Abstract
The type III secretion system (TTSS) is a key virulence mechanism of many important gram-negative bacterial pathogens. The TTSS is conserved among different bacterial pathogens, and mutations and deletions to the system significantly decrease virulence, making the TTSS an important potential therapeutic target. We have developed a high-throughput assay to search for inhibitors of the TTSS. We screened a commercial library of 20,000 small molecules for their ability to inhibit type III secretion by enteropathogenic Escherichia coli (EPEC). After discarding compounds that had no effect on secretion, inhibited bacterial growth, and/or caused degradation of EPEC-secreted proteins, the search was focused on a class of compounds that, while not direct inhibitors of type III secretion, inhibit expression of TTSS-related genes and other genes involved in virulence. This class of compounds does not affect bacterial viability or motility, indicating that it is not significantly affecting the expression of essential genes and is specific to virulence-associated genes. Transcriptional fusion assays confirmed that virulence-associated promoters were more sensitive to inhibition by this class of compounds. Overall, we have identified a class of compounds that can be used as a tool to probe the mechanism(s) that regulates virulence gene expression in EPEC.
The delivery of virulence factors directly into host cells to interfere with and alter host processes is a crucial step in bacterial virulence for several significant animal and plant pathogens (38, 41). The type III secretion system (TTSS) facilitates delivery of many bacterial effectors directly from the cytoplasm of gram-negative bacteria into host cells, thereby crossing bacterial inner membrane, peptidoglycan, and outer membrane and the host plasma membrane (reviewed in reference 16). Development of ways to interfere with this fundamental pathogenic mechanism could lead researchers to novel means for combating a variety of important gram-negative pathogens (reviewed in references 12 and 32).
The TTSS is present and highly conserved in many disease-causing gram-negative bacteria (16). TTSSs are also involved in the symbiotic relationship between Rhizobium species and legumes (35) and in the insect endosymbiont Sodalis glossinidius (4). Although not strictly pathogens, symbiotic bacteria establish intimate relationships with their hosts that resemble those that arise during pathogenesis, although the outcome is different. TTSS have not been found in nonpathogenic bacteria or in members of normal microbial flora of humans. While the TTSS has similarities to the flagellar assembly and export apparatus, there are substantial differences, supporting the concept of specifically targeting the type III apparatus for inhibition. A recent paper used a gene probe to detect type III genes as an indicator of virulence, and there was no interference due to genetic similarity between type III systems and the flagellar assembly apparatus (40).
Enteropathogenic Escherichia coli (EPEC) is a human pathogen responsible for outbreaks of diarrhea in both developing and developed countries (33). During infections, EPEC adheres to intestinal epithelial cells and forms actin-rich pedestals, through the binding of the outer membrane protein, intimin, to its receptor in the host. These actin-rich pedestals are called attaching and effacing (A/E) lesions (23, 31). Using a type III secretion apparatus, EPEC translocates effector proteins, called Esps (for E. coli-secreted proteins), into the host cell. This includes inserting a receptor for intimin, Tir (Translocated intimin receptor), into the host cell membrane, where it becomes tyrosine phosphorylated (5, 21). All of the known genes necessary for the A/E phenotype are localized within a 35.6-kb chromosomal region of EPEC strain E2348/69 containing 41 open reading frames (10, 27, 28). This region, designated LEE (locus of enterocyte effacement), is not found in normal flora E. coli or in E. coli K-12. The LEE-encoded genes are organized into five major operons (LEE1 to LEE5), two bicistronic operons (espGorf1 and grlRA), and four single genes (rorf3, cesF, map, and escD) (6, 29, 37). LEE1, LEE2, and LEE3 operons contain the type III apparatus genes; the LEE4 operon encodes the translocators and secreted Esp proteins. LEE5 encodes the secreted protein Tir; its chaperone, CesT; and intimin. Other genes distributed along the LEE, such as espG, espH, espF, and map, encode type III effectors. Recently a systematic deletion analysis was done of all 41 LEE genes of the related pathogen Citrobacter rodentium, and these mutants were functionally characterized in vitro and in a murine infection model (7). This work identified 33 virulence factors, including two virulence regulators and a switch for type III secretion.
Expression of EPEC's virulence factors encoded within the LEE is regulated by complex and multifactorial mechanisms. EPEC has a plasmid containing the per locus, which is involved in regulation of plasmid and chromosomal virulence genes (29, 34, 42). The LEE-encoded regulator (Ler) is a central regulator of the expression of the genes involved in forming the A/E lesion (1, 11, 29). Ler is a 15-kDa protein that exhibits amino acid similarity to the histone-like nucleoid structuring protein H-NS. Under inducing conditions, Ler binds to sequences in the vicinity of different LEE promoters and antagonizes the repression exerted by H-NS (1, 14; unpublished data). The activation of ler is controlled by another regulator encoded by the LEE, GrlA, which seems to act in a cascade fashion (7), and by a myriad of global regulators (reviewed in reference 2). Moreover, EPEC protein secretion is subject to environmental regulation, being induced under conditions similar to those in the gastrointestinal tract (6, 20).
