Abstract
The prevalence and mechanisms of macrolide resistance among 1,007 clinical pneumococcal isolates collected in Finland were investigated. Of these, 217 (21.5%) were resistant to erythromycin and 11% to clindamycin. Among the erythromycin-resistant isolates, mef(E) was present in 95 isolates (44%), mef(A) was present in 12 isolates (6%), and erm(B) was present in 90 isolates (41%). A double mechanism, mef(E) and erm(B), was detected in five isolates (2%). Ribosomal mutation was detected in 14 (6%) macrolide-resistant isolates in which no other determinant was found. Based on the telithromycin MICs, two groups of isolates were formed: 83.3% of the isolates belonged to a major group for which the telithromycin MIC range was ≤0.008 to 0.063 μg/ml, and 16.7% belonged to a minor group for which the telithromycin MIC range was 0.125 to 8 μg/ml. All except three isolates in the minor population carried a macrolide resistance gene.
Increasing resistance to macrolides among Streptococcus pneumoniae isolates is a worldwide problem. The proportion of resistant isolates ranges from 3 to 80% in different countries (2, 7, 20, 22, 23, 26, 33). Macrolide resistance is mediated by two main mechanisms in pneumococci: target site modification and drug efflux. The former is most often mediated by methylases encoded by the erm(B) gene, which is the most common methylase gene, or erm(A) [subclass erm(TR)], which is only infrequently found in pneumococci. Drug efflux is mediated by mef(A), which codes for an efflux pump (27). Two subtypes of mef efflux genes, mef(A) and mef(E), have been found in pneumococci (32, 40). These are variants of the same gene but are carried by different genetic elements (8, 36). An additional efflux mechanism, mediated by the msr(D) or the mel gene, has been found in genetic elements containing the mef gene (17, 38), but the significance of simultaneously carrying two efflux mechanisms is unknown. msr(D) and mel are homologues of the msr(A) gene found in staphylococci (38). Other possible mechanisms responsible for macrolide resistance in pneumococci include mutations in domain V or II of 23S rRNA or in genes coding for 50S ribosomal proteins L22 and L4 (27).
Telithromycin was the first ketolide introduced into clinical use. It is a semisynthetic derivative of erythromycin A composed of a 14-membered lactone ring, but the neutral sugar l-cladinose has been replaced by a keto group at position C-3. A C-11-C-12 carbamate side chain improves the affinity to ribosomes (1). According to present knowledge, telithromycin is effective against macrolide-resistant pneumococci, although some isolates may have elevated MICs to telithromycin (11, 12, 19, 24). Depending on the breakpoints and methods, the proportion of telithromycin nonsusceptibility has been reported to be 0.2% to 3.6% among macrolide-resistant pneumococci (11, 30).
The objectives of this study were to determine the prevalence of macrolide resistance in clinical isolates and the activity of telithromycin against clinical isolates and to investigate the molecular mechanisms of macrolide-resistant pneumococci.
(Preliminary results of this work have been presented at the 14th European Congress of Clinical Microbiology and Infectious Diseases, Prague, Czech Republic [P1475], and at the 4th International Symposium on Pneumococci and Pneumococcal Diseases, Helsinki, Finland [RES-40].)
MATERIALS AND METHODS
Pneumococcal isolates and susceptibility testing.
Pneumococal isolates (n = 1,007) were collected between May and December 2002 by a network of 24 Finnish Study Group for Antimicrobial Resistance (FiRe) laboratories, each of which was requested to send 50 consecutive pneumococcal isolates to the National Public Health Institute. Isolates were from both invasive sites (n = 129) and noninvasive sites (n = 878). The MICs for erythromycin, azithromycin, spiramycin, telithromycin, and clindamycin were determined by an agar plate dilution technique in a 5% CO2 atmosphere (35). Telithromycin was kindly provided by Sanofi Aventis (Romainville, France), while the other antimicrobials were purchased from their respective manufacturers. S. pneumoniae ATCC 49619 and Staphylococcus aureus ATCC 29213 were used as quality controls. CLSI (formerly NCCLS) breakpoints were used (31) for all antimicrobials except azithromycin, for which, due to the effect of the CO2 atmosphere, we used the following breakpoints: susceptibility, ≤1 mg/liter; intermediate, 2 mg/liter; and resistant, ≥4 mg/liter. Both intermediate and resistant isolates were taken into account when resistance percentages were calculated.
