Abstract
The Euregio Meuse-Rhine (EMR) is formed by the border regions of Belgium, Germany, and The Netherlands. Cross-border health care requires infection control measures, in particular since the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) differs among the three countries. To investigate the dissemination of MRSA in the EMR, 152 MRSA isolates were characterized by pulsed-field gel electrophoresis (PFGE), SCCmec typing, and multilocus sequence typing. PFGE revealed major clonal groups A, G, L, and Q, suggesting dissemination of MRSA in the EMR. Group A harbored mainly SCCmec type III and sequence types (STs) 239 and 241. The majority of the strains from group G harbored SCCmec type I and ST8 and ST247, whereas most strains from group L carried either SCCmec type IV or type I. Within group L, ST8 and ST228 were found, belonging to clonal complexes 8 and 5, respectively. Most strains from group Q included SCCmec type II and were sequence typed as ST225. Both ST225-MRSA-II and ST241-MRSA-III were novel findings in Germany. In addition, the SCCmec type of two isolates has not been described previously. One strain was classified as SCCmec type III but harbored the pls gene and the dcs region. Another strain was characterized as SCCmec type IV but lacked the dcs region. In addition, one isolate harbored both SCCmec type V and Panton-Valentine leukocidin. Finally, the SCCmec type of the strains was found to be correlated with the antibiotic susceptibility pattern.
Staphylococcus aureus is a potentially pathogenic bacterium that can cause various diseases such as postoperative wound infections and necrotizing pneumonia (20). S. aureus has a strong adaptive power to antibiotics. Since the introduction of methicillin in 1959, methicillin-resistant S. aureus (MRSA) strains have been isolated, first in the United Kingdom in 1961 and subsequently in other parts of the world. Although most of the MRSA strains are hospital acquired, community-acquired strains (CA-MRSA) have also recently been reported (32).
Resistance of MRSA strains to methicillin is determined by the presence of the mecA gene, which encodes the penicillin binding protein 2a. The mecA gene is localized on a mobile genetic element, which is designated the staphylococcal chromosomal cassette mec (SCCmec) (3, 14, 21). Currently, five main types of SCCmec (types I to V) are distinguished. SCCmec types I, II, and III are associated with hospital-acquired MRSA, whereas types IV and V are associated with CA-MRSA (12, 15). SCCmec types I, IV, and V exclusively encode resistance to β-lactam antibiotics. By contrast, SCCmec types II and III determine multiresistance, as these cassettes carry both integrated plasmid sequences (e.g., pT181 and pUB110) and transposons (e.g., Tn554) containing drug resistance genes. Besides the resistance genes on SCCmec, S. aureus can also carry drug resistance genes on other sites of its chromosome and on plasmids. Also situated on SCCmec are genes responsible for the regulation of the transcription of mecA: ΔmecR1 (on SCCmec types I, IV, and V) and mecR1 and mecI (on SCCmec types II and III) (13, 14, 15). For integration into and excision from the chromosome at a specific site (attBscc), genes encoding chromosomal cassette recombinases (ccr) are located within the SCCmec elements. These genes are designated ccrA1 and ccrB1 (in SCCmec type I), ccrA2 and ccrB2 (in SCCmec types II and IV), ccrA3 and ccrB3 (in SCCmec type III), and ccrC (in SCCmec type V) (6, 12, 15).
The Euregio Meuse-Rhine (EMR) is a region consisting of the Belgian provinces of Limbourg and Liège, the German-speaking region of Belgium, the region Aachen in Germany, and the southern part of the Dutch province of Limbourg, with an area of 10.478 km2. Cross-border patient mobility and free access to health care facilities within the European Union in general, and the EMR in particular, are important issues for patients, doctors, hospitals, sickness funds, and health care insurance companies. Each year, many thousands of the 3.7 million inhabitants of the EMR cross the border to visit a medical specialist or a hospital on the other side of the border. In an official publication of the European Commission (D. Byrne, Maastricht Conference on Cross-Border Health Care, Maastricht, The Netherlands, 8 June 2004), the EMR was therefore recently mentioned as a model region for the European Union in the field of cross-border health care and cross-border cooperation of hospitals and sickness funds. Nevertheless, an important issue of concern that is related to cross-border health care is the dissemination of multiresistant bacteria. In this regard, it is interesting to note that the three countries forming the EMR differ considerably in prevalence of MRSA isolated in hospitals (23.6%, 13.8%, and 0.6% in Belgium, Germany, and The Netherlands, respectively) (31). Consequently, the cross-border transfer of patients may have an important impact on the dissemination and prevalence of MRSA, in particular in cases where patients are transferred from countries with a relatively high prevalence to a country with a low prevalence. Therefore, we investigated the dissemination of MRSA isolates between hospitals from the EMR during the last 5 years. The MRSA isolates were subjected to pulsed-field gel electrophoresis (PFGE), SCCmec typing, and multilocus sequence typing (MLST) (2). As the presence of Panton-Valentine leukocidin (PVL) genes was suggested to be an important characteristic of CA-MRSA (33), the isolates were also subjected to a PVL-specific real-time PCR.