In all relevant model systems tested thus far, mutating or deleting part of the TTSS significantly decreases the virulence of the affected pathogen. In addition, every gene in the LEE region is required for full virulence (7). Presumably chemical compounds that specifically inhibit the type III system could prevent disease by impairing essential virulence properties within this system, although this hypothesis has not yet been formally tested. Although bacteria with mutations in type III secretion components are avirulent, it is not known if an inhibitor could reverse disease once it has started. While the present study was in progress, Wolf-Watz and colleagues screened a library of 9,400 compounds with a luciferase reporter assay in Yersinia pseudotuberculosis and identified several compounds that inhibited type III secretion (17). One compound affected bacterial motility and thus is not virulence-specific, while the other two are specific to the virulence-associated type III secretion system. The specific bacterial target(s) awaits further study.
We have screened an extensive number of marine and plant extracts and libraries of compounds for their ability to inhibit type III secretion of Esps by EPEC (25; reviewed in reference 12). In this study, we screened a commercial library of 20,000 molecules and identified a class of compounds that, while not direct inhibitors of type III secretion, inhibit the expression of type III-related LEE genes and other genes involved in the pathogenesis of EPEC.
MATERIALS AND METHODS
Bacterial strains.
EPEC strains (Table 1) were grown in Luria-Bertani (LB) broth as standing overnight cultures at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| EPEC E2348/69 | Prototypic O127:H6 EPEC strain | 8 |
| ΔescN | escN deleted from E2348/69 | 13 |
| ΔescC | escC deleted from E2348/69 | 13 |
| ΔescJ | escJ deleted from E2348/69 | N. Thomas, unpublished |
| EAEC 17-2 | Motile | T. Steiner |
| EAEC 17-2 | Nonmotile | T. Steiner |
| Plasmids | ||
| pescC-HSV | escC with HSV on C-terminal portion cloned into tet gene (BamHI-SalI) of pACYC184 | 13 |
| pKK232-8 | pBR322 derivative containing promoterless chloramphenicol acetyltransferase (cat) gene | Pharmacia |
| pLER-1179 | pKK232-8 derivative containing ler-cat transcriptional fusion | V. H. Bustamante, unpublished |
| pSEPZ-11 | pKK232-8 derivative containing LEE2-cat transcriptional fusion (pLEE2) | 1 |
| pSEPZ-17 | pKK232-8 derivative containing LEE2-cat transcriptional fusion (pLEE2-17) | 1 |
| pTIR394 | pKK232-8 derivative containing tir-cat transcriptional fusion | 37 |
| pLEE4-cat | pKK232-8 derivative containing LEE4-cat transcriptional fusion | Y. Martínez, unpublished |
| pEspC-cat | pKK232-8 derivative containing espC-cat transcriptional fusion | V. I. Martínez, unpublished |
| pCAT232 | pKK232-8 derivative containing bfpA-cat transcriptional fusion | 36 |
| pKptsH-cat | pKK232-8 derivative containing ptsH-cat transcriptional fusion | N. Olivares, unpublished |
| pKaroG-cat | pKK232-8 derivative containing aroG-cat transcriptional fusion | N. Olivares, unpublished |
| pKpyrC-cat | pKK232-8 derivative containing pyrC-cat transcriptional fusion | N. Olivares, unpublished |
ELISA.
The level of secretion of the EPEC-secreted protein EspB was monitored by enzyme-linked immunosorbent assay (ELISA) to assess the type III inhibition activity of a library of 20,000 compounds from Maybridge Plc (England). The compounds were dissolved in dimethyl sulfoxide (DMSO). EPEC from overnight standing culture was subcultured 1/10 in 200 μl of Dulbecco's modified Eagle medium (DMEM) containing a compound at 40 μM (final) or as indicated or with DMSO for 3 h in Immulon 2HB 96-well plates at 37°C in 5% CO2 to allow the EPEC-secreted proteins to adhere to the plate. The plates were then blocked overnight in 5% skim milk in phosphate-buffered saline (PBS) at 4°C. EspB was detected with mouse anti-EspB monoclonal antibody (MAb) (clone 2A11; 1/10,000; 1 h at room temperature) and a peroxidase-conjugated goat anti-mouse secondary antibody (Sigma; 1/5,000; 1 h at room temperature). The plates were washed with 0.1% Tween 20-PBS three times between reagent additions and five times before development. The ELISA was developed by adding 100 μl OPD solution (30 mg of o-phenylenediamine dihydrochloride [OPD] in 30 ml of 0.1 M citric acid, pH 4.5). The reaction was quenched with 100 μl of 3 N sulfuric acid and read at 492 nm in a TECAN Spectra-Fluor Plus plate reader. A duplicate plate of bacteria and compound or DMSO was also incubated for 3 h to monitor bacterial growth by reading the optical density at 600 nm (OD600). EspB secretion was normalized for bacterial growth in all experiments except for Fig. 3, where the effect of the compounds on bacterial growth was being assessed.