Detection of macrolide resistance mechanisms.
All erythromycin-resistant isolates (n = 217), 4 clindamycin-resistant isolates, and 41 randomly selected macrolide-susceptible isolates were investigated for the presence of the macrolide resistance genes mef(A/E), erm(B), and erm(TR) by a multiplex PCR method (16) with the primers described previously (16, 34, 39). Separate PCRs were run to differentiate efflux gene subclasses mef(A) and mef(E) in all mef-positive isolates, as well as to detect the presence of msr(D). The primers used for the detection of mef(A) and mef(E) have been described previously (5, 8). A modified primer pair was used for the detection of msr(D): 5′-CAGTTGGACGAAGTAACTCTG-3′ (forward primer) and 5′-CTCTTACGTTCTTCCTCTTTC-3′ (5). Testing for the detection of msr(D) was performed with 53 randomly selected isolates: 30 isolates with mef(E), 12 isolates with mef(A), 6 isolates with erm(B), and 5 susceptible isolates. The PCR run for mef(A), mef(E), and msr(D) included initial denaturation at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for mef(A) and msr(D) or 58°C for mef(E), and elongation at 72°C for 1 min. The magnesium concentration was 1.5 mM. All PCRs were run with a Whatman Biometra thermocycler (Biometra, Goettingen, Germany). Positive and negative controls were included in every run. Ribosomal mutations at positions 2058-2059 and 2611 of domain V of 23S rRNA (Escherichia coli numbering) and mutations in genes coding for 50S ribosomal proteins L4 and L22 were sought if the isolate was nonsusceptible to any of the antimicrobials tested and no known resistance gene was present. In addition, mutations were investigated in 13 randomly selected isolates: 7 with erm(B), 2 with mef(E), 2 with both erm(B) and mef(E), and 2 that showed a macrolide-susceptible phenotype. Mutations at positions 2058-2059 and 2611 of domain V of 23S rRNA were detected by a pyrosequencing technique (18, 37), and mutations in L4- and L22-coding genes were detected by sequencing (25, 42) with known primers (42). Primers for the detection of mutations at positions 2058 and 2059 have been described previously (18). The following primers were used for the detection of mutations at position 2611: for PCR, primers 5′-TGGGTTCAGAACGTCGTGAGA-3′ (forward primer) and 5′-GCGGTAAGTCCACTCTGGTC-3′ (reverse primer), and for pyrosequencing, primer 5′-CGTGAGACAGTTCGGTC-3′ (EMBL accession number AE0088386).
RESULTS
The prevalences of erythromycin, azithromycin, and clindamycin resistance were 21.5%, 22.3%, and 11.0%, respectively. The proportion of telithromycin-nonsusceptible isolates was not determined because of the lack of breakpoints for the method used here. Based on the telithromycin MICs, two groups of isolates were formed: a major group (83.3% of isolates) with an MIC range of ≤0.008 to 0.063 μg/ml and a minor group (16.7% of isolates) with an MIC in the range 0.125 to 8 μg/ml. All except three isolates in the minor group carried a macrolide resistance gene (Fig. 1). Of the 217 erythromycin-resistant isolates, 95 (44%) had mef(E), 12 (6%) had mef(A), and 90 (41%) had erm(B). Only one isolate had erm(TR). Five (2%) isolates carried both the erm(B) and the mef(E) genes. The erythromycin MICs in the mef(A)-positive isolates were higher than those in the mef(E) isolates (P = 0.002, Mann-Whitney U test). msr(D) was present in all mef-positive isolates tested but not in those with erm(B) or in susceptible isolates. Fourteen isolates (6%) in which no other mechanism was found had a mutation in domain V of 23S rRNA or in ribosomal protein L4 or L22 (Table 1). No macrolide resistance mechanisms were detected in susceptible isolates. The resistance mechanism remained unresolved in three isolates (Table 1). MIC data for isolates with different resistance mechanisms are summarized in Table 2.