MATERIALS AND METHODS
Clinical isolates.
One hundred fifty-two isolates of MRSA from individual patients, isolated between December 1999 and February 2004 from five geographically closely related hospitals in the EMR, were included in the study (Table 1). These included two Belgian hospitals (Hospital East-Limbourg, Genk, an 822-bed general hospital, and General Hospital Vesalius, Tongeren, a 355-bed general hospital), one German hospital (Universitätsklinikum Aachen, a tertiary 1,500-bed university hospital), and two Dutch hospitals (Atrium Medical Centre, Heerlen, an 811-bed general hospital, and University Hospital Maastricht, a tertiary 680-bed university hospital). The 152 MRSA isolates included 40 isolates from Belgium, 49 from Germany, and 63 from The Netherlands (Table 1). The isolates from the Belgian and German hospitals represented a random selection of 50% of the clinical isolates. The Dutch isolates were derived from surveillance cultures from patients at risk for MRSA carriage who were admitted to the two Dutch hospitals during the study period. During this period, infections with MRSA were not found in the two Dutch hospitals. All strains were identified as S. aureus by catalase and coagulase testing. Methicillin resistance was determined by the disk diffusion test with oxacillin concentration disks, according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (22), and by PCR amplification of the mecA gene (26).
TABLE 1.
Distribution of SCCmec types within MRSA isolates in the EMR, 1999-2004
| Country | No. (%) of:
|
Total (no. [%]) | ||||
|---|---|---|---|---|---|---|
| SCCmec I | SCCmec II | SCCmec III | SCCmec IV | NTa | ||
| Belgium | 24 (60) | 2 (5) | 1 (2.5) | 13 (32.5) | 0 (0) | 40 (100) |
| Germany | 8 (16) | 14 (29) | 18 (37) | 7 (14) | 2 (4) | 49 (100) |
| The Netherlands | 12 (19) | 2 (3) | 16 (25) | 31 (49) | 2 (3) | 63 (100) |
| Total | 44 (29) | 18 (12) | 35 (23) | 51 (34) | 4 (3) | 152 (100) |
NT, not typeable.
Reference strains.
MRSA strains COL, BK2464, ANS46, HDE288, and WIS were used as reference strains for SCCmec types I, II, III, IV, and V, respectively (15, 23). S. aureus strain 1206 was used as a positive control for the Tn554 PCR (35). For PFGE, reference strain S. aureus Ps 47 was used as a molecular weight marker, whereas MRSA strains BK2464, COL, HDE288, HU25, and PER34 were used as reference strains for the New York/Japan, archaic, pediatric, Brazilian, and Iberian clonal types, respectively (24).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed by microbroth dilution according to the NCCLS guidelines (22) for the following antibiotics: amikacin, amoxicillin, cefazolin, ciprofloxacin, clindamycin, co-trimoxazole, doxycycline, erythromycin, flucloxacillin, gentamicin, penicillin, rifampin, and vancomycin.
SCCmec typing.
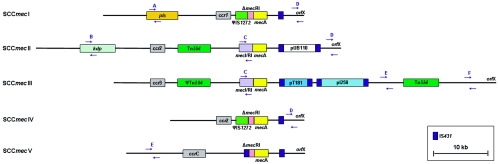
SCCmec typing was essentially carried out as described by Oliveira and de Lencastre (23), in which mecA and six different loci on SCCmec (Fig. 1) were amplified by PCR with the following modifications. PCR amplification of mecA sequences was carried out with primers mecA1 and mecA2 (Sigma Genosys, The Netherlands), resulting in a PCR product of 527 bp instead of 162 bp (26). PCR was performed in a volume of 50 μl containing 10 μl of a 0.5 McFarland suspension (1.5 × 108 CFU/ml) of the MRSA strain, 0.2 mM of each deoxynucleoside triphosphate (Amersham Biosciences, The Netherlands), 1× PCR buffer (QIAGEN, The Netherlands), 1.25 U HotStarTaq (QIAGEN, The Netherlands), and primers. The primer concentrations used were similar to those previously described (23), except for those of the mecA primers, which were 0.6 μM for both mecA1 and mecA2. The amplifications were performed on a GeneAmp PCR system, model 9600 (Applied Biosystems, The Netherlands), with the following program: 15 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 53°C, and 60 s at 72°C, followed by a postextension step of 10 min at 72°C. The PCR products were separated on 2% agarose gels in Tris-acetate-EDTA buffer, stained with ethidium bromide, and visualized with UV light by using a FluorChem imaging system (Alpha Innotech Corporation, The Netherlands).