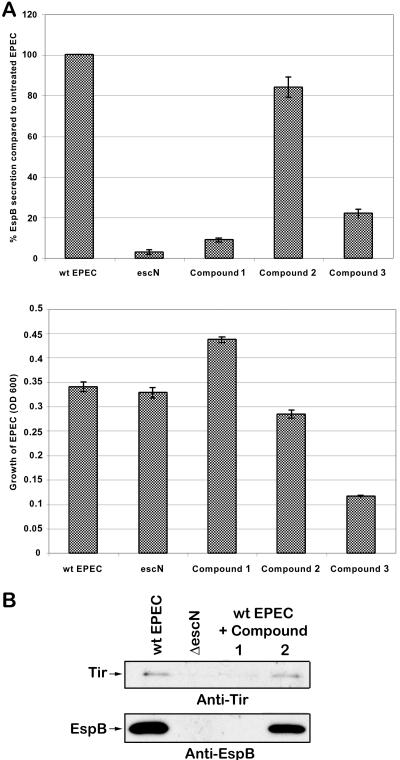
FIG. 3.
Screen for inhibitors of type III secretion by EPEC with no change in bacterial viability. (A) Graph of ELISA results monitoring percent EspB secretion compared to that in untreated EPEC. Bacteria were treated with the compounds at 40 μM for 3 h. The right panel graphs the viability of EPEC as measured by OD600. The results are shown ± standard deviation and are the average of three different experiments performed in triplicate. (B) Western blot for secreted proteins Tir and EspB in wt EPEC, ΔescN (type III apparatus mutant that does not secrete Tir or EspB), and wt EPEC treated with the compounds as described above. (C) Total secreted protein stained by Sypro-Ruby (Bio-Rad).
Monitoring protein degradation.
EPEC from overnight standing culture was subcultured 1/100 in 100 ml of DMEM for 5 h at 37°C in 5% CO2 (equivalent to an OD600 of 0.5). The culture was then harvested by centrifugation (5,000 × g, 10 min). The supernatant (secreted proteins) was filtered through a 0.2-μm bottle top filter to remove all bacteria and stored at −80°C.
Procedure for ELISA.
Filtered supernatant was incubated at 37°C in 5% CO2 for 3 h in Immulon 2HB 96-well plates with 40 μM compound or DMSO added as appropriate. The plates were then blocked overnight in 5% skim milk in PBS at 4°C. EspB was detected using the ELISA as described above.
Procedure for immunoblotting.
Filtered supernatant was incubated at 37°C in 5% CO2 for 5 h with 40 μM compound or DMSO. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer with β-mercaptoethanol was added, and samples (20 μl) were analyzed by SDS-PAGE.
Bacterial secreted proteins and whole-cell lysate.
EPEC from an overnight standing culture was subcultured 1/20 in 2 ml of DMEM for 5 h (equivalent to an OD600 of 0.9) at 37°C in 5% CO2 with 40 μM compound or DMSO. The culture was harvested by centrifugation (16,100 × g, 10 min). The supernatant (secreted proteins) was removed, and SDS sample buffer with β-mercaptoethanol added was normalized for OD600. The pellet was resuspended in ∼30 μl SDS sample buffer (normalized for OD600) with β-mercaptoethanol (bacterial lysate). Samples (20 μl for secreted proteins, 4 μl for bacterial lysate) were analyzed by SDS-PAGE.
Immunoblotting.
Samples were subjected to SDS-PAGE and transferred to nitrocellulose (pure nitrocellulose, 0.45 μm; Bio-Rad). Blots were blocked overnight at 4°C in 5% (wt/vol) skim milk-Tris-buffered saline with 1% Tween 20 (TBST) and incubated with primary antibody diluted in 1% skim milk-TBST for 1 h at room temperature. Blots were washed in TBST and then incubated in secondary antibody diluted in 1% skim milk-TBST for 1 h at room temperature. Blots were washed in TBST followed by detection with the ECL enhanced chemiluminescence reagent (Amersham Pharmacia Biotech). The following primary antibodies were used in the following amounts: mouse anti-EspB MAb clone 2A11, 1/50; mouse anti-Tir MAb clone 2A5, 1/50; rat anti-EscC, 1/2,500; rat anti-EscJ, 1/1,000; mouse anti-DnaK (Stressgen), 1/1,000; mouse anti-DnaJ (Stressgen), 1/1,000; rabbit anti-maltose-binding protein (MBP) (NEB), 1/500; mouse anti-herpes simplex virus (HSV) (Novagen), 1/1,000; and mouse anti-E. coli flagellin (Igen), 1/1,000. The following secondary antibodies were used in the following amounts: goat anti-mouse conjugated with horseradish peroxidase (HRP; Sigma), 1/5,000; goat anti-rat-HRP (BioCan Scientific), 1/5,000; and goat anti-rabbit-HRP (Sigma), 1/10,000.
Transcription assay.
Wild-type EPEC cells carrying the indicated transcriptional fusions (Table 1) were grown overnight in LB broth supplemented with ampicillin (100 μg/ml) and subcultured 1/50 in 10 ml of DMEM with the indicated amount of compound 1 or DMSO for 8 h at 37°C in 5% CO2. Samples were collected at an OD600 of 0.8. The chloramphenicol acetyltransferase microplate assay and protein determinations to calculate specific activity were performed as described previously (26, 36).
Motility assay.
Wild-type (wt) EPEC or positive (enteroaggregative E. coli [EAEC] 17-2 motile) or negative (EAEC nonmotile) controls were grown overnight in LB broth. Motility plates were prepared by pouring 5 ml of motility agar into each well of a six-well plate, adding a final concentration of 40 μM of compound in 200 μl of DMSO or DMSO alone, and allowing the plates to set. Wells were stabbed with the appropriate overnight culture and incubated for 18 h at 37°C.