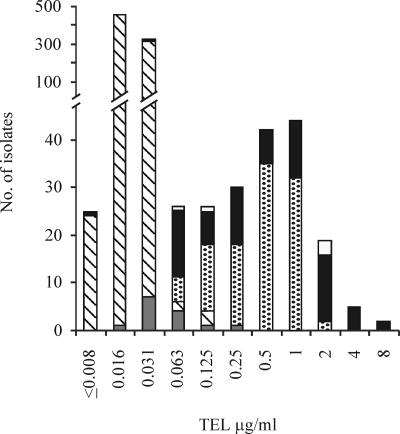
FIG. 1.
Distribution of telithromycin MICs of macrolide-susceptible and -resistant pneumococci harboring a macrolide resistance mechanism. Of the susceptible pneumococci, 41 isolates were randomly tested for the presence of a macrolide resistance determinant; none of them carried any macrolide resistance mechanism. Bars with slashes, isolates susceptible to erythromycin and azithromycin (n = 787); gray bars, isolates with mutations or undetermined mechanism (n = 17); spotted bars, isolates with mef(A) or mef(E) (n = 107); black bars, isolates with erm(B) or erm(TR) (n = 91); white bars, isolates with double mechanism erm(B) and mef(E) (n = 5).
TABLE 1.
Isolates with ribosomal mutations (including three isolates whose resistance mechanism remained unresolved) and their respective MICs to erythromycin, azithromycin, spiramycin, telithromycin, and clindamycin
| Isolate no. | Mutation in 23S rRNA gene, Domain V
|
Mutation in 50S ribosomal proteins
|
Resistance gene | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type and position | No. of mutated alleles total no. | L4 | L22 | ERY | AZM | SPI | TEL | CLI | ||
| 45 | A2059G | 1/4 | Wild | Wild | 32 | >128 | >128 | 0.031 | 2 | |
| 560 | A2059G | 1/4 | E30→K | Wild | 8 | >128 | 2 | 0.031 | 0.5 | |
| 561 | A2059G | 1/4 | E30→K | Wild | 32 | >128 | 128 | 0.031 | 1 | |
| 588 | A2059G | 2/4 | T94→I | Wild | 32 | >128 | >128 | 0.016 | 1 | |
| 904 | A2059G | 3/4 | Wild | Wild | 128 | >128 | >128 | 0.063 | 2 | |
| 1166 | A2059G | 4/4 | Wild | Wild | >128 | >128 | >128 | 0.031 | 4 | |
| 5 | C2611T | 4/4 | Wild | Wild | 0.25 | 8 | 4 | 0.063 | 4 | |
| 152 | C2611T | 4/4 | V205→G | A101→P | 0.125 | 4 | 2 | 0.031 | 2 | |
| 522 | C2611T | 4/4 | Wild | Wild | 0.125 | 8 | 4 | 0.031 | 2 | |
| 48 | Wild | 68E69 insertion | Wild | 1 | 4 | 4 | 0.031 | 0.25 | ||
| 438 | Wild | 68GQK69 insertion | Wild | 1 | 8 | 32 | 0.25 | 0.063 | ||
| 156 | Wild | S20→N | R22→C | 8 | 32 | 0.5 | 0.125 | 0.125 | ||
| 551 | Wild | S20→N | Wild | 2 | 64 | 32 | 0.063 | >128 | ||
| 545 | Wild | S20→N | Wild | mef(E) | 32 | 128 | 0.5 | 2 | 0.125 | |
| 837 | Wild | S20→N | Wild | mef(E) + erm(B) | >128 | >128 | >128 | 2 | >128 | |
| 843 | Wild | E30→K | Wild | >128 | >128 | >128 | 0.063 | >128 | ||
| 354 | Wild | Wild | Wild | 0.125 | 8 | 0.5 | 0.031 | 0.125 | ||
| 695 | Wild | Wild | Wild | 1 | 4 | 2 | 0.031 | 0.5 | ||
| 965 | Wild | Wild | Wild | 2 | 4 | 1 | 0.031 | 0.25 | ||
ERY, erythromycin; AZM, azithromycin; SPI, spiramycin; TEL, telithromycin; CLI, clindamycin.
