Abstract
A chromosomally encoded oxacillinase, OXA-69, was characterized from Acinetobacter baumannii AYE. β-Lactamase OXA-69 shared 97% amino acid identity with the recently described OXA-51 enzyme of A. baumannii and 62 and 56% amino acid identity with the carbapenem-hydrolyzing oxacillinases OXA-24 and OXA-23, respectively. Biochemical characterization of the purified OXA-69 revealed a narrow-spectrum hydrolysis profile but including, at a low level, imipenem and meropenem. By PCR and sequencing blaOXA-69-like genes were identified in all A. baumannii strains tested (n = 12), suggesting that this oxacillinase is naturally occurring in that species.
Acinetobacter baumannii is an opportunistic pathogen that may be an important threat due to its increasing multidrug resistance in nosocomial isolates, mostly from intensive care units (4). A. baumannii naturally produces a chromosomally encoded AmpC-type enzyme which is responsible for resistance to amino- and ureidopenicillins and narrow-spectrum cephalosporins and cephamycins and that may be overproduced (8, 15a, 34).
The oxacillin-hydrolyzing β-lactamases (oxacillinases) belong to class D of the β-lactamases (20). They usually hydrolyze oxacillin, methicillin, and cloxacillin better than benzylpenicillin, and their activity is inhibited by NaCl (20). Whereas most of the oxacillinases are plasmid mediated, several natural and chromosomally encoded oxacillinases have been reported in several environmental species (15, 27, 33) and also in clinically relevant gram-negative species, such as Pseudomonas aeruginosa, Aeromonas sp., and Ralstonia pickettii (2, 11, 12, 23, 32).
Since the first description of a carbapenem-hydrolyzing oxacillinase, in 1993 (25), several oxacillinases with a carbapenem-hydrolyzing activity have been reported in carbapenem-resistant isolates of A. baumannii (1, 5, 6, 7, 10, 13), but a precise knowledge of their distribution and of the natural β-lactamase content of A. baumannii in terms of OXA-type enzymes is still lacking.
A previous analysis of the β-lactamase content of the multidrug-resistant A. baumannii isolate AYE showed that it produced the extended-spectrum Ambler class A β-lactamase (ESBL) VEB-1, which is chromosome and integron located (31). Sequencing of the entire genome of this strain is in progress (D. Vallenet, personal communication), and in silico analysis identified a gene coding for a putative oxacillinase which is very similar, at the sequence level, to the recently described OXA-51 enzyme and its closely related variants (6, 7).
The aim of the present study was to characterize this putative β-lactamase to evaluate (i) the hydrolysis profile of this enzyme, (ii) the inducibility of its expression, and (iii) its distribution in a collection of A. baumannii strains from various geographical origins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. baumannii clinical isolate AYE expressing the ESBL VEB-1 has been studied previously (31). A. baumannii reference strains (CIP 70.10 and CIP7034T) and other A. baumannii isolates previously studied for their β-lactamase content were included in this study (Table 1). Escherichia coli DH10B and E. coli BL21(DE3) were used as hosts for cloning and expression experiments, respectively. The kanamycin-resistant pPCRBluntII- TOPO plasmid (Invitrogen Life Technologies, Cergy-Pontoise, France) was used as a cloning vector. The plasmid pET9a was used as an expression vector (Stratagene, Amsterdam, The Netherlands). The broad-host-range plasmid pAT801-RA (14) was used for transformation experiments in A. baumannii. A. baumannii CIP 70.10 and BM4547 reference strains were used as recipients for these experiments. Bacterial cultures were routinely grown in Trypticase soy (TS) broth at 37°C for 18 h, as indicated previously (13).
TABLE 1.