FIG. 1.
Schematic drawing of SCCmec types I to V. The major elements (ccr genes, IS431, IS1272, mecA, mecI, mecRI, orfX, pI258, pT181, pUB110, and Tn554) of the five SCCmec types are given, as are the six different loci (A to F) used for the typing of SCCmec according to the method of Oliveira and de Lencastre (23). The primers of the PCR for the six different SCCmec loci are indicated by arrows.
PCR for ccrAB and ccrC.
Most of the primers used for amplification of ccrAB and ccrC were used as described previously (11, 15). Primer β2, however, was replaced by a primer with the following sequence: 5′-ATTGCCTTGATAATAGCCTCT-3′ (primer β2a). The following reaction conditions were used: either 1.2 μM of forward primer β2a for ccrAB or 0.4 μM of forward primer γF for ccrC; 0.4 μM of reverse primer α2, α3, α4, or yR; 0.2 mM of each deoxynucleoside triphosphate; 1× PCR buffer; 2.5 U of HotStarTaq DNA polymerase; and 10 μl of a 0.5 to 1 McFarland suspension (1.5 × 108 to 3 × 108 CFU/ml) in a total volume of 50 μl. Amplification was performed on a GeneAmp PCR system, model 9600, by using the following program: 15 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C, followed by an extension step of 10 min at 72°C. PCR products were analyzed by electrophoresis through 1% agarose gels as described above.
PCR for Tn554.
The primers for Tn554 were used as described previously (16). The reaction conditions used were similar to those for the ccr PCR. Amplification was performed on a GeneAmp PCR system, model 9600, by using the following program: 15 min at 94°C, followed by 34 cycles of 1 min at 94°C, 1 min at 63°C, and 1 min at 72°C, followed by an extension step of 10 min at 72°C. PCR products were analyzed by electrophoresis through 1% agarose gels as described above.
PFGE.
PFGE was carried out essentially as described previously (11). The banding patterns were visualized with UV light by using a FluorChem imaging system. Subsequently, the patterns were analyzed with Dice comparison and unweighted-pair group matching analysis settings with GelCompar II 3.5 (Applied Maths, Sint-Martens-Latem, Belgium) according to the scheme of Tenover et al. (30). The position tolerance was set at 2.0%, and isolates with a similarity index of 0.80 or more were classified as a clonal group (5, 25).
MLST.
It has previously been shown that MRSA strains from one major clonal group, as demonstrated by PFGE, have either the same sequence type (ST) or STs that are related to a single clonal complex (5, 7, 25, 29). Therefore, two representative strains from each of the major clonal groups as obtained through PFGE (5, 7, 25, 29) were used for MLST (9). The primers used for MLST were identical to those described previously (9), with the exception of primers glpF-Dn and gmk-Up, which were replaced by primers glpF-Dna (5′-TGGTAAAATCGCATGTGCAATTC-3′) and gmk-Upa (5′-ATCGTTTTATCAGGACCATC-3′), respectively. The PCR products were sequenced using an ALFexpress II automatic sequencer (Amersham Biosciences, The Netherlands). Finally, the STs were determined by using the MLST database (http://www.mlst.net).
Real-time PCR for PVL.
PVL was detected with a real-time PCR method as described previously (8).
Statistical analysis.
The correlation between SCCmec type and antibiotic susceptibility pattern was determined by canonical discriminant analyses with the software package SPSS 11.0.1 (SPSS, Inc., The Netherlands). Canonical discriminant analyses are used for the investigation of one or more normally distributed interval independent variables (SCCmec type) and a categorical dependent variable (susceptibility pattern). This is a multivariate technique that considers the latent dimensions in the independent variables for predicting group membership in the categorical dependent variable.
RESULTS
Distribution of SCCmec types.
The SCCmec types (Fig. 1) could be determined for 148 of the 152 (97.4%) clinical isolates of MRSA. Only four of the five different types of SCCmec were found (I to IV [Table 1]) with the method described by Oliveira and de Lencastre (23). SCCmec types I and IV were predominant, with 29% and 34%, respectively, and types II and III were less common, with 12% and 23%, respectively. The different SCCmec types were not distributed similarly among the three countries from the EMR. In Belgium, SCCmec types I and IV predominated, whereas in Germany, the most common types were types II and III. In The Netherlands, the most frequently found types were types I, III, and IV (Table 1).