TABLE 2.
Macrolide resistance mechanisms and MIC data for erythromycin, azithromycin, spiramycin, telithromycin, and clindamycin
| Mechanism (no. of isolates) | MIC (μg/ml)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Erythromycin
|
Azithromycin
|
Spiramycin
|
Telithromycin
|
Clindamycin
|
|||||||||||
| Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | |
| mef(E) (95) | 0.5->128 | 8 | 64 | 1->128 | 32 | 128 | 0.125-1 | 1 | 1 | 0.031-2 | 0.5 | 1 | 0.031-0.25 | 0.125 | 0.25 |
| mef(A) (12) | 16-64 | 32 | 64 | 32->128 | 32 | 128 | 1 | 1 | 1 | 0.125-0.5 | 0.5 | 0.5 | 0.125-0.25 | 0.125 | 0.125 |
| erm(B) (90) | 2->128 | >128 | >128 | 2->128 | >128 | >128 | 1->128 | >128 | >128 | 0.008-8 | 0.25 | 2 | 0.125->128 | >128 | >128 |
| erm(B) + mef(E) (5) | >128 | >128 | >128 | 0.063-2 | 2 | >128 | |||||||||
| erm(A) (1) | >128 | >128 | >128 | 0.25 | >128 | ||||||||||
| Mutation only (14)a | 0.25->128 | 8 | >128 | 4->128 | 32 | >128 | 0.5->128 | 32 | >128 | 0.016-0.25 | 0.031 | 0.125 | 0.063->128 | 2 | >128 |
The individual genotypes and phenotypes of mutated isolates are presented in Table 1.
DISCUSSION
The prevalence of macrolide resistance among pneumococci in Finland doubled between 1999 and 2002 (www.ktl.fi/extras/fire). Currently, there are no signs that this worrying trend is slowing, despite recommendations to avoid the overuse of macrolides, and more effective measures such as encouraging the use of vaccines should be considered.
In this study we used a 5% CO2 supplement to confirm the proper growth of resistant isolates (15). The CO2 supplement may elevate macrolide, ketolide, and clindamycin MICs (6, 15). Despite the CO2 supplement, the results of this study can be considered reliable since 99% of the isolates with erythromycin MICs ≥0.05 μg/ml harbored a macrolide resistance determinant or had a mutation, thus reflecting the resistance category and genotype well. Moreover, none of the susceptible isolates carried macrolide resistance genes or mutations.
Two groups of pneumococci were formed on the basis of telithromycin MICs: a highly susceptible major group and a minor group of isolates in which the presence of macrolide resistance genes was associated with elevated telithromycin MICs. Nevertheless, the MICs were not constant among isolates with the same macrolide resistance determinant. This was especially true for erm(B)-positive isolates. It is not yet clear why some isolates carrying the same macrolide resistance determinant are fully susceptible to telithromycin but others are not. It may also be possible that true telithromycin resistance in pneumococci evolves in macrolide-resistant isolates that have a moderately elevated MIC to telithromycin.