Sources of A. baumannii isolates
| A. baumannii strain | Isolation
|
β-Lactamase content | MIC of IPMa (μg/ml) | Reference or source | |
|---|---|---|---|---|---|
| Yr | Location | ||||
| AYE | 2001 | Bicêtre, France | OXA-69, AmpC, VEB-1, OXA-10 | 1 | 31 |
| CH15 | 1997 | Rome, Italy | OXA-69, AmpC | 8 | This study |
| CH30 | 1998 | Ankara, Turkey | OXA-69, AmpC, OXA-58 | 32 | This study |
| CIP 70.10 | OXA-64, AmpC | 0.5 | CIPb | ||
| BM4547 | Paris, France | OXA-64, AmpC | 1 | 17 | |
| CLA-1 | 2001 | Bicêtre, France | OXA-66, AmpC, OXA-40 | >32 | 13 |
| MK8560 | 1999 | Warsaw, Poland | OXA-66, AmpC | 16 | This study |
| BAR | 2004 | Toulouse, France | OXA-66, AmpC | 8 | This study |
| CH13 | 1997 | Barcelona, Spain | OXA-71, AmpC | 1 | This study |
| SDF | Marseilles, France | OXA-75 | 0.25 | 16 | |
| AMA-1 | 1999 | Bicêtre, France | OXA-76, AmpC, PER-1 | 2 | 29 |
| CIP7034T | OXA-77, AmpC | 0.5 | CIPb | ||
IPM, imipenem.
Pasteur Institute strain collection.
Antimicrobial agents and MIC determinations.
The antimicrobial agents and their sources have been described elsewhere (26). MICs were determined by an agar dilution technique as previously described (26). Results of susceptibility testing were recorded according to the guidelines of the CLSI (formerly NCCLS) (22).
Cloning and PCR experiments.
For each PCR experiment, 500 ng of total DNA was used in a standard PCR. Using total DNA of A. baumannii strains, PCR amplifications of the blaOXA-69-like genes were performed with the external primers OXA-69A (5′-CTAATAATTGATCTACTCAAG-3′) and OXA-69B (5′-CCAGTGGATGGATGGATAGATTATC-3′), giving rise to a 975-bp fragment. The PCR product obtained with total DNA of A. baumannii AYE as template was subsequently cloned in the pPCRBluntII-TOPO plasmid, resulting in recombinant plasmid pTOPO-OXA-69, and transformed in E. coli DH10B as previously described (13). E. coli strains harboring recombinant plasmids were selected onto amoxicillin (15 μg/ml)- and kanamycin (30 μg/ml)-containing TS agar plates. In parallel, the same 975-bp insert was introduced into the SmaI-restricted plasmid pAT801-RA, giving rise to plasmid pAT-OXA-69, and transformed as described previously (14) into electrocompetent A. baumannii CIP 70.10 and into its point mutant derivative A. baumannii BM4547 (Table 1), which overproduces the AdeABC efflux pump (17). Transformants were selected on ticarcillin (15 μg/ml)- and rifampin (25 μg/ml)-containing plates.
The recombinant plasmid used for overexpression of β-lactamase OXA-69 was constructed as follows: a 922-bp PCR-generated fragment using primers containing NdeI and BamHI restriction sites, respectively (OXA-69Nde, 5′-CTTATAAGTCATATGAACATTAAAGC-3′, and OXA-69Bam, 5′-CTCTATAAAAAGGGATCCGGGCTA-3′) was cloned into pPCRBluntII-TOPO plasmid according to the manufacturer's instructions. The insert of the latter plasmid was removed with NdeI-BamHI and cloned into NdeI/BamHI-restricted pET9a expression vector (Stratagene), giving rise to plasmid to pET-OXA-69.
DNA sequencing and protein analysis.
PCR-generated fragments, purified by using Qiaquick PCR purification spin columns (QIAGEN), and the inserts of the recombinant plasmids were sequenced on both strands on an ABI3100 automated sequencer (Applied Biosystems, Les Ulis, France). The nucleotide and the deduced protein sequences were analyzed with software available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Dendrograms were derived from a multiple sequence alignment by a parsimony method using the phylogeny package PAUP (Phylogenetic Analysis Using Parsimony), version 3.0 (35).
IEF analysis and induction studies.