The SCCmec types of 4 (DOM068, DOM083, DOM114, and DOM012) of the 152 MRSA isolates (3%) could not be determined according to the method described by Oliveira and de Lencastre (23). DOM068 was found to contain loci A, C, D, and E, as well as F (Table 2), which is uncommon to the prototypes of the five known SCCmec types, as shown in Fig. 1. Another isolate, DOM083, was also found to have a unique SCCmec organization, possessing loci C, D, E, and F (Table 2). The SCCmec structure of isolates DOM068 and DOM083 was further characterized by determination of the presence of two other loci from the SCCmec cassette, i.e., ccr and Tn554. Both strains were found to possess Tn554 as well as ccrAB3, indicating that their SCCmec cassettes most strongly resemble the type III cassette (Table 2).
TABLE 2.
Results of non-typeableb SCCmec within MRSA isolates
The cassette of another strain, DOM114, was found to contain only a single locus from the six loci that were defined by Oliveira and de Lencastre (23), i.e., locus E (Table 2). Since DOM114 was also found to possess the ccrC gene, its SCCmec cassette can be classified as type V (Table 2).
The fourth “nontypeable” isolate, DOM012, lacked all six loci as defined by Oliveira and de Lencastre (23). However, this strain was found to contain ccrAB2, which is characteristic for both type II and type IV cassettes. Since the DOM012 cassette did not contain Tn554, it was classified as type IV (Table 2).
PFGE analyses.
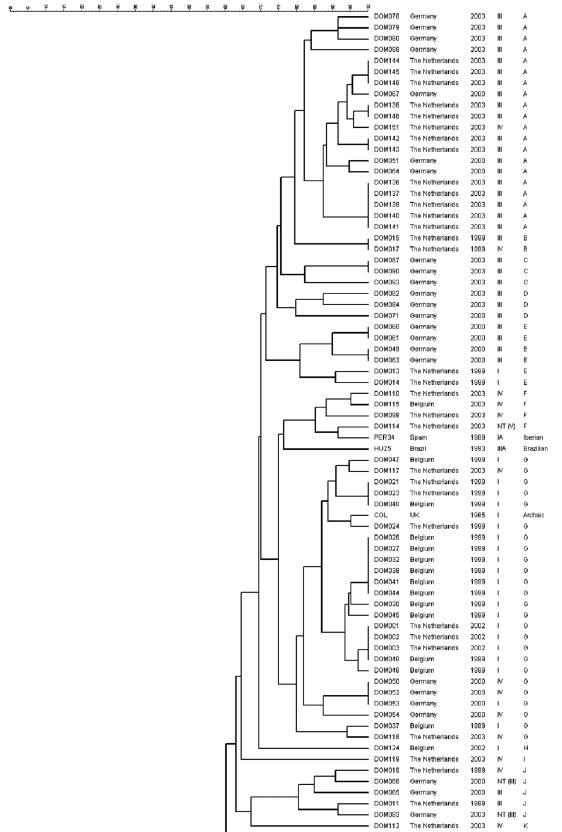
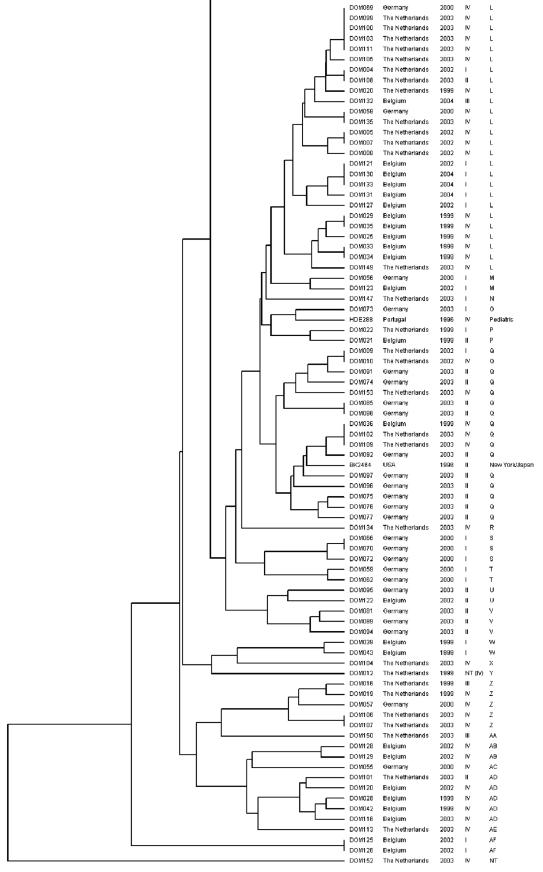
Each of the MRSA strains was subjected to analysis by PFGE. On the basis of the PFGE patterns, a dendrogram was constructed (Fig. 2). One of the isolates (DOM152) could not be typed due to repeated difficulties with the isolation of DNA from this strain. A total of 32 clonal groups (A to AF) were distinguished, of which four were major clonal groups (A, G, L, and Q). Major clonal group A was closely related to the clonal groups B, C, D, and E. Taken together, these groups included 34 of the 152 MRSA isolates (22%). A large majority of these 34 strains (88%), isolated from both Germany and The Netherlands, harbored SCCmec type III.