RESULTS
Initial screen for inhibitors of type III secretion by EPEC.
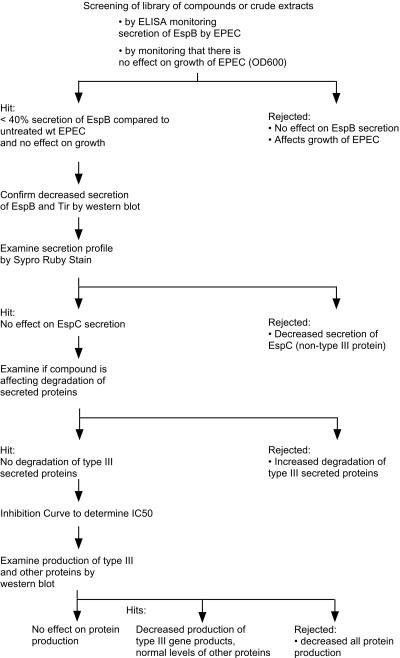
We developed a high-throughput assay to test extracts from a variety of sources, including marine organisms and plants, in a search for inhibitors of the TTSS (25). The assay is based on an ELISA to monitor if compounds or extracts affect secretion of one of the type III secreted proteins, EspB. Figure 1 shows a flowchart of the experimental strategy: monitoring EspB secretion by ELISA, monitoring growth of EPEC, confirming the effect on secretion by Western blotting and Sypro-Ruby protein stain, examining the degradation of type III secreted proteins, and examining production of type III and other proteins. Positive hits are those compounds or extracts that reduce EspB secretion while not affecting growth of EPEC.
FIG. 1.
Flowchart of assays to screen libraries of small molecules for their ability to inhibit type III secretion by EPEC.
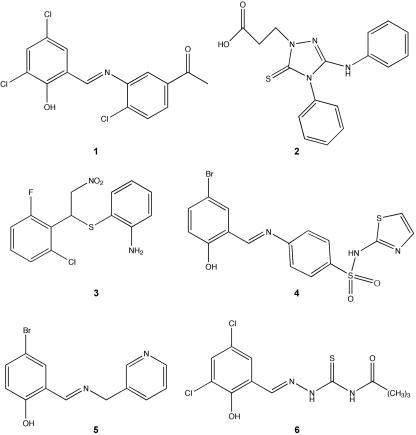
We screened 20,000 compounds from a library of small molecules (Maybridge Plc, England) for their ability to inhibit type III secretion of Esps by EPEC without affecting growth or general secretion of the bacterium. While the majority of compounds tested (99%) had no effect on EPEC, 64 compounds (0.32%) affected its growth and 30 compounds (0.15%) decreased EspB secretion without affecting growth. Compound 2 is an example of a compound that had no effect on EPEC, while compound 3 is an example of one that affected bacterial growth (Fig. 2 and 3). Compound 1 showed decreased EspB and Tir secretion (EspB by ELISA, Western blotting, and Sypro-Ruby protein staining; Tir by Western blotting), while not affecting bacterial growth (Fig. 3).
FIG. 2.
Structure of compounds from the Maybridge library of small molecules discussed in this study. Compound 1 decreased production of type III system-associated and other virulence-associated proteins, compound 2 had no observed effect on type III secretion or viability, compound 3 affected bacterial viability, and compounds 4 to 6 have the salicylideneaniline substructure and act like compound 1.
By examining bacterial growth supernatants by Sypro-Ruby (Bio-Rad)-stained protein polyacrylamide gels, we examined how these compounds affect secretion of the highly abundant EspB and EspD as well as their effect on EspC secretion. EspC is an EPEC non-type III secreted protein (22). EspC is non-LEE encoded and is a member of the autotransporter family (30, 39; reviewed in reference 15). While EPEC grown in compound 2 had similar amounts of EspC to untreated EPEC, EPEC grown in compound 1 had much reduced levels of EspC (Fig. 3C).
Compound 1 does not induce the breakdown of EspB.
In previous screens for inhibitors of type III secretion, we had found that some compounds affect the stability or increase the degradation of the type III secreted proteins (S. Ruschkowski and B. B. Finlay, unpublished observations). A compound that decreased the stability of the type III secreted proteins would appear to be a false positive since it would have less EspB measured by type III ELISA, Western blotting, and gel staining. The autotransporter EspC is an extremely stable protein, as evidenced by its resistance to degradation by proteinase K (22), and thus is not expected to be affected by a compound that decreases the stability or increases the degradation of secreted proteins. We therefore examined if any of these compounds affected the stability of the type III secreted proteins and the autotransporter EspC.
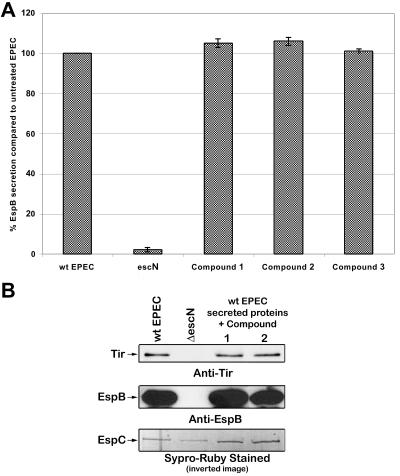
Bacterial supernatant containing secreted proteins from wild-type EPEC was incubated with a compound at 37°C, and the amount of EspB was determined by ELISA and Western blotting (Fig. 4). Compounds 1 and 2 did not affect the stability of EspB and Tir in the bacterial supernatant. The stability of EspC was not affected by any of the compounds tested, as expected (Fig. 4B, bottom).