Isoelectric focusing (IEF) analysis was performed with a pH 3.5 to 9.5 Ampholine polyacrylamide gel (Amersham Pharmacia Biotech) with culture extracts of the A. baumannii isolates and of E. coli DH10B harboring a plasmid (pTOPO-OXA-69). The inducibility of the β-lactamase contents from the ESBL-producing A. baumannii AYE and the ceftazidime-susceptible A. baumannii isolate SDF (Table 1) was tested in TS broth at 37°C using imipenem (0.25, 0.5, and 1 μg/ml) and cefoxitin (125, 250, and 500 μg/ml) as β-lactam inducers as described previously (12, 23), and hydrolysis was measured with 200 μM of imipenem as substrate. The β-lactamase activity was defined as 1 unit of enzyme that hydrolyzed 1 μmol of imipenem per min. The total protein content was measured with bovine albumin as the standard (DC protein assay kit; Bio-Rad Laboratories).
β-Lactamase purification, relative molecular mass determination, and N-terminal sequencing.
Induction of an exponentially growing culture of E. coli BL21(DE3)(pET-OXA-69) with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was performed at 37°C for 5 h in TS broth. Four liters of this culture was pelleted and resuspended in 30 ml of 20 mM Tris-H2SO4 buffer (pH 9). The protein extracts were purified as described previously (11, 28) with some modifications. Briefly, culture extracts were subjected to several purification steps including ion-exchange chromatography with a Q-Sepharose column first with 20 mM Tris-HCl buffer (pH 9), followed by chromatography on an S-Sepharose column equilibrated with 20 mM BisTris buffer (pH 6.5). Elution of the β-lactamase was performed with a linear K2SO4 gradient (0 to 500 mM). Peaks of β-lactamase activities were pooled and dialyzed with 50 mM phosphate buffer (pH 7.0). The protein content was measured by the Bio-Rad DC protein assay, and the specific activities of the crude extract and the purified β-lactamase from E. coli BL21(DE3)(pET-OXA-69) were compared. The protein purification rate and the relative molecular mass of β-lactamase OXA-69 were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) analysis (11). We also performed a PAGE analysis in order to estimate the apparent molecular weight of OXA-69. Protein transfer onto a polyvinylidene difluoride membrane and N-terminal Edman sequencing were performed as previously described (11).
Kinetic studies.
Purified β-lactamase was used for determination of kinetic parameters (kcat and Km) performed at 30°C in a reaction buffer made of 100 mM Tris-H2SO4, 300 mM K2SO4 (pH 7.0), in which NaHCO3 was added to a final concentration of 10 mM in order to avoid biphasic kinetics (18, 24). The initial rates of hydrolysis of β-lactams were determined with a UV spectrophotometer as previously described (13). Fifty percent inhibitory concentration (IC50) was determined as the clavulanate, the tazobactam, or the sulbactam concentration that reduced the hydrolysis rate of 100 μM of nitrocefin by 50% under conditions in which the enzyme was preincubated with various concentrations of inhibitor for 3 min at 30°C before addition of the substrate (13). The effect of divalent cations on the enzymatic activity was investigated by adding ZnSO4 and CuSO4 to the reaction buffer at a final concentration of 50, 100, 200, or 500 μM.
Genotypic comparison.
Genotyping was performed to analyze clonal diversity of the isolates. Pulsed-field gel electrophoresis was carried out after digestion of whole-cell DNAs with ApaI restriction enzyme, as previously described (31).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL/GenBank nucleotide sequence database under the accession numbers AY859527 for OXA-69, AY859528 for OXA-71, and AY859529 for OXA-75.
RESULTS AND DISCUSSION
Susceptibility testing.
MICs of β-lactams for A. baumannii AYE showed resistance to penicillins, expanded-spectrum cephalosporins, and aztreonam, as previously reported (Table 2) (31). Addition of clavulanic acid and tazobactam significantly restored the β-lactam activity for ticarcillin (Table 2), which was consistent with the expression of β-lactamase VEB-1. In addition, A. baumannii AYE overproduced a naturally occurring cephalosporinase due to ISAba1 insertion upstream of its blaAmpC gene (C. Héritier, L. Poirel, and P. Nordmann, submitted for publication).
TABLE 2.