FIG. 2.
Dendrogram of the 152 clinical MRSA isolates and five reference clones. The five columns on the right represent MRSA isolate code, country of origin, year of isolation, SCCmec type, and clonal group, respectively. NT, not typeable.
The second major clonal group, group G, contained 26 of the 152 MRSA isolates (17%), of which 21 (81%) harbored SCCmec type I and 5 (19%) harbored SCCmec type IV. Most isolates from this group originated from Belgium and The Netherlands. Group G was also found to include reference strain COL, representative for the archaic clone, which is one of the six major MRSA clones spread worldwide (2). Clonal group F, which was related to group G, comprised only four isolates (3%), three of which harbored SCCmec type IV and one of which harbored SCCmec type V. The reference strain for the Iberian clone (2), PER34, was linked to clonal group F.
The third major clonal group, group L, included 26 of the 152 MRSA isolates (17%). Of these, 18 (69%) contained SCCmec type IV, 6 (23%) contained SCCmec type I, 1 (4%) contained SCCmec type II, and 1 (4%) contained SCCmec type III. Most of the strains were isolated in Belgium and The Netherlands. None of the reference strains were linked to this clonal group.
The fourth major group, group Q, contained 16 of the 152 MRSA isolates (11%). Ten (63%) of these carried SCCmec type II, whereas five (31%) contained SCCmec IV and one (6%) contained SCCmec type I. All except one of the strains from this group were isolated in either Germany or The Netherlands. Group Q also included reference strain BK2464, which is a representative of the New York/Japan clone (2).
MLST analyses.
To investigate which major MRSA clones from which clonal complexes are disseminated in the EMR, two representative strains from each major clonal PFGE group were subjected to MLST. As shown in Table 3, MLST identified six different sequence types (ST8, -225, -228, -239, -241, and -247) that belonged to two clonal complexes (CC5 and CC8). In major clonal group A, two different STs, which both belong to CC5, were found: (i) ST239-MRSA-III, a single-locus variant (SLV) of ST8 and representative for the Brazilian clone, and (ii) ST241-MRSA-II, an SLV of ST239-MRSA-III (at locus yqiL). The two STs that were found in major clonal group G are different SLVs of a single ST, i.e., ST250; ST247-MRSA-I is an SLV at the gmk locus, and ST8-MRSA-I is an SLV at the yqiL locus of ST250 (10). Interestingly, the strains from major clonal group L (DOM111 and DOM131) were typed as ST8-MRSA-IV and ST228-MRSA-I, respectively, which belong to different clonal complexes (CC8 and CC5, respectively). Both strains from major clonal group Q were typed as ST225-MRSA-II, an SLV at the tpi locus of ST5 (10).
TABLE 3.
Typing results of the four major clonal groups
| Code | Country | Major clonal group | SCCmec type | MLST | ST | CCa |
|---|---|---|---|---|---|---|
| DOM078 | Germany | A | III | 2-3-1-1-4-4-30 | 241 | 8 |
| DOM141 | The Netherlands | A | III | 2-3-1-1-4-4-3 | 239 | 8 |
| DOM038 | Belgium | G | I | 3-3-1-12-4-4-16 | 247 | 8 |
| DOM053 | Germany | G | I | 3-3-1-1-4-4-3 | 8 | 8 |
| DOM111 | The Netherlands | L | IV | 3-3-1-1-4-4-3 | 8 | 8 |
| DOM131 | Belgium | L | I | 1-4-1-4-12-24-29 | 228 | 5 |
| DOM077 | Germany | Q | II | 1-4-1-4-12-25-10 | 225 | 5 |
| DOM092 | Germany | Q | II | 1-4-1-4-12-25-10 | 225 | 5 |
CC, clonal complex.
Prevalence of PVL.
