Abstract
We investigated the sequence and the genetic context of the erm(T) gene in six inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus (formerly S. bovis biotype II.2) isolates. In all isolates, the erm(T) genes were flanked by two IS1216V-like elements with the same polarity and were found to be inserted in the chromosome.
Streptococcus gallolyticus subsp. pasteurianus, formerly known as Streptococcus bovis biotype II.2 (10), can be a cause of endocarditis in elderly people and of septicemia and meningitis in newborns (1, 2, 14). We previously found that erm(T) was present in inducible erythromycin-resistant isolates of this species (12). In this study, we determined the sequences of erm(T) and its flanking regions from six inducible erythromycin-resistant S. gallolyticus subsp. pasteurianus isolates and investigated the genetic support of the erm(T)-containing elements.
Bacterial strains.
Six clinical isolates of erm(T)-positive, inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus were studied. The six isolates were from blood cultures and collected during the period 2000 to 2003 at the Bacteriology Laboratory, National Taiwan University Hospital, a 2,000-bed teaching hospital in northern Taiwan. One erythromycin-susceptible reference strain (ATCC 43144) of S. gallolyticus subsp. pasteurianus and two isolates of erm(T)-negative S. gallolyticus subsp. pasteurianus [one was erythromycin resistant due to the presence of an erm(B) gene, and the other was erythromycin susceptible] were used as negative controls. The isolates were identified by the API system (bioMérieux Vitek, France), and identification was confirmed by 16S rRNA gene sequences.
Nucleotide sequence of erm(T) and flanking regions in isolate NTUH-7421.
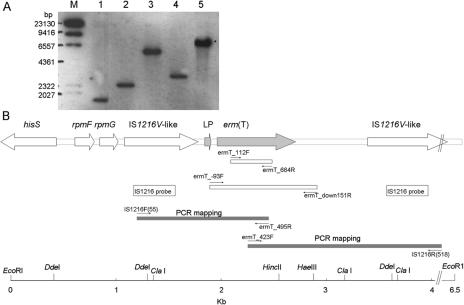
We have previously determined the partial erm(T) sequence in the erythromycin-resistant S. gallolyticus subsp. pasteurianus NTUH-7421 isolate (12). In this study, we determined the sequence of the entire erm(T) gene and its flanking regions by using a long accurate PCR in vitro cloning kit (Takara Shuzo Co. Ltd., Japan). The protocol had been described previously (13). Briefly, a Southern blot analysis (13) was performed with the DNA of NTUH-7421, which was digested with a panel of restriction enzymes and detected with a digoxigenin-labeled erm(T)-specific probe prepared by PCR amplification of erm(T) by using primers ermT_112F and ermT_684R (Table 1). Probe labeling and detection were carried out by using a commercial kit (Roche Diagnostics GmbH, Penzberg, Germany). This erm(T)-specific probe hybridized to a 6.5-kb EcoRI genomic fragment of NTUH-7421 (Fig. 1A). After ligating the EcoRI-digested DNA fragments with cassette adapters, the amplification was performed with cassette primers (C1 for the first PCR and C2 for the second PCR) supplied by the manufacturer and a target gene-specific primer, either ermT_112F or ermT_684R (Table 1). The PCR conditions for long accurate PCR were as described previously (13). Amplification fragments were subsequently sequenced on both strands by an Applied Biosystems Model ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.) with a Taq BigDye-Deoxy Terminator cycle sequencing kit (Applied Biosystems), according to the manufacturer's instructions.
TABLE 1.
PCR primers used in this study
| Description | Primer name | Sequence (5′ to 3′) | Target nucleotide position (nucleotide range) | Annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|---|
| erm(T) probe shown in Fig. 1 | ermT_112F | GGTTCAGGGAAAGGTCATTTCAC | erm(T) ∼112-134 | 52 | 573 |
| ermT_684R | GCTAATATTGTTAAAATCGTCAATTCC | erm(T) ∼684-658 | |||
| erm(T) probe shown in Fig. 3 and 4 | ermT_−93F | CAACACAGTTCATTATCAACC | erm(T) ∼−93-−73 | 57 | 978 |
| ermT_down151R | CATGGAAAGTAATTGCCG | erm(T)-down ∼134-151 | |||
| IS1216V probe | IS1216F(55) | CCGTGGGCTACTATCTTCGTT | IS1216V ∼56-76 | 52 | 487 |
| IS1216R(518) | AATTTATTGCGTCTCTTTACTGGA | IS1216V ∼542-519 | |||
| PCR detection of left IS1216V | IS1216F(55) | CCGTGGGCTACTATCTTCGTT | IS1216V ∼56-76 | 52 | 1,317 |
| ermT_495R | TGGATGAAAGTATTCTCTAGGG | erm(T) ∼495-474 | |||
| PCR detection of right IS1216V | ermT_423F | ACTAGCACTATTTTTAATGACAGAAGTTGA | erm(T) ∼423-452 | 52 | 1,601 |
| IS1216R(518) | AATTTATTGCGTCTCTTTACTGGA | IS1216V ∼542-519 |
FIG. 1.