MICs of β-lactams for A. baumannii AYE, E. coli DH10B(pTOPO-OXA-69), A. baumannii CIP 70.10(pAT-OXA-69), and A. baumannii BM4547(pAT-OXA-69) and reference strains E. coli DH10B, A. baumannii CIP 70.10, and A. baumannii BM4547
| β-Lactama | MIC (μg/ml) for strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A. baumannii AYE | E. coli DH10B (pTOPO-OXA-69) | E. coli DH10B | A. baumannii CIP 70.10 (pAT-OXA-69) | A. baumannii CIP 70.10 (pOXA-40)b | A. baumannii CIP 70.10 | A. baumannii BM4547 (pAT-OXA-69) | A. baumannii BM4547 | |
| Ampicillin | >512 | 16 | 4 | 64 | >256 | 32 | 64 | 64 |
| Ampicillin + CLA | >512 | 8 | 4 | 16 | >256 | 8 | 16 | 16 |
| Ticarcillin | >512 | 8 | 4 | 16 | >256 | 8 | 32 | 16 |
| Ticarcillin + CLA | 32 | 8 | 4 | 16 | >256 | 8 | 32 | 16 |
| Piperacillin | 256 | 4 | 2 | 16 | >256 | 16 | 16 | 16 |
| Piperacillin + TZB | 8 | 4 | 2 | 4 | >256 | 4 | 4 | 4 |
| Cephaloridine | >512 | 4 | 4 | >512 | >512 | >512 | >512 | >512 |
| Cephalothin | >512 | 4 | 4 | >512 | >512 | >512 | >512 | >512 |
| Cefuroxime | >512 | 4 | 4 | 64 | 64 | 64 | 64 | 64 |
| Ceftazidime | >512 | 0.06 | 0.06 | 2 | 4 | 2 | 4 | 4 |
| Cefotaxime | 512 | 0.06 | 0.06 | 8 | 16 | 8 | >32 | >32 |
| Cefepime | 512 | 0.06 | 0.06 | 1 | 2 | 1 | >32 | >32 |
| Cefpirome | 512 | 0.06 | 0.06 | 2 | 4 | 2 | >32 | >32 |
| Aztreonam | >512 | 0.06 | 0.12 | 64 | 64 | 64 | 64 | 64 |
| Imipenem | 1 | 0.06 | 0.06 | 0.5 | 4 | 0.5 | 1 | 1 |
| Meropenem | 1 | 0.06 | 0.06 | 0.5 | 4 | 0.5 | 1 | 1 |
CLA, clavulanic acid (2 μg/ml); TZB, tazobactam (4 μg/ml).
Data are from reference 14.
Cloning and expression of the blaOXA-69 β-lactamase gene.
A blaOXA-like gene (whose product is similar to the recently described OXA-51 enz yme and its closely related variants [6, 7]) was detected in the A. baumannii isolate AYE chromosome during the ongoing sequencing project. The G+C content of this gene named blaOXA-69 was 39.3%, which compares with the G+C content of the A. baumanni genome (38.8%) (P. E. Fournier, personal communication). Cloning experiments using specific primers and PCR showed E. coli DH10B(pTOPO-OXA-69) producing an active enzyme that was named OXA-69. This recombinant strain exhibited only reduced susceptibility to several β-lactams; this phenotype corresponded to expression of a narrow-spectrum oxacillinase, although the resistance phenotype conferred by an oxacillinase may be different in E. coli and in A. baumannii (Table 2). The recombinant strain remained susceptible to imipenem (MIC of 0.06 μg/ml). Once overexpressed in E. coli BL21(DE3), the cloned β-lactamase conferred reduced susceptibility to penicillins unchanged after clavulanic acid addition (data not shown).
DNA sequence analysis of the 975-bp insert of pTOPO-OXA-69 revealed an open reading frame of 825 bp encoding a 274-amino-acid preprotein with a relative molecular mass of 27.7 kDa. IEF analysis of cultures of E. coli DH10B(pTOPO-OXA-69) revealed a pI value of 8.4, whereas a corresponding signal was not identified from the A. baumannii strain.
Genetic environment of blaOXA-69.
