetje J., Schoot R. The SAFE Procedure: A Practical Stopping Heuristic for Active Learning-Based Screening in Systematic Reviews and Meta-Analyses. System. Rev. 2023;13:81. doi: 10.21203/rs.3.rs-2856011/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark J., Glasziou P., Del Mar C., Bannach-Brown A., Stehlik P., Scott A.M. A full systematic review was completed in 2 weeks using automation tools: a case study. J. Clin. Epidemiol. 2020;121:81–90. doi: 10.1016/j.jclinepi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Bouhouita-Guermech S., Gogognon P., Bélisle-Pipon J.-C. Specific challenges posed by artificial intelligence in research ethics. Front. Artif. Intell. 2023;6 doi: 10.3389/frai.2023.1149082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khlaif Z.N., Mousa A., Hattab M.K., Itmazi J., Hassan A.A., Sanmugam M., Ayyoub A. The Potential and Concerns of Using AI in Scientific Research: ChatGPT Performance Evaluation. JMIR Med. Educ. 2023;9 doi: 10.2196/47049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupps W.J., Jr. Artificial intelligence and academic publishing. J. Cataract Refract. Surg. 2023;49:655–656. doi: 10.1097/j.jcrs.0000000000001223. [DOI] [PubMed] [Google Scholar]
- 22.Alowais S.A., Alghamdi S.S., Alsuhebany N., Alqahtani T., Alshaya A.I., Almohareb S.N., Aldairem A., Alrashed M., Bin Saleh K., Badreldin H.A., et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med. Educ. 2023;23:689. doi: 10.1186/s12909-023-04698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marletta S., Eccher A., Martelli F.M., Santonicco N., Girolami I., Scarpa A., Pagni F., L'Imperio V., Pantanowitz L., Gobbo S., et al. Artificial intelligence-based algorithms for the diagnosis of prostate cancer: A systematic review. Am. J. Clin. Pathol. 2024;161:526–534. doi: 10.1093/ajcp/aqad182. [DOI] [PubMed] [Google Scholar]
- 24.Koteluk O., Wartecki A., Mazurek S., Kołodziejczak I., Mackiewicz A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. J. Pers. Med. 2021;11 doi: 10.3390/jpm11010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazarika I. Artificial intelligence: opportunities and implications for the health workforce. Int. Health. 2020;12:241–245. doi: 10.1093/inthealth/ihaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosny A., Parmar C., Quackenbush J., Schwartz L.H., Aerts H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stokel-Walker C. AI bot ChatGPT writes smart essays - should professors worry? Nature. 2022 doi: 10.1038/d41586-022-04397-7. [DOI] [PubMed] [Google Scholar]
- 28.Thorp H.H. ChatGPT is fun, but not an author. Science. 2023;379:313. doi: 10.1126/science.adg7879. [DOI] [PubMed] [Google Scholar]
- 29.Ganjavi C., Eppler M.B., Pekcan A., Biedermann B., Abreu A., Collins G.S., Gill I.S., Cacciamani G.E. Publishers’ and journals’ instructions to authors on use of generative artificial intelligence in academic and scientific publishing: bibliometric analysis. BMJ. 2024;384 doi: 10.1136/bmj-2023-077192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini M., Rasmussen L.M., Resnik D.B. Using AI to write scholarly publications. Account. Res. 2024;31:715–723. doi: 10.1080/08989621.2023.2168535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison H., Griffin S.J., Kuhn I., Usher-Smith J.A. Software tools to support title and abstract screening for systematic reviews in healthcare: an evaluation. BMC Med. Res. Methodol. 2020;20:7. doi: 10.1186/s12874-020-0897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalil H., Ameen D., Zarnegar A. Tools to support the automation of systematic reviews: a scoping review. J. Clin. Epidemiol. 2022;144:22–42. doi: 10.1016/j.jclinepi.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Wagner G., Lukyanenko R., Paré G. Artificial intelligence and the conduct of literature reviews. J. Inf. Technol. 2021;37:209–226. doi: 10.1177/02683962211048201. [DOI] [Google Scholar]
