Abstract
In enterococci, intrinsic low-level resistance to gentamicin does not abolish synergism with a cell wall-active antibiotic while high-level resistance due to acquired aminoglycoside-modifying enzymes does. To study the impact of intermediate levels of resistance to gentamicin (64 < MIC < 500 μg/ml), we selected in vitro three consecutive generations of mutants of Enterococcus faecalis JH2-2 with MICs of gentamicin at 128 μg/ml for G1-1477, 256 μg/ml for G2-1573, and 512 μg/ml for G3-1688. E. faecalis 102, which is highly resistant to gentamicin by enzymatic inactivation was used as control. In in vitro killing curves experiments, gentamicin concentrations allowing bactericidal activity and synergism in combination with amoxicillin increased from 4 μg/ml (1/16th the MIC), 16 μg/ml (one-eighth the MIC), 64 μg/ml (one-quarter the MIC), and 256 μg/ml (one-half the MIC) for strains JH2-2, G1-1477, G2-1573 and G3-1688, respectively. As expected, no bactericidal effect of the combination or synergism could be obtained with strain 102. In rabbits with aortic endocarditis caused by strain G1-1477 or G2-1573, combination therapy with amoxicillin and gentamicin was significantly more active than amoxicillin alone (P < 0.05) but not in those infected with the strains G3-1688 and 102. Thus, intermediate levels of resistance to gentamicin was not associated with a loss of a beneficial effect of the gentamicin-amoxicillin combination in vivo even though higher concentrations of gentamicin were necessary to achieve in vitro synergism. Therefore, the use of an MIC of 500 μg/ml as a clinical cutoff limit to predict in vivo benefit of the combination remains a simple and effective tool.
Optimal antimicrobial therapy for serious enterococcal infections requires the use of synergistic bactericidal combinations of a cell wall-active agent, such as penicillin or a glycopeptide, with an aminoglycoside. Enterococcus faecalis, the most frequently isolated enterococcal species, is intrinsically resistant to low levels of aminoglycosides due to inefficient active transport of aminoglycosides across the cytoplasmic membrane. However, a bactericidal effect is observed when amoxicillin is combined with gentamicin even on strains with MICs of gentamicin ranging from 4 to 64 μg/ml (18). By contrast, no synergistic bactericidal effect is observed in combining a cell wall-active antibiotic with gentamicin on strains with high-level resistance to aminoglycosides (MIC > 500 μg/ml) such as those which have acquired genes encoding aminoglycoside-modifying enzymes by horizontal transfer (19).
However, Dressel at al (9). have already reported the lack of synergism between amoxicillin and gentamicin in three clinical isolates of enterococci, despite gentamicin MICs below 500 μg/ml (8 and 16 μg/ml). The aminoglycoside resistance mechanism was not described in the report. Chow et al. (4, 5) also described E. faecalis harboring a novel gentamicin resistance gene, aph(2")-Ic, which compromised ampicillin-gentamicin synergism despite a gentamicin MIC of 256 μg/ml.
We previously reported the emergence of E. faecalis mutants with intermediate levels of resistance to gentamicin, defined as MICs ranging from 64 to 500 μg/ml, both in vitro and in rabbits with experimental endocarditis during treatments containing gentamicin (1, 16). To our knowledge, the impact of the intermediate level of resistance to gentamicin on the gentamicin-amoxicillin synergism is poorly described in vitro and it has never been studied in vivo. In order to investigate this point, we selected in vitro E. faecalis mutants with intermediate levels of gentamicin resistance and we studied the synergistic effect of amoxicillin and gentamicin on these mutants, in vitro and in experimental endocarditis.
MATERIALS AND METHODS
Bacterial strains and media.
E. faecalis JH2-2 is susceptible to amoxicillin and has low-level resistance to gentamicin (MIC, 32 to 64 μg/ml) (13). In vitro, three successive step mutants resistant to gentamicin were selected on brain heart infusion (BHI) (Difco, Serlabo, Bonneuil sur Marne, France) agar containing gentamicin in concentrations twice the MIC of the parent strain. Frequencies of isolation of mutants were approximately 10−5, 10−8, and 10−9 for the first, second and third steps, respectively.