Genetic organization of erm(T) resistance element and flanking regions in S. gallolyticus subsp. pasteurianus NTUH-7421. (A) Southern blot hybridization of erm(T) probe to S. gallolyticus subsp. pasteurianus NTUH-7421 genomic DNA after digestion with restriction enzymes ClaI, DdeI, HaeIII, HindII, and EcoRI (lanes 1 to 5, respectively). Lane M, DNA marker (digoxigenin-labeled DNA Molecular Weight Marker II′ [Roche]). (B) Genetic organization of erm(T) resistance element and flanking regions in S. gallolyticus subsp. pasteurianus NTUH-7421. Arrows represent putative open reading frames. The restriction sites are also shown.
By use of this strategy, the nucleotide sequence of a 4,107-bp fragment containing the entire erm(T) and its flanking regions was determined (Fig. 1B). The DNA sequence analysis revealed that the erm(T) gene of NTUH-7421 consists of 735 nucleotides, with a very low G+C content (25.4%), and is preceded by an AGGAG ribosome binding site consensus sequence and by a 60-nucleotide leader peptide-encoding gene sequence. The leader peptide-encoding gene sequence encodes a 19-amino-acid peptide, MGIFSIFVINTVHYQPNKK, which was 100% identical to that of Lactobacillus reuteri erm(T) (11) but differed from that of staphylococcal erm(C) (6) by one amino acid (Fig. 2). The erm(T) of NTUH-7421 had 99% identity with that of plasmid pGT633 from L. reuteri 100-63 or a tylosin-resistant Lactobacillus sp. (11, 15).
FIG. 2.
Alignment of the leader peptide-encoding sequences of erm(T) in S. gallolyticus subsp. pasteurianus NTUH-7421, erm(GT) in Lactobacillus species (GenBank accession number M64090), and staphylococcal erm(C) (GenBank accession number M17990). A dash indicates an identical nucleotide and a dot indicates a gap. TAA (stop codon) is underlined.
The inducible erythromycin-resistant phenotype in S. gallolyticus subsp. pasteurianus NTUH-7421 is probably dependent on the presence of an intact leader peptide sequence located immediately upstream to erm(T). This is supported by the finding of Tannock et al. that the erm(T) in the plasmid pGT633 of L. reuteri contained additional tandem duplication of 26-bp direct repeats, which may lead to constitutive expression of erythromycin resistance (11).
Two identical IS1216V-like elements were found on both sides of erm(T), with the same polarity (Fig. 1B). The IS1216V sequence from S. gallolyticus is nearly identical (with only one nucleotide difference) to that of a vancomycin-resistant Enterococcus faecium (3) (GenBank accession number L40841).
Upstream from the left IS1216V-like element, three open reading frames were detected (Fig. 1B). By comparing with the sequences in database, the best matches for the products of these open reading frames were the rpmF putative ribosomal protein L32 of Streptococcus pyogenes MGAS315 (GenBank accession number AE014172) (98% identity), the rpmG putative ribosomal protein L33 of S. pyogenes MGAS315 (GenBank accession number AE014172) (98% identity), and the HisS putative histidyl-tRNA synthetases of Streptococcus thermophilus CNRZ1066 (GenBank accession number CP000024) (95% sequence identity).
The region downstream of erm(T) of S. gallolyticus subsp. pasteurianus NTUH-7421 was very similar (93% nucleotide identity) to that of a broad-host-range plasmid containing erm(T) from a tylosin-resistant Lactobacillus sp. over a length of about 300 bp (15). The fragment after the 300-bp sequence and before the other IS1216V had no significant homology with known sequences.
Identification of erm(T) and IS1216V-like element in other clinical isolates.