- 34.van de Schoot R., de Bruin J., Schram R., Zahedi P., de Boer J., Weijdema F., Kramer B., Huijts M., Hoogerwerf M., Ferdinands G., et al. An open source machine learning framework for efficient and transparent systematic reviews. Nat. Mach. Intell. 2021;3:125–133. doi: 10.1038/s42256-020-00287-7. [DOI] [Google Scholar]
- 35.van Dijk S.H.B., Brusse-Keizer M.G.J., Bucsán C.C., van der Palen J., Doggen C.J.M., Lenferink A. Artificial intelligence in systematic reviews: promising when appropriately used. BMJ Open. 2023;13 doi: 10.1136/bmjopen-2023-072254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armijo-Olivo S., Craig R., Campbell S. Comparing machine and human reviewers to evaluate the risk of bias in randomized controlled trials. Res. Synth. Methods. 2020;11:484–493. doi: 10.1002/jrsm.1398. [DOI] [PubMed] [Google Scholar]
- 37.de la Torre-López J., Ramírez A., Romero J.R. Artificial intelligence to automate the systematic review of scientific literature. Computing. 2023;105:2171–2194. doi: 10.1007/s00607-023-01181-x. [DOI] [Google Scholar]
- 38.Kung J. Elicit (product review) J. Can. Health Libr. Assoc. 2023;44:15. doi: 10.29173/jchla29657. [DOI] [Google Scholar]
- 39.Burns J. The Effortless Academic; 2024. SciSpace: An All-In-One AI Tool for Literature Reviews. [Google Scholar]
- 40.Guimaraes N.S., Ferreira A.J.F., Ribeiro Silva R.C., de Paula A.A., Lisboa C.S., Magno L., Ichiara M.Y., Barreto M.L. Deduplicating records in systematic reviews: there are free, accurate automated ways to do so. J. Clin. Epidemiol. 2022;152:110–115. doi: 10.1016/j.jclinepi.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 41.McKeown S., Mir Z.M. Considerations for conducting systematic reviews: A follow-up study to evaluate the performance of various automated methods for reference de-duplication. Res. Synth. Methods. 2024;15:896–904. doi: 10.1002/jrsm.1736. [DOI] [PubMed] [Google Scholar]
- 42.Forbes C.A.-O., Greenwood H., Carter M., Clark J. Automation of duplicate record detection for systematic reviews: Deduplicator. Syst. Rev. 2024;13:206. doi: 10.1186/s13643-024-02619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Cai W., Zhang Y. Proceedings of the 2014 IEEE International Conference on Data Mining. IEEE Computer Society; 2014. Stability-Based Stopping Criterion for Active Learning. [Google Scholar]
- 44.Ishibashi H., Hino H. Stopping Criterion for Active Learning Based on Error Stability. arXiv. 2021 doi: 10.48550/arXiv.2104.01836. Preprint at. [DOI] [Google Scholar]
- 45.Callaghan M.W., Müller-Hansen F. Statistical stopping criteria for automated screening in systematic reviews. Syst. Rev. 2020;9:273. doi: 10.1186/s13643-020-01521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muthu S. The efficiency of machine learning-assisted platform for article screening in systematic reviews in orthopaedics. Int. Orthop. 2023;47:551–556. doi: 10.1007/s00264-022-05672-y. [DOI] [PubMed] [Google Scholar]
- 47.Liu J.J.W., Ein N., Gervasio J., Easterbrook B., Nouri M.S., Nazarov A., Richardson J.D. Usability and agreement of the SWIFT-ActiveScreener systematic review support tool: Preliminary evaluation for use in clinical research. PLoS One. 2024;19 doi: 10.1371/journal.pone.0291163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J.J.W., Ein N., Gervasio J., Easterbrook B., Nouri M.S., Nazarov A., Richardson J.D. Usability and Accuracy of the SWIFT-ActiveScreener: Preliminary evaluation for use in clinical research. medRxiv. 2023 doi: 10.1101/2023.08.24.23294573. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gates A., Vandermeer B., Hartling L. Technology-assisted risk of bias assessment in systematic reviews: a prospective cross-sectional evaluation of the RobotReviewer machine learning tool. J. Clin. Epidemiol. 2018;96:54–62. doi: 10.1016/j.jclinepi.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Marshall I.J., Kuiper J., Wallace B.C. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. J. Am. Med. Inform. Assoc. 2016;23:193–201. doi: 10.1093/jamia/ocv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling C., Zhao X., Lu J., Deng C., Zheng C., Wang J., Chowdhury T., Li Y., Cui H., Zhang X. Domain specialization as the key to make large language models disruptive: A comprehensive survey. arXiv. 2023 doi: 10.48550/arXiv.2305.18703. Preprint at. [DOI] [Google Scholar]