At each step, one colony was selected at random and named G1-1477, G2-1573, and G3-1688, respectively. E. faecalis 102, which is highly resistant to gentamicin (MIC > 2,000 μg/ml) by production of a bifunctional enzyme, 6′-N-aminoglycoside acetyltransferase-2"-O-aminoglycoside phosphotransferase [AAC-(6′)-APH(2"]) was used as the control in experimental endocarditis (7). Escherichia coli DH5a(pAM6306) (5), E. coli NC95 (23), E. coli KHE5-2 (15), and E. faecium JH7 (8) were also used to detect genes encoding aminoglycoside-modifying enzymes (see below). Brain heart infusion broth and agar were used in all experiments. All incubations were at 37°C.
Determination of MICs.
MICs were determined by the agar dilution method (20, 21). Serial twofold dilutions of antibiotic were added in brain-heart infusion agar (Difco, Serlabo, Bonneuil sur Marne, France) in order to obtain the following final concentrations of gentamicin ranged from 4 to 2048 mg/ml. Inocula of each E. faecalis strain grew overnight in brain-heart infusion broth at 37°C. Plates were inoculated with a Steers replicator device delivering approximately 104 to 105 CFU per spot (10 μl) and were incubated at 37°C for 24 h. Assays were performed at least in duplicate. The MIC was defined as the lowest antibiotic concentration resulting in complete inhibition of visible growth. The stability of mutant was investigated in vitro. Each strain was incubated overnight in BHI broth at 37°C, then plated on BHI agar and incubated for 24 h. Thereafter, one CFU was resuspended in 10 ml of BHI broth, and 0.1 ml of the overnight culture was plated on BHI agar. This sequence was reported two times. Gentamicin MIC was determined for 5 random colonies of the last plate.
Test for synergy.
Killing curve analysis was used to study bactericidal synergy between gentamicin and amoxicillin over time in vitro. Exponentially growing E. faecalis was diluted in glass tubes containing 10 ml of BHI broth (pH 7.0; Difco) to obtain a starting inoculum of 107 CFU/ml, as previously described (16). Amoxicillin concentration was one-half the MIC of the study strain in all experiments, and gentamicin concentrations ranged from one-half to 1/16th the MIC in series of experiments performed on each of the strains tested. To count the surviving bacteria, aliquots of 0.1 ml were taken after 0, 3, 6, and 24 h of incubation and were diluted in water from 10−2 to 10−8 depending of the inoculum load. Then, 0.1 ml of the undiluted and diluted inocula was plated on BHI agar and incubated at 37°C for 24 h (20). The minimal accurately countable number of CFU per milliliter was 1 log10.
Synergism was defined as a ≥2-log10 decrease in CFU/ml between the combination of amoxicillin plus gentamicin and amoxicillin alone, the most active constituent, after 24 h of incubation. A bactericidal effect was defined as a ≥3-log10 decrease in CFU/ml between the initial counts and those observed after 24 h of incubation. Drug carryover effect was insignificant in our experiments because cultures for killing curve tests were diluted from minimum 10-fold minimum to 10−8-fold before count of surviving bacteria.
DNA amplification of genes encoding aminoglycoside-modifying enzymes.
DNA extracted from each strain tested using a Magna Pure LC (Roche, Mannheim, Germany). Genes aph(2")-Ib, aph(2")-Ic, aph(2")-Id; aadE, and aac(6′)-aph(2") encoding aminoglycoside-modifying enzymes were detected by amplification in a I-cycler (Bio-Rad, Marnes-la-coquette, France) using specific primers, as described (5, 6, 15).
PCR products were electrophoresed in agarose gels (2%, W/v) containing ethidium bromide (0.5 μg ml−1) and visualized under UV irradiation.
Experimental endocarditis.
Aortic endocarditis was induced in female New Zealand White rabbits (2.5 to 3 kg) by insertion of a polyethylene catheter through the right carotid artery into the left ventricle, as previously described (1, 16). Twenty-four hours after catheter insertion, rabbits were inoculated by the ear vein with 107 CFU of E. faecalis. The catheter was left in place throughout the experiment. Forty-eight hours after inoculation, animals received amoxicillin (50 mg/kg three times a day) or gentamicin (2 mg/kg three times a day) or both, intramuscularly for 3 days. Control animals were sacrificed 48 h after inoculation (start of therapy). Treated animals were sacrificed by an intravenous injection of pentobarbital 8 h after the last antibiotic injection. At the time of sacrifice, the heart was removed and the left side chambers were examined, vegetations were excised, pooled for each rabbit, weighed, and homogenized in 1 ml of sterile distilled water. Living bacteria were counted on agar. The results were expressed as log10 CFU per gram of vegetation. The difference of bacterial concentrations in vegetations between animals treated with amoxicillin alone or plus gentamicin (ΔCFU) was calculated for each strain to evaluate the benefit of antibiotic association.