Only 2 (DOM103 and DOM114) of the 152 MRSA isolates (1.3%) contained PVL. Although both strains were isolated in The Netherlands in 2003, they differed in both SCCmec type and PFGE type. Strain DOM103 was classified within major clonal group L, harboring SCCmec type IV, whereas strain DOM114 was classified within group F, carrying SCCmec type V (Fig. 2).
Correlation between SCCmec type and antibiotic susceptibility pattern.
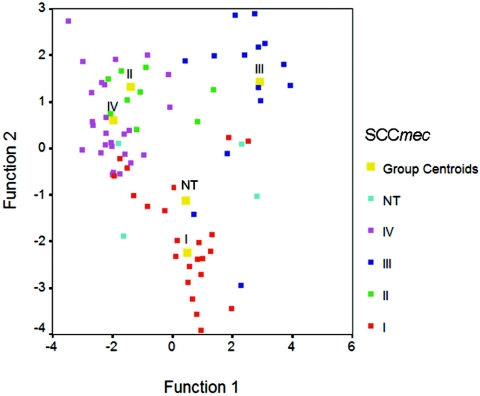
The antibiotic resistance patterns for the SCCmec types are presented in Table 4. Only the non-β-lactam antibiotics are presented in this table, since all MRSA strains were resistant to the four β-lactam antibiotics tested, i.e., amoxicillin, cefazolin, flucloxacillin, and penicillin. To investigate if a correlation exists between the SCCmec type and the antibiotic susceptibility pattern of MRSA isolates, canonical discriminant analyses were performed. As shown in Fig. 3, the SCCmec types were centered around the four group centroids, indicating that SCCmec type and susceptibility pattern were indeed correlated. More specifically, the antibiotic susceptibility pattern had a predictive value of 84.1% for SCCmec type I, 83.3% for SCCmec type II, 85.7% for SCCmec type III, and 86.3% for SCCmec type IV (Table 5). As shown in Table 4 and Fig. 3, the correlation is more pronounced for SCCmec types II, III, and IV than for SCCmec type I.
TABLE 4.
Distribution of the non-β-lactam antibiotic resistance patterns among the 152 MRSA strains and their corresponding SCCmec types
| SCCmec type | No. of strains | No. (%) of resistant MRSA strainsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMK | CIP | CLI | COT | DOX | ERY | GEN | RIF | VAN | ||
| I | 44 | 43 (98) | 42 (96) | 32 (73) | 42 (96) | 17 (39) | 38 (86) | 42 (96) | 7 (16) | 0 (0) |
| II | 18 | 17 (94) | 17 (94) | 17 (94) | 3 (17) | 1 (6) | 17 (94) | 1 (6) | 1 (6) | 0 (0) |
| III | 35 | 35 (100) | 33 (94) | 30 (86) | 34 (97) | 32 (91) | 34 (97) | 32 (91) | 6 (17) | 0 (0) |
| IV | 51 | 34 (67) | 40 (78) | 11 (22) | 10 (20) | 0 (0) | 28 (55) | 22 (43) | 0 (0) | 0 (0) |
| NTb | 4 | 3 (75) | 2 (50) | 1 (25) | 2 (50) | 2 (50) | 1 (25) | 3 (75) | 2 (50) | 0 (0) |
AMK, amikacin; CIP, ciprofloxacin; CLI, clindamycin; COT, co-trimoxazole; DOX, doxycycline; ERY, erythromycin; GEN, gentamicin; RIF, rifampin; VAN, vancomycin.
NT, not typeable.
FIG. 3.
Statistical analyses of SCCmec types and antibiotic susceptibility patterns. The discriminant function 1 and function 2 are latent variables that are created as a linear combination of discriminating variables. NT, not typeable.
TABLE 5.
Correlation between the SCCmec types and the antibiotic susceptibility patterns of the 152 MRSA strains
| SCCmec type | Predicted SCCmec type (no. [%])a
|
Total (no. [%]) | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV | NTb | ||
| I | 37 (84.1) | 0 (0) | 2 (4.5) | 5 (11.4) | 0 (0) | 44 (100) |
| II | 0 (0) | 15 (83.3) | 1 (5.6) | 1 (5.6) | 1 (5.6) | 18 (100) |
| III | 3 (8.6) | 1 (2.9) | 30 (85.7) | 1 (2.9) | 0 (0) | 35 (100) |
| IV | 0 (0) | 7 (13.7) | 0 (0) | 44 (86.3) | 0 (0) | 51 (100) |
| NTb | 0 (0) | 0 (0) | 1 (25) | 1 (25) | 2 (50) | 4 (100) |
Predicted from susceptibility pattern.
NT, not typeable.