The proportions of different macrolide resistance determinants recorded here were similar to those from a previous Finnish study on invasive pneumococci (33) and resemble those in North America and Scotland, where the efflux mechanism is the most prevalent (2, 13, 21). This is in contrast to the situation in Europe, where erm(B) dominates (7, 21, 29, 30). There have recently been reports of pneumococci carrying a double mechanism, both erm(B) and mef(E) (3, 14, 28). The spread of similar strains is considered of great concern, since they are often multiresistant and are clonally related (14, 28). In a recent report on the global situation, the prevalence of isolates having both the erm(B) and the mef(E) genes was 7% (14). In our study, only 2% of isolates carried a double mechanism; and in those isolates, the mef(E) subtype was always present together with erm(B).
mef(A) and mef(E) have 90% similarity at the nucleotide level and are considered variants of the same gene, mef(A) (36); but because they are carried in different genetic elements in pneumococci, they should be differentiated (8). In addition, there are epidemiological and phenotypic differences between these subtypes (2, 8, 17). For instance, it has been reported that mef(A) isolates have higher MICs to erythromycin than mef(E) isolates (2). A similar observation was recorded in this study. mef(E) is the prevailing efflux gene subtype in the United States, Asia, and South Africa (5) and, according to this study, also in Finland. mef(A) has been more frequently reported in other parts of Europe (2, 5, 29, 32).
The proportion of isolates with mutations in this study was relatively high (6%) compared to that indicated in a recent report on the global prevalence (10). The most frequent mutation in our study was an A2059G change in domain V of 23S rRNA, which has been reported to be one of the most common mutations in pneumococci (9, 10). An A2059G transition leads to modification at the erythromycin binding site, which causes resistance to 14-, 15-, and 16-membered macrolides and elevated MICs to clindamycin but not to telithromycin (12, 41, 42). Position C2611 is another common site where mutations have been found in the pneumococcus (10, 42). The C2611U mutation has been described in laboratory strains of pneumococci obtained after serial passage on azithromycin or clindamycin (4), and it was only recently found in clinical isolates of S. pneumoniae (9). In our study, the isolates with this mutation shared a similar phenotype, being susceptible to erythromycin but not to azithromycin or clindamycin. The MICs to these agents were only slightly elevated, however.
Six new mutations that, to the best of our knowledge, have not previously been described were found in this study. Two of these mutations (68E69 and 68GQK69 insertions) were located in the highly conserved region 63LPWRQKGTGRAR74 of the L4 protein, where mutations conferring macrolide resistance have been described previously (9, 35, 42). The possible role of these mutations, as well as the role of other new mutations (T94I and V205G in L4 or R22C and A101P in L22), in conferring macrolide resistance awaits experimental confirmation.
In conclusion, the level of erythromycin resistance is increasing in Finland. The dominant macrolide resistance mechanism is an efflux mechanism caused by either mef(E) or mef(A). Although telithromycin has good activity against pneumococci, the significance of macrolide-resistant isolates having an elevated MIC to telithromycin should be further investigated.
Acknowledgments
This work was supported by grant 73351 from the Academy of Finland. We thank Helena Seppälä for valuable comments in preparing the manuscript and Anna-Liisa Lumiaho, Erkki Nieminen, Anne Nurmi, and Tuula Randell for excellent technical assistance. We thank Sanofi Aventis for providing telithromycin.