Antibiotic plasma concentration.
Serum peak and trough antibiotic concentrations were determined in the ear venous blood in non infected rabbits after a single i.m. gentamicin (2 mg/kg) or amoxicillin (50 mg/kg) injection 0.5 and 8 h after antibiotic injection by fluorescence polarization immunoassay (detection limit of 0.3 mg/liter) (AxSYM System; Abbott Diagnostics, Rungis, France) and by high performance liquid chromatography (detection limit of 0.2 mg/liter), respectively (14).
Statistics.
In order to investigate a benefit of the amoxicillin/gentamicin combination versus amoxicillin alone, we compared for each strain the efficacy of amoxicillin alone, the most effective single-drug regimen, to the amoxicillin/gentamicin combination.
Bacterial counts in vegetations are expressed as means ± standard deviation. The statistical significance of differences in vegetation titers was determined by analysis of variance followed by Fisher's test for multiple comparison (12).
RESULTS
MICs.
The MIC of amoxicillin was 0.5 μg/ml for all strains. The MICs of gentamicin were 64, 128, 256, 512, and >2,000 μg/ml against JH2-2, G1-1477, G2-1573, G3-1688, and E. faecalis 102, respectively. All mutants were stable after four subcultures on BHI agar.
In vitro synergy between amoxicillin and gentamicin.
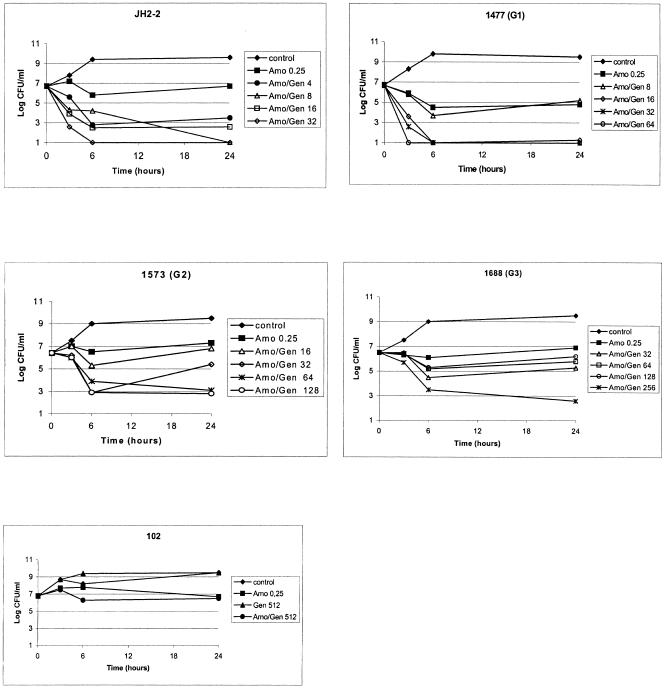
Gentamicin alone at one-half, one-quarter, one-eighth, and 1/16th the MIC for all E. faecalis strains was ineffective (data not shown). Amoxicillin alone at a concentration of one-half the MIC was not bactericidal against any of the strains studied (Fig. 1). The amoxicillin concentration used was 0.25 μg/ml (one-half the MIC) because no synergism with gentamicin was detectable with an amoxicillin concentration above or equal to the MIC (0.5 μg/ml) (data not shown). The amoxicillin and gentamicin combination was bactericidal and synergistic against JH2-2 even at low gentamicin concentrations (4 μg/ml: 1/16th the MIC). The gentamicin concentrations required to observed bactericidal activity and synergism in combination with amoxicillin increased from 1/16th to one-eighth, one-quarter, and one-half of the MIC for strains JH2-2, G1-1477, G2-1573 and G3-1688, respectively. For each strain, the same gentamicin concentration was required to obtain synergism and bactericidal activity combined with amoxicillin (Fig. 1 and Table 1). As expected, no bactericidal effect and no synergism were observed with the amoxicillin-gentamicin combination against strain E. faecalis 102 (Fig. 1e).