DISCUSSION
Monitoring the dissemination of MRSA in the EMR is important, since known and novel MRSA clones may spread from country to country through cross-border patient care. In particular, the spread of MRSA harboring either SCCmec type II or type III, which encodes multiresistance, could pose a serious threat to health care facilities.
In this study, 152 MRSA strains were characterized, isolated in hospitals from the EMR between 1999 and 2004. Typing of the strains by PFGE revealed four major clonal groups, suggesting dissemination of MRSA in the EMR. The strains that were classified within major clonal group A and within the closely related, minor clonal groups B to E comprised 22% of all isolates. Two representative strains from group A were also typed by MLST and were classified as ST239-MRSA-III and ST241-MRSA-III. Both STs form part of CC8 (10). Although strains with the signature of ST239-MRSA-III were previously found in both Germany and The Netherlands, this study is the first to report the presence of ST241-MRSA-III in Germany (10).
The second major clonal group, group G, included MRSA strains harboring mainly SCCmec type I (Fig. 2). MLST of two representative clones from group G revealed two different STs, which were classified within the same clonal complex (CC8), i.e., ST8-MRSA-I and ST247-MRSA-I. Both STs have previously been found in the countries from the EMR: ST8-MRSA-I in Belgium and The Netherlands and ST247-MRSA-I in Belgium and Germany (7, 10, 36).
MRSA strains from major clonal group L harbored mainly SCCmec types IV and I. From this group, two strains with different SCCmec types (one strain of type IV and one of type I) were selected for analysis by MLST. Thus, these strains were classified as ST8-MRSA-IV and ST228-MRSA-I, respectively. Interestingly, these STs belong to different clonal complexes, i.e., CC8 and CC5, respectively. Although these clonal complexes were more closely related to each other than they are to other clonal complexes (19), the finding of strains from different MLST clonal complexes within a single PFGE clonal group was novel for S. aureus. Nevertheless, this finding was in line with previous reports that demonstrated the higher discriminatory power of MLST over PFGE for bacterial species other than S. aureus, such as Vibrio cholerae, Salmonella, and Listeria monocytogenes (17, 18, 28). Both ST8-MRSA-IV and ST228-MRSA-I were found previously in the EMR countries: ST8-MRSA-IV in Germany and The Netherlands and ST228-MRSA-I in Belgium and Germany (10, 36).
MRSA strains from the fourth major clonal group, group Q, harbored mainly SCCmec type II. Two representatives from this group were both typed as ST225-MRSA-II, an ST belonging to CC5. Although ST225-MRSA-II has previously been found in the United States (29), the finding of this ST in Germany is novel. Since ST225-MRSA-II is an SLV at the tpi locus of strain ST5-MRSA-II, which was previously found in Belgium (7), ST225-MRSA-II may be derived from ST5-MRSA-II. Recent studies reported the finding of ST22-MRSA-IV and ST45-MRSA-IV in Belgium and ST45-MRSA-I and ST45-MRSA-IV in The Netherlands (7, 34). These STs, however, were not found in this study.
For 4 of the 152 MRSA isolates (3%) the SCCmec type could not be determined using the method described by Oliveira and de Lencastre (23). The percentage of nontypeable SCCmec cassettes was low compared to that from other studies, in which 10 to 15% could not be typed (4, 11). Since two of the nontypeable MRSA strains (DOM068 and DOM083) were found to possess ccrAB3, they could be considered SCCmec type III strains. However, compared to the type III cassette prototype (Fig. 1), strain DOM068 contained two additional loci, locus A (pls gene) and locus D (dcs region). Strains with an organization of SCCmec loci similar to that for DOM068 have not been reported yet. In contrast, a strain with an additional locus D as opposed to the type III prototype, as seen in strain DOM083, has previously been described by Aires de Sousa and de Lencastre (1).
Another nontypeable strain, DOM012, was found to contain only a single SCCmec locus apart from mecA. This locus, ccrAB2, was present in the prototype cassettes of both type II and type IV. However, since strain DOM012 possessed neither mecI (locus C) nor Tn554, its gene cassette had a higher similarity with the cassette of type IV than with that of type II. Also, the resistance to β-lactam antibiotics found was in line with SCCmec type IV. We therefore concluded that strain DOM012 carries SCCmec type IV but lacks locus D.
The low prevalence of PVL was in accordance with previous studies (8, 27). Both strains (DOM103 and DOM114) were very likely not related, as they were classified within different clonal groups and harbored different SCCmec types. DOM114 is to our knowledge the first reported PVL-positive MRSA strain carrying SCCmec type V.