FIG. 4.
Compound 1 does not induce the breakdown of type III secreted proteins. To determine if compound 1 affects the stability of the type III secreted proteins or the autotransporter EspC, bacterial supernatant containing secreted proteins from wild-type EPEC was incubated with 40 μM compound. (A) The amount of EspB was determined by ELISA. The results are shown ± standard deviation and are the average of three different experiments performed in triplicate. (B) The amounts of Tir and EspB were determined by Western blot analysis, and the amount of EspC was monitored by Sypro-Ruby protein stain (Bio-Rad).
Compound 1 reduced the amount of type III secreted proteins Tir and EspB inside EPEC.
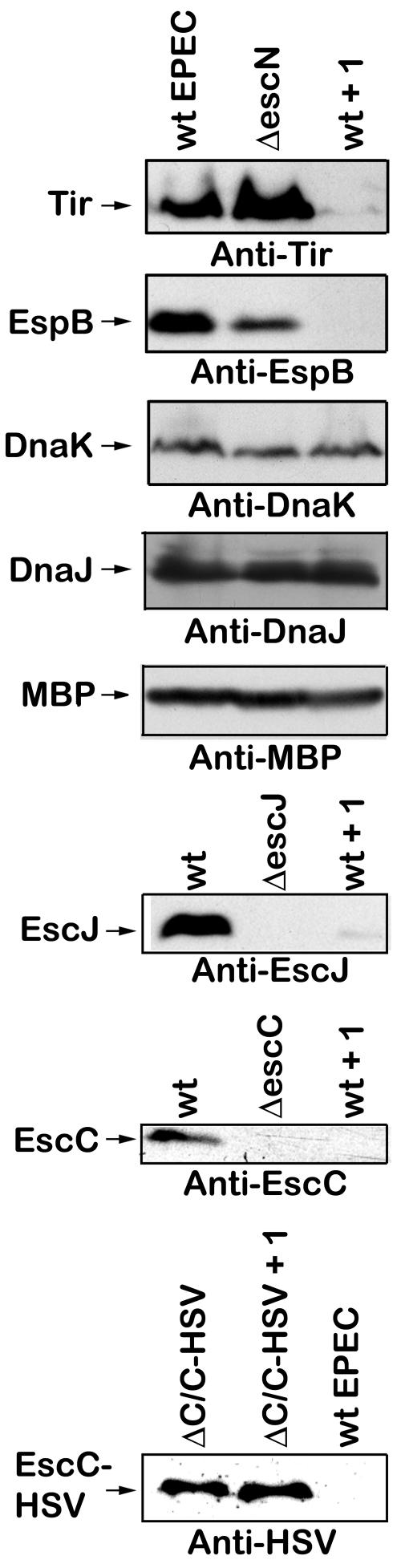
Compound 1 reduced levels of EspB and Tir in culture supernatants, and this was not due to growth inhibition or decreased protein stability (Fig. 3 and 4). The IC50 (the concentration of compound 1 that caused a 50% inhibition of secretion of EspB) was calculated to be 15 μM (data not shown). Compound 1 could have reduced secreted proteins because it had an effect on the production of these proteins by EPEC. Thus the levels of the type III secreted proteins Tir and EspB were examined inside the bacteria by Western blotting of a whole bacterial lysate (Fig. 5, first two panels). Compound 1 reduced the amount of Tir and EspB inside the bacteria to barely detectable levels. DnaK and DnaJ are abundant heat shock proteins that function as chaperones inside the bacterial cytoplasm (24). Maltose binding protein is an abundant periplasmic protein (19). However, compound 1 does not alter the amount of DnaK, DnaJ, or MBP produced by EPEC (Fig. 5, third panel), suggesting that compound 1 does not have a generalized effect on protein synthesis.
FIG. 5.
Compound 1 reduced production of both LEE-encoded EPEC-secreted proteins and type III apparatus components but not the production of non-virulence-associated proteins. Wild-type EPEC was grown in DMEM for 5 h with 40 μM compound 1 (wt + 1) or DMSO. The ΔescN, ΔescJ, and ΔescC type III secretion apparatus mutants were grown in DMEM for 5 h with the corresponding volume of DMSO. ΔC/C-HSV (short for ΔescC/pescC-HSV) is the escC type III apparatus mutant complemented in trans with a plasmid containing HSV-tagged escC under the control of a tetracycline resistance gene promoter and was grown in DMEM for 5 h with 40 μM compound 1 or DMSO. The bacteria were harvested by centrifugation and lysed in SDS sample buffer. The levels of Tir, EspB, DnaK, DnaJ, MBP, EscJ, EscC, and EscC-HSV were determined by Western blot analysis.
Compound 1 reduced production of both LEE-encoded EPEC-secreted proteins and type III apparatus components.