FIG. 1.
Time-kill curves for amoxicillin (Amo) at one-half the MIC and gentamicin (Gen) at one-half, one-quarter, one-eighth, and 1/16th the MIC against strains of Enterococcus faecalis with increased levels of resistance to gentamicin.
TABLE 1.
Gentamicin concentrations required to achieve synergy and bactericidal activity in combination with amoxicillin against E. faecalis strains with increased levels of resistance to gentamicin
| Therapeutic level of gentamicina | Gentamicin concn (μg/ml) required [fraction of MIC]
|
|||
|---|---|---|---|---|
| JH2-2 | 1477 (G1) | 1573 (G2) | 1688 (G3) | |
| MIC | 64 | 128 | 256 | 512 |
| Concn for synergy | 4 [1/16] | 16 [1/8] | 64 [1/4] | 256 [1/2] |
| Concn for bactericidal activity | 4 [1/16] | 16 [1/8] | 64 [1/4] | 256 [1/2] |
Synergy was defined as a 2 log10 CFU/ml reduction of the amoxicillin/gentamicin combination in comparison with amoxicillin alone after 24 h of incubation. Bactericidal activity was defined as a 3 log10 CFU/ml reduction of the amoxicillin/gentamicin combination after 24 h of incubation. Amoxicillin was used at a concentration of 0.25 μg/ml (0.5 times the MIC). Gentamicin concentrations are expressed as absolute values and as a fraction of the gentamicin MIC.
DNA amplification of genes encoding aminoglycoside-modifying enzymes.
The five genes aph(2")Ib, aph(2")-Ic, aph(2")-Id, aadE, and aac(6′)-aph(2") encoding all known aminoglycoside-modifying enzymes associated with gentamicin resistance in enterococci, were not detected in the G1-1477, G2-1573, and G3-1688 mutants (data not shown).
Experimental endocarditis.
Peak and trough serum levels were 28 ± 10 μg/ml and 0.32 ± 0.20 μg/ml, respectively, for amoxicillin and 4.65 ± 0.33 μg/ml and <0.2 μg/ml, respectively, for gentamicin. Values are the mean ± standard deviation of dosages in six and five rabbits for the amoxicillin and gentamicin regimens, respectively.
As shown in Table 2, no significant differences were observed between bacterial counts in the vegetations of control rabbits infected with either of the four strains studied, suggesting the absence of impact on virulence, at least in this model. Amoxicillin was significantly more effective in rabbits infected by strain JH2-2 than in those infected with any other strain (P < 0.05). Gentamicin alone was not effective against any strains in comparison with controls. The bacterial reduction obtained with the combination of amoxicillin plus gentamicin compared with amoxicillin alone varied from 0 to 1.78 log10 CFU/g according to the study strain (P< 0.05) (Table 2). The activity of the combination was significantly different from that of amoxicillin alone for the two first steps mutants G1-1477 (P = 0.036) and G2-1573 (P = 0.024), with intermediate levels of resistance to gentamicin but not for E. faecalis G3-1688 and E. faecalis 102, the two strains with high levels of resistance to gentamicin.
TABLE 2.
Activity of gentamicin alone or in combination with amoxicillin in experimental endocarditis due to E. faecalisa
| Regimen | Mean log10 CFU/g of vegetation ± SD for strain:
|
||||
|---|---|---|---|---|---|
| JH2-2 (MIC = 64 μg/ml)b | 1477 (MIC = 128 μg/ml) | 1573 (MIC = 256 μg/ml) | 1688 (MIC = 512 μg/ml) | 102 (MIC > 2,000 μg/ml) | |
| None (controls, start of therapy) | 8.74 ± 0.97 (n = 5) | 7.76 ± 1.22 (n = 4) | 9.66 ± 1.10 (n = 4) | 8.62 ± 1.27 (n = 4) | 9.35 ± 0.45 (n = 4) |
| Amoxicillin | 5.98 ± 1.66 (n = 7) | 8.20 ± 1.39c (n = 6) | 8.36 ± 0.40c (n = 8) | 7.53 ± 1.15c (n = 7) | 7.90 ± 0.39c (n = 8) |
| Gentamicin | 8.27 ± 0.89 (n = 7) | 8.82 ± 1.09 (n = 8) | 9.57 ± 1.41 (n = 4) | 9.77 ± 1.02 (n = 4) | 9.52 ± 0.40 (n = 4) |
| Amoxicillin + gentamicin | 5.02 ± 1.82 (n = 7) | 6.42 ± 1.55d,e (n = 8) | 7.21 ± 1.09d,e (n = 8) | 7.29 ± 0.85 (n = 8) | 7.91 ± 0.42 (n = 8) |
| ΔCFU, amox/assoce | 0.96 ± 1.82 | 1.78 ± 1.55f | 1.15 ± 1.09f | 0.24 ± 0.85 | −0.01 ± 0.42 |
Results show the mean ± standard deviation log10 CFU of aortic vegetations per gram after 3 days of treatment with amoxicillin at 50 mg/kg and/or gentamicin at 2 mg/kg intramuscularly three times a day. Controls and gentamicin groups were not statistically different between strains. The number of rabbits infected with the strain and treated with the regimen corresponding to the column and the line af the table is shown in parentheses.