In contrast to several acquired oxacillinase genes, the blaOXA-69 gene was not associated with integron or transposon structures (20). The sequence identified upstream of blaOXA-69 encoded an FsxA-like protein corresponding to a putative suppressor of F exclusion. Interestingly, a gene encoding a putative acetyltransferase likely playing a role in aminoglycoside resistance and sharing 71% amino acid identity with an orthologue in Acinetobacter sp. ADP1 was identified 71 bp downstream of the blaOXA-69 gene.
Sequence analysis of β-lactamase OXA-69.
The five first amino acids of the mature protein OXA-69, identified by N-terminal sequencing, were S-P-Y-I-V. OXA-69 contained the conserved motifs of serine β-lactamases such as S-T-F-K at positions 70 to 73 and the YGN motif at positions 144 to 146 class D β-lactamase numbering (9). As observed for OXA-51-like oxacillinases (6, 7), the valine residue of the S-X-V triad at positions 118 to 120 was replaced by an isoleucine in OXA-69. A K-S-G motif was identified at positions 216 to 218 as observed in the OXA-40-like and OXA-58 enzymes but not in other oxacillinases (13, 30).
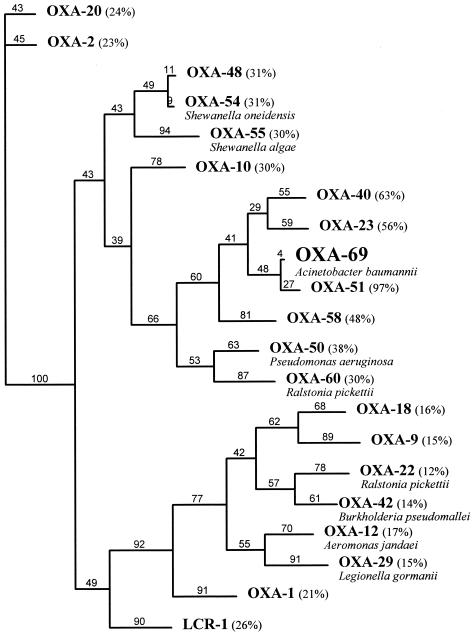
The common motif of oxacillinases, Q-X-X-X-L, usually found at class D β-lactamase positions 176 to 180, was replaced by the motif E-A-Q-F-A (glutamate-alanine-glycine-phenylalanine-alanine) in OXA-69, which is similar to the motif identified in the naturally occurring oxacillinase OXA-60 from R. pickettii, which possesses some carbapenemase activity (12) (Fig. 1).
FIG. 1.
Dendrogram obtained for several oxacillinases by parsimony analysis (35). Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The number of changes is indicated above each branch, and the percent values at branching points (underlined) refer to the number of times that a particular node was found in 100 bootstrap replications. The distance along the vertical axis has no significance. Numbers in parentheses indicate percentages of amino acid identity with the β-lactamase OXA-69. Bacterial species names are for those oxacillinases that are naturally occurring.
β-Lactamase OXA-69 shared 97, 63, 59, and 56% amino acid identity with the carbapenem-hydrolyzing oxacillinases OXA-51, OXA-40, OXA-58, and OXA-23 from A. baumannii, respectively (6, 7, 10, 13, 30). β-Lactamase OXA-69 shared its highest amino acid identity with several oxacillinases known to hydrolyze imipenem (Fig. 1).
Biochemical properties of OXA-69.
We observed two forms of the enzyme by PAGE analysis, with apparent molecular masses of ca. 27 and 55 kDa, which may correspond to monomeric and dimeric forms of OXA-69, respectively. After purification, specific activity of β-lactamase OXA-69 against 100 μM nitrocefin was 375 mU/mg of protein, and its purity was estimated to be >95% by sodium dodecyl sulfate-PAGE analysis.
Kinetic parameters of purified β-lactamase OXA-69 indicated that it had a hydrolysis profile that included benzylpenicillin, ampicillin, ticarcillin, piperacillin, imipenem, and meropenem (Table 3). Nitrocefin was the better substrate whereas OXA-69 did not hydrolyze ceftazidime, cefotaxime, cefepime, or aztreonam. The very weak catalytic efficiency (kcat/Km) of OXA-69 for most β-lactams resulted from its low affinity (high Km values) for these substrates, by contrast with data reported for the closely related OXA-51 (7).