- 52.Wilkins D. Automated title and abstract screening for scoping reviews using the GPT-4 Large Language Model. arXiv. 2023 doi: 10.48550/arXiv.2311.07918. Preprint at. [DOI] [Google Scholar]
- 53.Ambrose R., William T., Ben W., Abdullah P., Munira E., Mark S., Xingyi S. Bio-SIEVE: Exploring Instruction Tuning Large Language Models for Systematic Review Automation. arXiv. 2023 doi: 10.48550/arXiv.2308.06610. Preprint at. [DOI] [Google Scholar]
- 54.Torres J., Muligan C., Jorge J., Moreira C. PROMPTHEUS: A Human-Centered Pipeline to Streamline SLRs with LLMs. arXiv. 2024 doi: 10.48550/arXiv.2410.15978. Preprint at. [DOI] [Google Scholar]
- 55.Kusa W., Knoth P., Hanbury A. Proceedings of the 32nd ACM International Conference on Information and Knowledge Management. Association for Computing Machinery; 2023. CRUISE-Screening: Living Literature Reviews Toolbox. [Google Scholar]
- 56.Goudman L., Russo M., Pilitsis J.G., Eldabe S., Duarte R.V., Billot M., Roulaud M., Rigoard P., Moens M. Treatment modalities for patients with Persistent Spinal Pain Syndrome Type II: A systematic review and network meta-analysis. Commun. Med. 2025;5:63. doi: 10.1038/s43856-025-00778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goudman L., Demuyser T., Pilitsis J.G., Billot M., Roulaud M., Rigoard P., Moens M. Gut dysbiosis in patients with chronic pain: a systematic review and meta-analysis. Front. Immunol. 2024;15 doi: 10.3389/fimmu.2024.1342833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callens J., Lavreysen O., Goudman L., De Smedt A., Putman K., Van de Velde D., Godderis L., Ceulemans D., Moens M. Does rehabilitation improve work participation in patients with chronic spinal pain after spinal surgery: a systematic review. J. Rehabil. Med. 2025;57 doi: 10.2340/jrm.v57.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Callens J., Moens M., Goudman L. 2024. Return to work after electrical neurostimulation for chronic pain: a systematic review and meta-analysis. [Google Scholar]
- 60.Muka T., Glisic M., Milic J., Verhoog S., Bohlius J., Bramer W., Chowdhury R., Franco O.H. A 24-step guide on how to design, conduct, and successfully publish a systematic review and meta-analysis in medical research. Eur. J. Epidemiol. 2020;35:49–60. doi: 10.1007/s10654-019-00576-5. [DOI] [PubMed] [Google Scholar]
- 61.Sterne J.A.C., Savović J., Page M.J., Elbers R.G., Blencowe N.S., Boutron I., Cates C.J., Cheng H.Y., Corbett M.S., Eldridge S.M., et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366 doi: 10.1136/bmj.l4898. [DOI] [PubMed] [Google Scholar]
- 62.Sanderson S., Tatt I.D., Higgins J.P.T. Tools for assessing quality and susceptibility to bias in observational studies in epidemiology: a systematic review and annotated bibliography. Int. J. Epidemiol. 2007;36:666–676. doi: 10.1093/ije/dym018. [DOI] [PubMed] [Google Scholar]
- 63.Wells G., Shea B., O'connell D., Peterson J., Welch V., Losos M., Tugwell P. Ottawa Health Research Institute; 2004. Quality Assessment Scales for Observational Studies. [Google Scholar]
- 64.Downs S.H., Black N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J. Epidemiol. Community Health. 1998;52:377–384. doi: 10.1136/jech.52.6.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landis J.R., Koch G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets used in this article were previously collected and described, with sources described in the key resources table.
-
•
The software and code used in this study are available in https://www.r-project.org/. Autonomous generated codes used in the STAR Methods section of the text are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.