Gentamicin MIC.
P < 0.05 versus JH2-2.
P < 0.05 versus amoxicillin alone.
Difference in CFU between amoxicillin alone versus the combination.
Significantly different from zero (P < 0.05).
DISCUSSION
Currently, it is considered that, in ampicillin-susceptible enterococci, gentamicin MICs lower than 500 μg/ml are predictive of (i) in vitro synergy between a cell wall-active agent and gentamicin at concentration such as 4 μg/ml, which is low enough to be achievable in clinical practice, and (ii) of a beneficial effect of the combination in vivo (3, 9). There are, however, some controversies about amoxicillin concentrations used to detect synergism with gentamicin in enterococci. Some authors recommend concentrations of at least the MIC of amoxicillin or more (17, 18), while others use amoxicillin at half the MIC or less (4, 9, 22). We used an amoxicillin concentration of half the MIC because it was the first low concentration allowing us to point out a synergism defined according to the American Society for Microbiology recommendations. All experiments with intermediate-level resistance to gentamicin in enterococci are made with amoxicillin concentration of one-half and one-quarter the MIC (4, 9).
The use of 500 μg/ml as a limit to predict synergism in vitro between amoxicillin and gentamicin was not appropriate in our experiments. Indeed, gentamicin at a concentration of 4 μg/ml did not allow us to detect synergism in vitro between amoxicillin and gentamicin against the first-step mutant G1-1477 and the second-step mutant G2-1573, despite a MIC of gentamicin below 500 μg/ml. Synergism was observed when increased concentrations of gentamicin were used in all three step-mutants including G3-1688, whose MIC of gentamicin was above 500 μg/ml. Each step of acquired gentamicin resistance required a relatively higher gentamicin concentration to achieve synergism and bactericidal activity in combination with amoxicillin, ranging from one-eighth the MIC for JH2-2 to one-half the MIC for 1688 (G3) (Fig. 1 and Table 1). However, these gentamicin concentrations are not achievable in humans.
By contrast, and as expected, no synergism was observed between amoxicillin and gentamicin against strain 102, which is highly resistant to gentamicin by means of production of the aminoglycoside-modifying enzymes AAC(6′) and APH(2′). Some authors reported a single clinical isolate of E. faecalis among 18 strains with gentamicin MICs of 256 μg/ml by aph(2")-Ic expression, which expressed no synergism with amoxicillin in vitro (4). Dressel et al. (9) found three clinical E. faecalis among 4,411 isolates of Enterococcus spp. expressing low-level resistance to gentamicin (MIC at 8 and 16 μg/ml), which had lost synergism with amoxicillin in vitro. In our experience, among 100 clinical isolates of Enterococcus spp., only one strain had a gentamicin MIC between 64 and 512 μg/ml and the same phenotype of aminoglycoside resistance as our JH2-2 derivatives (data not shown).
The precise mechanism of gentamicin resistance is not yet characterized in mutants G1-1477, G2-1573, and G3-1688. However, it is probably nonenzymatic since (i) PCR results for the five known genes encoding for acquired aminoglycoside-modifying enzymes were negative, and (ii) the characteristics of the technique used to obtain the mutants is not designed to select resistant clones by means of acquisition of resistance genes by horizontal transfer. Ongoing experiments in our laboratory indicate that resistance might be due to modifications in permeability limiting gentamicin uptake.