Although antibiotic resistance in S. aureus can also be determined by sequences other than SCCmec, a correlation of approximately 85% was found between antibiotic susceptibility pattern and SCCmec type. Rapid identification of the SCCmec type of MRSA isolates by PCR could therefore be useful to predict the antibiotic susceptibility pattern of isolates and, consequently, guide the choice of antibiotics used for treatment. Hence, the identification of the SCCmec type of clinical isolates might contribute to prevent the unnecessary use of vancomycin, which is needed only in case of MRSA isolates harboring SCCmec type II or III.
In summary, MRSA strains belonging to clonal complexes 5 and 8 were disseminated in the EMR and several “new” types were found: both ST225-MRSA-II and ST241-MRSA-III were novel findings in the German part of the EMR. Furthermore, one strain was classified as SCCmec type III, but contained the pls gene and the dcs region, and another strain was characterized as SCCmec type IV, but lacked the dcs region. One isolate harboring both SCCmec type V and PVL was found.
.
Acknowledgments
The study was partly performed within the framework of the Interreg-III project “Cross-Border Health Care in the Euregio Meuse Rhine.”
We thank H. de Lencastre and D. C. Oliveira from The Rockefeller University, New York, N.Y., for providing the reference strains for SCCmec typing; T. Ito from the Juntendo University, Tokyo, Japan, for providing the reference strain (WIS) for SCCmec V; and W. Wannet from the National Institute of Public Health and Environment (RIVM) in Bilthoven, The Netherlands, for the MRSA cluster 28 strain. We are grateful to Monique Coomans and Alexandra Heinzmann for excellent technical assistance with the MIC determinations, Peter Terporten for the statistical analysis, and Erik Beuken for his help with DNA sequencing.
REFERENCES
- 1.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40:101-111. [DOI] [PubMed] [Google Scholar]
- 3.Berger-Bächi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165-171. [DOI] [PubMed] [Google Scholar]
- 4.Chung, M., G. Dickinson, H. De Lencastre, and A. Tomasz. 2004. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J. Clin. Microbiol. 42:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, P. M. Giffard, and Australian Group for Antimicrobial Resistance. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 7.Denis, O., A. Deplano, C. Nonhoff, R. De Ryck, R. de Mendonca, S. Rottiers, R. Vanhoof, and M. J. Struelens. 2004. National surveillance of methicillin-resistant Staphylococcus aureus in Belgian hospitals indicates rapid diversification of epidemic clones. Antimicrob. Agents Chemother. 48:3625-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deurenberg, R. H., C. Vink, C. Driessen, M. Bes, N. London, J. Etienne, and E. E. Stobberingh. 2004. Rapid detection of Panton-Valentine leukocidin from clinical isolates of Staphylococcus aureus strains by real-time PCR. FEMS Microbiol. Lett. 240:225-228. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanssen, A.-M., G. Kjeldsen, and J. U. Ericson Sollid. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 13.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updates 6:41-52. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, L. B., N. Frimodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 17.Kotetishvili, M, O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. G. Morris, Jr. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay, J. A., and M. T. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 20.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 21.Mongkolrattanothai, K., S. Boyle, M. D. Kahana, and R. S. Daum. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 37:1050-1058. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing; ninth informational supplement M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Roth, E., F. Lorenzo-Diaz, N. Batista, A. Moreno, and S. Mendez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Predari, S. C., M. Ligozzi, and R. Fontana. 1991. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob. Agents Chemother. 35:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevost, G., P. Couppie, P. Prevost, S. Gayet, P. Petiau, B. Cribier, H. Monteil, and Y. Piemont. 1995. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J. Med. Microbiol. 42:237-245. [DOI] [PubMed] [Google Scholar]
- 28.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiemersma, E. W., S. L. A. M. Bronzwaer, O. Lyytikäinen, J. E. Degener, P. Schrijnemakers, N. Bruinsma, J. Monen, W. Witte, H. Grundmann, and European Antimicrobial Resistance Surveillance System participants. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg. Infect. Dis. 10:1627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Broek, P. J. 2003. Staphylococcus aureus, a successful pathogen. Ned. Tijdschr. Geneeskd. 147:1045-1048. (In Dutch.) [PubMed] [Google Scholar]
- 33.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wannet, W. J., E. Spalburg, M. E. Heck, G. N. Pluister, R. J. Willems, and A. J. De Neeling. 2004. Widespread dissemination in The Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J. Clin. Microbiol. 42:3077-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westh, H., D. M. Hougaard, J. Vuust, and V. T. Rosdahl. 1995. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob. Agents. Chemother. 39:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witte, W. 2004. International dissemination of antibiotic resistant strains of bacterial pathogens. Infect. Genet. Evol. 4:187-191. [DOI] [PubMed] [Google Scholar]