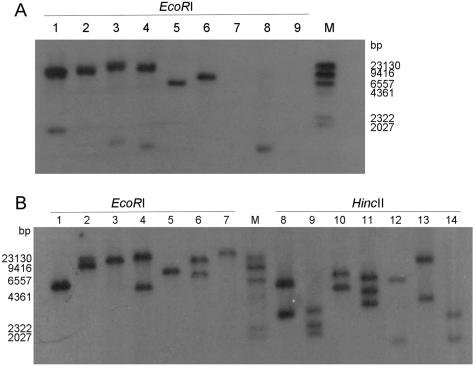
To determine whether IS1216V is present on sites other than the neighborhood of erm(T) and to determine whether the other five erm(T)-containing isolates contain similar structures, genomic DNAs were digested with EcoRI or HincII and hybridized to the erm(T)-specific probe (see above) or an IS1216V-specific probe obtained from NTUH-7421 by PCR with primers described in Table 1. The erm(T)- and IS1216V-specific probes apparently hybridized to the same EcoRI fragment (Fig. 3A, lane 1, and B, lane 1, respectively) in NTUH-7421, confirming the linkage between erm(T) and IS1216V in this isolate. Moreover, in this isolate, the IS1216V-specific probe hybridized to only two HincII fragments, indicating that IS1216V is present only in the neighborhood of erm(T). In the remaining five erm(T)-containing isolates, the erm(T)- and IS1216V-specific probes apparently hybridized to the same EcoRI fragments (Fig. 3). However, additional bands hybridizing to the IS1216V probe in erm(T)-positive and erm(T)-negative isolates were observed (Fig. 3B), indicating that it was not specific to erm(T).
FIG. 3.
Southern blot hybridization of erm(T) and IS1216V probe to S. gallolyticus subsp. pasteurianus strains. (A) Hybridization with an erm(T)-specific probe. Lanes 1 to 6 show erm(T)-positive isolates. Lane 1, NTUH-7421; lane 2, NTUH-8819; lane 3, NTUH-7499; lane 4, NTUH-3004; lane 5, NTUH-1043; lane 6, NTUH-4807; lanes 7 and 8, erm(T)-negative clinical isolates NTUH-1443 and NTUH-4046, respectively; lane 9, S. gallolyticus subsp. pasteurianus ATCC 43144; lane M, DNA marker (digoxigenin-labeled DNA Molecular Weight Marker II′ [Roche]). (B) Hybridization with IS1216V-specific probe. Lanes 1 to 6 and 8 to 13, erm(T)-positive isolates, as in lanes 1 to 6 in panel A. Lanes 7 and 14, erm(T)-negative clinical isolate NTUH-1443.
We further confirmed that the regions flanking the erm(T) gene were identical in all isolates by PCR mapping and sequencing, which were carried out with the primers listed in Table 1 and Fig. 1.
IS1216-like modules were found to be associated with antibiotic resistance determinants in enterococcal strains. Enterococcus hirae S185R was reported to have a plasmid-borne pbp3r gene linked to erm(AM), aadE, and the tnp of IS1216V (9). The IS1216V element is also part of the Tn1546-like elements in vancomycin-resistant enterococci (3, 16). Furthermore, IS1216V was proposed to mediate the horizontal spread of the vancomycin resistance transposon Tn5506 in E. faecium (4). IS1216V is a member of the ISS1 family, which includes elements known to be involved in cointegration and recombination processes in Lactococcus lactis (8). To our best knowledge, IS1216V in S. gallolyticus or related species has not been previously reported.
Genetic support of erm(T).
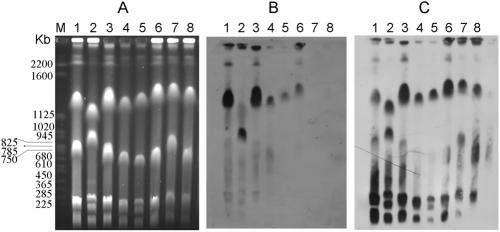
To determine the location of the erm(T) genetic element in S. gallolyticus, the DNAs of S. gallolyticus isolates were digested with I-CeuI (Fig. 4A) and then hybridized with either a 16S rRNA gene probe or the erm(T) probe (Fig. 4B and C). Chromosomal DNA from S. gallolyticus was prepared as described previously (7). Pulsed-field gel electrophoresis was performed at 200 V and 14°C with a CHEF-DRII apparatus (Bio-Rad Laboratories), with the pulse times ranging from 60 to 120 s for 24 h.
FIG. 4.
Localization of erm(T) on I-CeuI-generated chromosome fragments of S. gallolyticus subsp. pasteurianus isolates. (A) I-CeuI fragment restriction patterns separated by pulsed-field gel electrophoresis. (B) Hybridization with a probe specific for the erm(T) gene. (C) Hybridization with a 16S rRNA gene-specific probe. Lane M, molecular size standard (Saccharomyces cerevisiae chromosomal DNA). Lanes 1 to 6 show erm(T)-positive isolates. Lane 1, NTUH-7421; lane 2, NTUH-8819; lane 3, NTUH-7499; lane 4, NTUH-3004; lane 5, NTUH-1043; lane 6, NTUH-4807; lanes 7 and 8, erm(T)-negative clinical isolates NTUH-1443 and NTUH-4046, respectively.