Adequate and efficient antibiotic therapy rests largely on the knowledge of in vitro bacterial drug susceptibility. Extrapolation to in vivo efficacy takes into account pharmacokinetic and pharmacodynamic parameters but remains empirical and requires some interpretation of in vitro susceptibility testing. That our mutants were isogenic allowed to determine if testing synergism in vitro could be used to predict synergism in vivo in experimental endocarditis model in rabbits. We chose this model because it is recognized to predict treatment efficacy in humans (10). A key figure of the model is that it allows to mimic antibiotic concentrations in humans. Indeed plasma concentrations of gentamicin obtained in rabbits with 2 mg/kg every 8 h were similar to those obtained in humans treated with 1 mg/kg every 8 h. In both instances peak concentrations were of 3 to 5 μg/ml, which is what is recommended for the treatment of enterococcal endocarditis in humans (11).
In the rabbit model, the amoxicillin plus gentamicin combination was significantly more effective than amoxicillin alone against JH2-2 derivatives with intermediate levels of resistance to gentamicin (i.e., G1-1477 and G2-1573). This was in contrast with what we observed in vitro, where we showed that the combination of amoxicillin with gentamicin at 4 μg/ml was not synergistic. An explanation for this discrepancy could be the increased bactericidal efficacy of gentamicin in vivo due to the presence of human or rabbit serum, as previously described in vitro and in experimental endocarditis for E. faecalis (16). Another explanation may be the amoxicillin concentrations achieved in vivo, which remains above the MIC for at least 7 h of the 8 during the dosing interval, in opposition with the sub-MIC concentrations used in vitro.
As expected, the amoxicillin-gentamicin combination failed to show any benefit in vivo compared to amoxicillin alone not only against strain 102, but also against strain G3-1688, which is also highly resistant to gentamicin but does not produce any aminoglycoside-modifying enzymes. Even if no current recommendations to perform killing curves on a routine basis are available, it seems that the results of killing curve experiments performed in vitro using a fixed concentration of gentamicin (i.e., 4 μg/ml) do not reliably predict the occurrence of in vivo synergism between amoxicillin and gentamicin in strains with increased levels of gentamicin resistance. The simple detection of a high level of gentamicin resistance (i.e., MIC > 500 μg/ml) remains a more accurate tool. However, it should be noted that, even if no statistical benefit of the combination was detected in vivo for strain G3-1688, the strain clearly behaved differently from strain 102, both in vitro and in vivo. Indeed, synergisms was totally abolished for the latter strain while still achievable in vitro and with intermediate expression in vivo for G3-1688. This difference was probably related to the presence of an aminoglycoside-modifying enzyme in strain 102. Indeed, it has been shown that production of aminoglycoside-modifying enzymes suppressed all aminoglycoside activity in experimental endocarditis, and thus synergy, even when produced at low levels (2).
To conclude, our data suggest that an intermediate level of resistance to gentamicin (64 < MIC < 500 μg/ml) in strains that do not possess the aph(2")-Ic gene is not associated with a loss of synergy between amoxicillin and gentamicin in vivo, despite higher requirements in gentamicin concentrations to achieve synergy in vitro. Therefore, in clinical practice, the use of an MIC of 500 μg/ml as the cutoff limit in predicting the in vivo benefit of the combination in E. faecalis infections remains a simple and effective tool.
Acknowledgments
We gratefully acknowledge Patrice Courvalin for providing the strains E. faecalis 102, Escherichia coli DH5α(pAM6306), E. coli NC95, E. coli KHE5-2, and E. faecium aadE and Michel Arthur for helpful technical assistance.