The members of the Finnish Study Group for Antimicrobial Resistance in 2002 were as follows: Anja Kostiala Thompson and Merja Rautio (Jorvi Hospital, Espoo); Risto Renkonen and Anna Muotiala (MedixDiacor Laboratory Service, Espoo); Martti Vaara and Petteri Carlson (Helsinki University Central Hospital, Helsinki); Hannele Somer (Mehiläinen Hospital, Helsinki); Anni Virolainen-Julkunen (Yhtyneet Laboratoriot Oy, Helsinki); Jukka Korpela and Ritva Heikkilä (Central Hospital of Kanta-Häme, Hämeenlinna); Suvi-Sirkku Kaukoranta and Heikki Kaukoranta (Central Hospital of North-Karelia, Joensuu); Antti Nissinen (Central Hospital of Keski-Suomi, Jyväskylä); Pekka Ruuska (Central Hospital of Kainuu, Kajaani); Henrik Jägerroos (Central Hospital of Lapland, Rovaniemi); Martti Larikka (Central Hospital of Länsi-Pohja, Kemi); Simo Räisänen (Central Ostrobothnian Hospital District, Kokkola); Ulla Larinkari (Central Hospital of Kymenlaakso, Kotka); Marja-Leena Katila and Ulla Kärkkäinen (Kuopio University Hospital, Kuopio); Hannu Sarkkinen and Pauliina Kärpänojan (Central Hospital of Päijät-Häme, Lahti); Maritta Kauppinen and Seppo Paltemaa (Central Hospital of South-Karelia, Lappeenranta); Päivi Kärkkäinen (Mikkeli Central Hospital, Mikkeli; Savonlinna Central Hospital, Savonlinna); Ilmo Pietarinen (Deaconess Institution in Oulu, Oulu); Markku Koskela (Oulu University Hospital, Oulu); Sini Pajarre (Central Hospital of Satakunta, Pori); Sinikka Oinonen and Virpi Ratia (Central Hospital of Seinäjoki, Seinäjoki); Paul Grönroos (Koskiklinikka, Tampere); Risto Vuento and Oili Liimatainen (Tampere University Hospital, Tampere); Maj-Rita Siro (Health Center Pulssi, Turku); Erkki Eerola and Raija Manninen (University of Turku, Turku); Olli Meurman (Turku University Central Hospital, Turku); Marko Luhtala (Central Hospital of Vaasa, Vaasa); Pentti Huovinen and Katrina Lager (National Public Health Institute, Turku).
REFERENCES
- 1.Ackermann, G., and A. C. Rodloff. 2003. Drugs of the 21st century: telithromycin (HMR 3647)—the first ketolide. J. Antimicrob. Chemother. 51:497-511. [DOI] [PubMed] [Google Scholar]
- 2.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean, D. C., and J. D. Klena. 2002. Prevalence of erm(A) and mef(B) erythromycin resistance determinants in isolates of Streptococcus pneumoniae from New Zealand. J. Antimicrob. Chemother. 50:597-599. [DOI] [PubMed] [Google Scholar]
- 4.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, M. M., S. Doktor, R. Flamm, and D. Shortridge. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J. Clin. Microbiol. 42:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, T. A., L. M. Kelly, M. R. Jacobs, and P. C. Appelbaum. 2000. Antipneumococcal activity of telithromycin by agar dilution, microdilution, E test, and disk diffusion methodologies. J. Clin. Microbiol. 38:1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decousser, J. W., P. Pina, F. Viguier, F. Picot, P. Courvalin, and P. Allouch. 2004. Invasive Streptococcus pneumoniae in France: antimicrobial resistance, serotype, and molecular epidemiology findings from a monthly national study in 2000 to 2002. Antimicrob. Agents Chemother. 48:3636-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doktor, S. Z., V. D. Shortridge, J. M. Beyer, and R. K. Flamm. 2004. Epidemiology of macrolide and/or lincosamide resistant Streptococcus pneumoniae clinical isolates with ribosomal mutations. Diagn. Microbiol. Infect. Dis. 49:47-52. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, D. J., S. Douthwaite, I. Morrissey, S. Bakker, J. Poehlsgaard, L. Jakobsen, and D. Felmingham. 2003. Macrolide resistance by ribosomal mutation in clinical isolates of Streptococcus pneumoniae from the PROTEKT 1999-2000 study. Antimicrob. Agents Chemother. 47:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, D. J., and D. Felmingham. 2004. Activities of telithromycin against 13,874 Streptococcus pneumoniae isolates collected between 1999 and 2003. Antimicrob. Agents Chemother. 48:1882-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell, D. J., I. Morrissey, S. Bakker, and D. Felmingham. 2002. Molecular characterization of macrolide resistance mechanisms among Streptococcus pneumoniae and Streptococcus pyogenes isolated from the PROTEKT 1999-2000 study. J. Antimicrob. Chemother. 50(Suppl. S1):39-47. [DOI] [PubMed] [Google Scholar]