To investigate if compound 1 had an effect on all type III system-related proteins, the amounts of two type III apparatus components, EscJ and EscC, were examined in whole bacterial lysates. Compound 1 reduced the amounts of EscJ and EscC inside the bacteria (Fig. 5, fourth and fifth panels). However, when EPEC was transformed with a plasmid containing herpes simplex virus (HSV)-tagged escC under the control of a tetracycline resistance gene promoter (13), compound 1 did not affect the levels of HSV-tagged EscC (Fig. 5, last panel). This, combined with the DnaK, DnaJ, and MBP results, suggests that compound 1 specifically inhibits type III system- and/or virulence-associated products under endogenous transcriptional/translational control.
Virulence-associated promoters are more sensitive to transcriptional inhibition by compound 1.
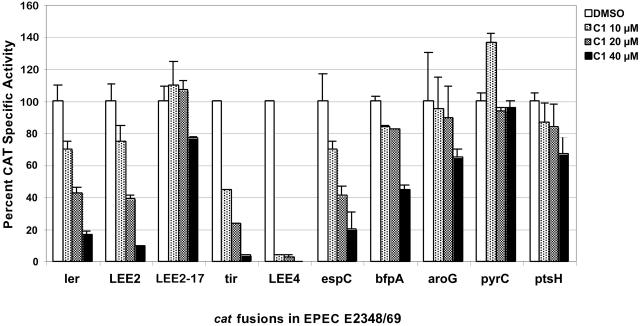
Compound 1 could be affecting the transcription of type III system-associated genes, the translation of type III system-associated genes, or the stability/turnover of type III system-associated proteins. The effect of compound 1 on transcription of type III system-associated and nonrelated genes was examined in EPEC by growing bacteria containing the appropriate transcriptional fusions (Fig. 6). As mentioned above, the LEE encodes a TTSS, its secreted proteins, and the bacterial outer membrane adhesin intimin. Increasing concentrations of compound 1 reduced the transcriptional activity of ler (LEE-encoded regulator), LEE2 (type III apparatus), LEE4 (EspA, EspD, EspB, etc.), and tir (LEE5) promoters, with LEE4 promoter being dramatically repressed even at 10 μM. LEE2-17 is a constitutive version of the LEE2 promoter and was included as a control. As expected, LEE2-17 transcription was not significantly reduced by compound 1. While EspC is not secreted by the TTSS (22), it is also regulated by Ler (9, 22). espC was similarly down-regulated by compound 1. BfpA is the structural subunit of the bundle-forming pili, and its expression is regulated by PerA (also called BfpT) (26). Expression of bfpA was affected by compound 1, but to a much lesser degree than other virulence genes. In contrast, non-virulence-related promoters, like, for example, the one directing the expression of the housekeeping gene pyrC, were not affected, even at a concentration of compound 1 of 40 μM, and others, like the aroG and ptsH promoters, were only moderately affected at high concentrations of compound 1 (Fig. 6). The expression of all virulence-related promoters was more sensitive to compound 1.
FIG. 6.
Virulence-associated promoters are more sensitive to transcriptional inhibition by compound 1. Wild-type EPEC cells carrying the indicated transcriptional fusions (Table 1) were grown in DMEM at 37°C in 5% CO2 for 8 h with the indicated amount of compound 1 (C1) or DMSO. Chloramphenicol acetyltransferase (CAT) microplate assays and protein determinations to calculate specific activity were performed and graphed as a percentage of CAT specific activity compared to that in untreated EPEC for each transcriptional fusion. The results are shown ± standard deviation and are the average of at least three different experiments performed in duplicate.
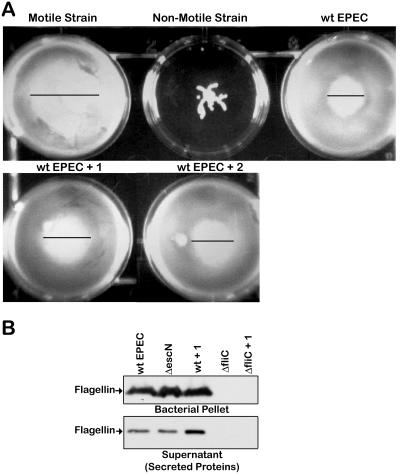
Compound 1 does not affect bacterial motility or flagellin expression or secretion.
Since the type III secretion apparatus and the flagellar assembly and export apparatus share many similarities, the effect of compound 1 on bacterial motility and flagellin expression and secretion was examined (Fig. 7). Compound 1 does not alter EPEC's motility, as evidenced by the zones of migration being the same in treated and untreated EPEC (Fig. 7A). Moreover, compound 1 did not alter the amount of flagellin expressed by EPEC or secreted by EPEC. Overall this strongly indicates that compound 1 is specific for virulence-associated products.
FIG. 7.
Compound 1 does not affect the motility or the level of flagellin of EPEC. (A) Strains of bacteria were stabbed into motility agar containing 40 μM compound 1 (+ 1) or compound 2 (+ 2) and grown for 18 h at 37°C. The positive control is a motile strain of EAEC, and the negative control is a nonmotile strain of EAEC. (B) Wild-type EPEC, the ΔescN type III mutant, or the ΔfliC flagellin filament subunit mutant were grown in DMEM for 5 h with 40 μM compound or DMSO as indicated. The bacteria were harvested by centrifugation and lysed in SDS sample buffer to yield the bacterial pellet, and the culture supernatant yielded the secreted proteins. The level of flagellin in the samples was determined by Western blot analysis.