REFERENCES
- 1.Aslangul, E., M. Baptista, B. Fantin, F. Depardieu, M. Arthur, P. Courvalin, and C. Carbon. 1997. Selection of glycopeptide-resistant mutants of VanB-type Enterococcus faecalis BM4281 in vitro and in experimental endocarditis. J. Infect. Dis. 175:598-605. [DOI] [PubMed] [Google Scholar]
- 2.Caulin, E., A. Coutrot, C. Carbon, and E. Collatz. 1996. Resistance to amikacin and isepamicin in rabbits with experimental endocarditis of an aac(6′)-Ib-bearing strain of Klebsiella pneumoniae susceptible in vitro. Antimicrob. Agents Chemother. 40:2848-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-589. [DOI] [PubMed] [Google Scholar]
- 4.Chow, J. W., S. M. Donabedian, D. B. Clewell, D. F. Sahm, and M. J. Zervos. 1998. In vitro susceptibility and molecular analysis of gentamicin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 32:141-146. [DOI] [PubMed] [Google Scholar]
- 5.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Clewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, N. C., O. Olsvik, J. M. Swenson, C. A. Spiegel, and F. C. Tenover. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courvalin, P., C. Carlier, and E. Collatz. 1980. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J. Bacteriol. 143:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courvalin, P. M., W. V. Shaw, and A. E. Jacob. 1978. Plasmid-mediated mechanisms of resistance to aminoglycoside-aminocyclitol antibiotics and to chloramphenicol in group D streptococci. Antimicrob. Agents Chemother. 13:716-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dressel, D. C., M. A. Tornatore-Reuscher, C. R. Boschman, V. Stosor, T. Zembower, M. J. Postelnick, G. A. Noskin, and L. R. Peterson. 1999. Synergistic effect of gentamicin plus ampicillin on enterococci with differing sensitivity to gentamicin: a phenotypic assessment of NCCLS guidelines. Diagn. Microbiol. Infect. Dis. 35:219-225. [DOI] [PubMed] [Google Scholar]
- 10.Fantin, B., and C. Carbon. 1992. In vivo antibiotic synergism: contribution of animal models. Antimicrob. Agents Chemother. 36:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavalda, J., P. J. Cardona, B. Almirante, J. A. Capdevila, M. Laguarda, L. Pou, E. Crespo, C. Pigrau, and A. Pahissa. 1996. Treatment of experimental endocarditis due to Enterococcus faecalis using once-daily dosing regimen of gentamicin plus simulated profiles of ampicillin in human serum. Antimicrob. Agents Chemother. 40:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godfrey, K. 1985. Statistics in practice. Comparing the means of several groups. N. Engl. J. Med. 313:1450-1456. [DOI] [PubMed] [Google Scholar]
- 13.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jehl, F., C. Gallion, and H. Monteil. 1990. High-performance liquid chromatography of antibiotics. J. Chromatogr. 531:509-548. [DOI] [PubMed] [Google Scholar]
- 15.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2")-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefort, A., M. Arthur, L. Garry, C. Carbon, P. Courvalin, and B. Fantin. 2000. Bactericidal activity of gentamicin against Enterococcus faecalis in vitro and in vivo. Antimicrob. Agents Chemother. 44:2077-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopardo, H. A., M. E. Venuta, and E. A. Rubeglio. 1995. Penicillin resistance and aminoglycoside-penicillin synergy in enterococci. Chemotherapy 41:165-171. [DOI] [PubMed] [Google Scholar]
- 18.Moellering, R. C., Jr., O. M. Korzeniowski, M. A. Sande, and C. B. Wennersten. 1979. Species-specific resistance to antimicrobial synergism in Streptococcus faecium and Streptococcus faecalis. J. Infect. Dis. 140:203-208. [DOI] [PubMed] [Google Scholar]
- 19.Moellering, R. C., Jr., B. E. Murray, S. C. Schoenbaum, J. Adler, and C. B. Wennersten. 1980. A novel mechanism of resistance to penicillin-gentamicin synergism in Streptococcus faecalis. J. Infect. Dis. 141:81-86. [DOI] [PubMed] [Google Scholar]
- 20.Pearson, R. D., R. T. Steigbigel, H. T. Davis, and S. W. Chapman. 1980. Method of reliable determination of minimal lethal antibiotic concentrations. Antimicrob. Agents Chemother. 18:699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steers, E., E. L. Foltz, B. S. Graves, and J. Riden. 1959. An inocula replicating apparatus for routine testing and bacterial susceptibility to antibiotics. Antibiot. Chemother. (Basel) 9:307-311. [PubMed] [Google Scholar]
- 22.Torres, C., J. M. Torres, C. Tenorio, and F. Baquero. 1993. Synergistic activity of tobramycin-netilmicin against Enterococcus faecalis producing 6′-aminoglycoside-acetyltransferase, 2"-aminoglycoside-phosphotransferase enzyme. Eur. J. Clin. Microbiol. Infect. Dis. 12:646-648. [DOI] [PubMed] [Google Scholar]
- 23.Tsai, S. F., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2")-Id, in Enterococcus spp. Antimicrob. Agents Chemother. 42:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]