- 14.Farrell, D. J., I. Morrissey, S. Bakker, L. Morris, S. Buckridge, and D. Felmingham. 2004. Molecular epidemiology of multiresistant Streptococcus pneumoniae with both erm(B)- and mef(A)-mediated macrolide resistance. J. Clin. Microbiol. 42:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fasola, E. L., S. Bajaksouzian, P. C. Appelbaum, and M. R. Jacobs. 1997. Variation in erythromycin and clindamycin susceptibilities of Streptococcus pneumoniae by four test methods. Antimicrob. Agents Chemother. 41:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueira-Coelho, J., M. Ramirez, M. J. Salgado, and J. Melo-Cristino. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug Resist. 10:31-36. [DOI] [PubMed] [Google Scholar]
- 17.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 18.Haanpera, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton-Miller, J. M., and S. Shah. 2002. Activity of ketolide ABT-773 (cethromycin) against erythromycin-resistant Streptococcus pneumoniae: correlation with extended MLSK phenotypes. J. Antimicrob. Chemother. 50:907-913. [DOI] [PubMed] [Google Scholar]
- 20.Hoban, D., K. Waites, and D. Felmingham. 2003. Antimicrobial susceptibility of community-acquired respiratory tract pathogens in North America in 1999-2000: findings of the PROTEKT surveillance study. Diagn. Microbiol. Infect. Dis. 45:251-259. [DOI] [PubMed] [Google Scholar]
- 21.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibilities to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyde, T. B., K. Gay, D. S. Stephens, D. J. Vugia, M. Pass, S. Johnson, N. L. Barrett, W. Schaffner, P. R. Cieslak, P. S. Maupin, E. R. Zell, J. H. Jorgensen, R. R. Facklam, and C. G. Whitney. 2001. Macrolide resistance among invasive Streptococcus pneumoniae isolates. JAMA 286:1857-1862. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 24.Jalava, J., J. Kataja, H. Seppala, and P. Huovinen. 2001. In vitro activities of the novel ketolide telithromycin (HMR 3647) against erythromycin-resistant Streptococcus species. Antimicrob. Agents Chemother. 45:789-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jalava, J., M. Vaara, and P. Huovinen. 2004. Mutation at the position 2058 of the 23S rRNA as a cause of macrolide resistance in Streptococcus pyogenes. Ann. Clin. Microbiol. Antimicrob. 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlov, R. S., T. M. Bogdanovitch, P. C. Appelbaum, L. Ednie, L. S. Stratchounski, M. R. Jacobs, and B. Bozdogan. 2002. Antistreptococcal activity of telithromycin compared with seven other drugs in relation to macrolide resistance mechanisms in Russia. Antimicrob. Agents Chemother. 46:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGee, L., K. P. Klugman, A. Wasas, T. Capper, and A. Brink. 2001. Serotype 19f multiresistant pneumococcal clone harboring two erythromycin resistance determinants (erm(B) and mef(A)) in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morosini, M. I., R. Canton, E. Loza, M. C. Negri, J. C. Galan, F. Almaraz, and F. Baquero. 2001. In vitro activity of telithromycin against Spanish Streptococcus pneumoniae isolates with characterized macrolide resistance mechanisms. Antimicrob. Agents Chemother. 45:2427-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing; 14th informational supplement. NCCLS document M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.Oster, P., A. Zanchi, S. Cresti, M. Lattanzi, F. Montagnani, C. Cellesi, and G. M. Rossolini. 1999. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob. Agents Chemother. 43:2510-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pihlajamaki, M., J. Jalava, P. Huovinen, and P. Kotilainen. 2003. Antimicrobial resistance of invasive pneumococci in Finland in 1999-2000. Antimicrob. Agents Chemother. 47:1832-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pihlajamaki, M., T. Kaijalainen, P. Huovinen, and J. Jalava. 2002. Rapid increase in macrolide resistance among penicillin non-susceptible pneumococci in Finland, 1996-2000. J. Antimicrob. Chemother. 49:785-792. [DOI] [PubMed] [Google Scholar]
- 35.Pihlajamaki, M., J. Kataja, H. Seppala, J. Elliot, M. Leinonen, P. Huovinen, and J. Jalava. 2002. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 46:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363-365. [DOI] [PubMed] [Google Scholar]
- 38.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]