DISCUSSION
We screened a commercial library of 20,000 small molecules for their ability to inhibit type III secretion by EPEC. After discarding compounds that had no effect, inhibited bacterial growth, and/or caused degradation of EPEC-secreted proteins, the characterization was focused on one compound. Compound 1 decreased the amount of secreted proteins not by interfering with the type III secretion apparatus but through affecting the amounts of type III secretion-associated products (Tir, EspB, EscJ, and EscC) and of the non-LEE-encoded virulence protein (EspC), while not affecting the production of proteins not associated with virulence (DnaK, DnaJ, MBP, and flagellin) or that are expressed under the control of a constitutive promoter (HSV-tagged EscC).
Compound 1 is composed of a halogenated salicylaldehyde derivative condensed with a 3-aminoacetophenone derivative via a Schiff base linkage (Fig. 2). An interesting aspect of this study is that in our screen of the small molecule library, we identified three other compounds which also contain this salicylideneaniline substructure (Fig. 2, compounds 4 to 6). These three compounds act like compound 1 in reducing the levels of virulence-associated proteins but not other proteins (data not shown), suggesting that the salicylideneaniline moiety is of primary importance for the observed biological activity of our lead structure. More surprisingly, a recent paper that was published while the present study was under way identified a similar structure (18). Kauppi et al. (18) screened a chemical library for inhibition of transcription of type III system-related genes in Yersinia pseudotuberculosis and identified a molecule with a similar Schiff base core that decreased reporter transcription and effector protein secretion. However, this molecule also affected motility of the bacterium, suggesting that it was not only inhibiting the TTSS but also the flagellar assembly apparatus. Notably compound 1 and the other three similar compounds identified in the current study did not affect bacterial motility (Fig. 7).
Furthermore, as briefly mentioned in the introduction, screening of this chemical library for inhibition of Yersinia transcription with a luciferase reporter assay yielded two other molecules that inhibited the transcription of type III system-associated genes and effector protein secretion with little effect on bacterial growth (18). One of these molecules, which has no structural similarity to compound 1, appears to be acting on regulatory elements in Yersinia's type III pathway (17, 18). Overall, combined with the present study, these results are evidence in support of TTSS-specific inhibitors.
The genes encoding the proteins whose production was affected by compound 1, including espC, have in common that they are transcriptionally regulated by Ler (9). This suggested that compound 1 could be specifically affecting virulence gene expression at the transcriptional level, while not affecting bacterial viability. The transcriptional fusion studies confirmed that the transcriptional activity of virulence-associated promoters is more sensitive to inhibition by compound 1, since the expression of all the Ler-dependent promoters tested was inhibited at least 60% even at 20 μM compound 1, while the rest of the promoters were not significantly affected at this concentration (Fig. 6).
As outlined in the introduction, EPEC's gene regulation is complex and not fully understood (2, 7). The expression of the LEE-encoded positive regulator Ler, which is central for the expression of the virulence genes encoded within and outside the LEE, is controlled by a number of global regulators such as IHF, Fis, H-NS, BipA, Per, and QseA (reviewed in reference 2). Ler relieves the repression of most LEE promoters by displacing H-NS or interfering with the formation of H-NS-mediated nucleoprotein complexes (1, 14). A new twist to EPEC's regulation was recently reported (7). The LEE also encodes a positive regulator of Ler called GrlA (global regulator of LEE activator) and a negative regulator named GrlR (global regulator of LEE repressor). GrlR most likely regulates LEE expression by modulating GrlA activity (7), as suggested by the ability of GrlR and GrlA to interact (3).
Compound 1 inhibits the transcription of ler; thus, the effect on the transcription of other LEE genes and espC is likely due to a reduction in Ler synthesis and/or Ler function, instead of being a direct effect on the Ler-dependent promoters. This was also suggested by the lack of inhibition of the expression of a constitutive version of the LEE2 promoter (Fig. 6). Compound 1 had an inhibitory effect on the transcription of bfpA only at the highest concentration, which suggests that PerA, the transcriptional activator of bfpA and/or the bfpA promoter, is not as sensitive to this compound as the Ler-dependent promoters. Another potential target of compound 1 is GrlA, which acts as a specific positive regulator of ler expression in attaching and effacing pathogens (7). Alternatively, compound 1 may enhance the negative role of GrlR in LEE gene expression. However, at this stage we can't rule out the possibility that compound 1 interferes with ler transcription at a different level, for example, by selectively altering the interaction of the RNA polymerase or global regulators with a particular subset of promoters including ler.
Expression of all Ler-regulated genes was reduced between 60 and 97% in the presence of 20 μM of compound 1, while the expression of all controls was unaltered. While virulence-associated genes were inhibited 80 to 100% at the highest concentration of compound 1 tested, the transcription of the non-virulence genes aroG and ptsH, but not the housekeeping gene pyrC, was moderately reduced (Fig. 6). However, since bacterial viability is not reduced, motility is not affected, and the level of non-virulence-associated proteins is not altered by treatment with compound 1, this mild effect on some non-virulence promoters is not critical for the bacteria or for the expression of essential genes. It is very clear from the secretion and expression data that compound 1 is having a significant and dramatic effect on the expression of Ler-dependent genes that encode virulence factors within (EspB, Tir, type III apparatus) and outside (EspC) the LEE.