In all isolates, the chromosomal bands recognized by the erm(T) probe (Fig. 4B) were also recognized by the 16S rRNA probe (Fig. 4C), revealing a chromosomal location of the erm(T) element in all isolates. The flanking hisS, rpmF, and rpmG-like sequences located upstream from the erm(T) element in strain NTUH-7421 further suggested a chromosomal location for the erm(T) gene in this isolate.
Concluding remarks.
To our best knowledge, erm(T) has been found only in Lactobacillus and the bovis group of streptococci (5, 11, 12, 15). The presence of erm(T) on S. gallolyticus subsp. pasteurianus is not only found in Taiwan. Lee et al. also reported similar rates of erm(T) in their isolates (5). The presence of erm(T) and IS1216V in S. gallolyticus subsp. pasteurianus suggests that genetic exchange might occur between S. gallolyticus and other gram-positive bacteria, such as Lactobacillus or Enterococcus.
Nucleotide sequence accession number.
The erm(T)-containing 4,107-bp nucleotide sequence from S. gallolyticus subsp. pasteurianus strain NTUH-7421 was deposited in GenBank under accession number AY894138.
Acknowledgments
This work was supported by grant NSC 93-2314-B-002-279 from the National Science Council of Taiwan.
REFERENCES
- 1.Clarridge, J. E., III, S. M. Attorri, Q. Zhang, and J. Bartell. 2001. 16S ribosomal DNA sequence analysis distinguishes biotypes of Streptococcus bovis: Streptococcus bovis biotype II/2 is a separate genospecies and the predominant clinical isolate in adult males. J. Clin. Microbiol. 39:1549-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant, R. J., T. R. Whitehead, and J. E. Orr. 2000. Streptococcus bovis meningitis in an infant. J. Clin. Microbiol. 38:462-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handwerger, S., and J. Skoble. 1995. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 39:2446-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heaton, M. P., L. F. Discotto, M. J. Pucci, and S. Handwerger. 1996. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 171:9-17. [DOI] [PubMed] [Google Scholar]
- 5.Lee, R. A., P. C. Y. Woo, A. P. C. To, S. K. P. Lau, S. S. Y. Wong, and K.-Y. Yuen. 2003. Geographical difference of disease association in Streptococcus bovis bacteraemia. J. Med. Microbiol. 52:903-908. [DOI] [PubMed] [Google Scholar]
- 6.Mayford, M., and B. Weisblum. 1990. The ermC leader peptide: amino acid alterations leading to differential efficiency of induction by macrolide-lincosamide-streptogramin B antibiotics. J. Bacteriol. 172:3772-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polzin, K. M., and M. Shimizu-Kadota. 1987. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J. Bacteriol. 169:5481-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raze, D., O. Dardenne, S. Hallut, M. Martinez-Bueno, J. Coyette, and J.-M. Ghuysen. 1998. The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob. Agents Chemother. 42:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlegel, L., F. Grimont, E. Ageron, P. A. D. Grimont, and A. Bouvet. 2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53:631-645. [DOI] [PubMed] [Google Scholar]
- 11.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 12.Teng, L. J., P. R. Hsueh, S. W. Ho, and K. T. Luh. 2001. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan Antimicrob. Agents Chemother. 45:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng, L. J., P. R. Hsueh, Y. H. Wang, H. M. Lin, K. T. Luh, and S. W. Ho. 2001. Determination of Enterococcus faecalis groESL full-length sequence and application for species identification. J. Clin. Microbiol. 39:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripodi, M. F., L. E. Adinolfi, E. Ragone, E. Durante Mangoni, R. Fortunato, D. Iarussi, G. Ruggiero, and R. Utili. 2004. Streptococcus bovis endocarditis and its association with chronic liver disease: an underestimated risk factor. Clin. Infect. Dis. 38:1394-1400. [DOI] [PubMed] [Google Scholar]
- 15.Whitehead, T. R., and M. A. Cotta. 2001. Sequence analyses of a broad host-range plasmid containing ermT from a tylosin-resistant Lactobacillus sp. isolated from swine feces. Curr. Microbiol. 43:17-20. [DOI] [PubMed] [Google Scholar]
- 16.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. A. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]