Although the aim of this study was to identify direct inhibitors of the TTSS, we found an inhibitor of expression of virulence-associated products. We have identified a class of compounds that can potentially be used therapeutically, as well as being used as a tool for elucidating the intricate regulatory systems that modulate EPEC's virulence. Investigations into both the bacterial target(s) for this compound and whether this compound works on related pathogens such as enterohemorrhagic E. coli, the causative agent of illness associated with hamburger, are under way.
Acknowledgments
A.G. was supported by doctoral research awards from the Medical Research Council of Canada/Canadian Institutes for Health Research (CIHR), the Imperial Order of the Daughters of the Empire, and the Michael Smith Foundation for Health Research. B.B.F. and J.L.P. are Howard Hughes Medical Institute (HHMI) International Research Scholars. B.B.F. is a CIHR Distinguished Investigator and the Peter Wall Distinguished Professor. Operating grants from CONACyT, DGAPA and HHMI to J.L.P. and the Canadian Bacterial Disease Network to B.B.F supported this work.
The authors thank Sharon Ruschkowski and Markus Stein for developing the initial type III secretion assay; Amelia Gracey for technical assistance in improving the assay; Nikhil Thomas for kindly providing the anti-EscJ and anti-EscC antibodies and ΔescJ strain; Ted Steiner for providing strains of motile and nonmotile EAEC; and Norma Olivares, Enrique Merino, Verónica Martínez, and Ygnacio Martínez-Laguna for providing transcriptional fusions. The authors thank Samantha Gruenheid, Bruce Vallance, and Ted Steiner for their advice and Wanyin Deng, Nat Brown, and Corey Stephenson for their input and for critically reading the manuscript.
REFERENCES
- 1.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, S. C., R. D. Haigh, P. P. E. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creasey, E. A., R. M. Delahay, S. Daniell, and G. Frankel. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic E. coli. Microbiology 148:2093-2106. [DOI] [PubMed] [Google Scholar]
- 4.Dale, C., S. A. Young, D. T. Haydon, and S. C. Welburn. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. USA 98:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deibel, C., S. Kramer, T. Chakraborty, and F. Ebel. 1998. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol. Microbiol. 28:463-474. [DOI] [PubMed] [Google Scholar]
- 6.Deng, W., Y. Li, P. R. Hardwidge, E. A. Frey, R. A. Pfuetzner, S. Lee, S. Gruenheid, N. C. Strynadka, J. L. Puente, and B. B. Finlay. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vázquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ahsman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg, M. S., S. B. Calderwood, A. Donohue-Rolfe, G. T. Keusch, and J. B. Kaper. 1990. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infect. Immun. 58:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. A. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier, A., and B. B. Finlay. 2002. The hunt for type III secretion system inhibitors as potential antimicrobials. ASM News 68:383-387. [Google Scholar]
- 13.Gauthier, A., J. L. Puente, and B. B. Finlay. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowkles, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauppi, A. M., R. Nordfelth, U. Hägglund, H. Wolf-Watz, and M. Elofsson. 2003. Salicylanilides are potent inhibitors of type III secretion in Yersinia. Adv. Exp. Med. Biol. 529:97-100. [DOI] [PubMed] [Google Scholar]
- 18.Kauppi, A. M., R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241-249. [DOI] [PubMed] [Google Scholar]
- 19.Kellermann, O., and S. Szmelcman. 1974. Active transport of maltose in Escherichia coli K12: involvement of “periplasmic” maltose binding protein. Eur. J. Biochem. 47:139-149. [DOI] [PubMed] [Google Scholar]
- 20.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 65:2606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, B., and B. B. Finlay. 1995. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA 92:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberek, K., C. Georgopoulos, and M. Zylicz. 1988. Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl. Acad. Sci. USA 85:6632-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linington, R. G., M. Robertson, A. Gauthier, B. B. Finlay, R. van Soest, and R. J. Andersen. 2002. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4:4089-4092. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island of enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 29.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 30.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, S., M. F. Feldman, and G. Cornelis. 2001. The type III secretion system of Gram-negative bacteria: a potential therapeutic target? Expert Opin. Ther. Targets 5:327-339. [DOI] [PubMed] [Google Scholar]
- 33.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porter, M. E., P. Mitchell, A. J. Roe, A. Free, D. G. E. Smith, and D. L. Gally. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54:1117-1133. [DOI] [PubMed] [Google Scholar]
- 35.Preston, G. M., N. Bertrand, and P. B. Rainey. 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol. Microbiol. 41:999-1014. [DOI] [PubMed] [Google Scholar]
- 36.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein, M., B. Kenny, M. A. Stein, and B. B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuber, K., J. Frey, A. P. Burnens, and P. Kuhnert. 2003. Detection of type III secretion genes as a general indicator of bacterial virulence. Mol. Cell Probes 17:25-32. [DOI] [PubMed] [Google Scholar]
- 41.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 42.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]