TABLE 3.
Kinetic parameters of purified β-lactamase OXA-69a
| Substrate | kcat (10−3 s−1) | Km (μM) | kcat/Km (10−3 mM−1 · s−1) | kcat/Km (%)b |
|---|---|---|---|---|
| Benzylpenicillin | 200 | 710 | 300 | 100 |
| Ampicillin | 60 | 240 | 250 | 85 |
| Ticarcillin | 300 | 2,800 | 110 | 40 |
| Piperacillin | 200 | 4,000 | 50 | 20 |
| Nitrocefin | 400 | 130 | 3,200 | 1,100 |
| Cephalothin | 4 | 190 | 20 | 7 |
| Cephaloridine | 50 | 2,500 | 20 | 7 |
| Ceftazidime | NDc | |||
| Cefepime | ND | |||
| Aztreonam | ND | |||
| Imipenem | 100 | 3,600 | 30 | 10 |
| Meropenem | 60 | 4,500 | 10 | 3 |
| Oxacillin | 200 | 3,700 | 60 | 20 |
Data are the means of three independent experiments. Standard deviations were within 10% of the means.
Percentage is calculated as compared to that of benzylpenicillin, taken as 100%.
ND, no detectable hydrolysis (<0.01 s−1).
No biphasic kinetics were obtained for all the substrates tested. No significant effect of divalent cations on the activity of OXA-69 was observed. Hydrolysis of oxacillin was detected at a low level as for other carbapenem-hydrolyzing oxacillinases (13). Hydrolysis of expanded-spectrum cephalosporins was not detected, and hydrolysis of cephalothin and cephaloridine was of a very low level. Interestingly, OXA-69 hydrolyzed imipenem and meropenem, as observed for several naturally produced oxacillinases such as OXA-50 from P. aeruginosa, OXA-54 from Shewanella oneidensis, OXA-55 from Shewanella algae, and OXA-60 from R. pickettii (11, 12, 15, 27). Nevertheless, the ability of OXA-69 to hydrolyze carbapenems was very weak, since low affinities for imipenem and meropenem were found with Km values of 3,600 and 4,500 μM, respectively, resulting in low catalytic efficiencies for these substrates. By contrast, β-lactamases OXA-40 identified in A. baumannii and OXA-60 from R. pickettii possessed a high affinity for imipenem but did not hydrolyze meropenem and OXA-50 from P. aeruginosa possessed a good affinity for imipenem but a low affinity for meropenem, whereas OXA-58 identified from A. baumannii possessed very high affinities for both carbapenems. Surprisingly, the Km value of imipenem reported for OXA-51 was much lower than that of OXA-69 (7).
Inhibition studies as measured by IC50s showed that OXA-69 activity was weakly inhibited by clavulanic acid (100 μM), tazobactam (40 μM), and sulbactam (30 μM) as found for most of the oxacillinases. OXA-69 activity was well inhibited by NaCl as for most of the oxacillinases (IC50 = 7.5 mM) (20).
Distribution of blaOXA-69-like genes in A. baumannii isolates.
Analysis of the pulsed-field gel electrophoresis patterns of the ApaI-restricted DNAs of several A. baumannii clinical isolates and reference strains of various geographical origins showed that these strains were not clonally related (data not shown). Among 12 studied strains, an OXA-69-like sequence was identified, making this gene a feature of A. baumannii. The OXA-69-like sequences identified differed by no more than nine residues (Table 4). The genetic variability of OXA-69-like sequences in A. baumannii was low, as found for the naturally produced β-lactamases OXA-22- and OXA-60-like of R. pickettii (12, 23) and OXA-50-like of P. aeruginosa (11).
TABLE 4.
Comparison of amino acid sequences of the chromosome-encoded oxacillinases of A. baumannii isolates
| A. baumannii strain(s) | OXA | Amino acid at class D β-lactamase positiona
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 15 | 23 | 25 | 35 | 48 | 53 | 88 | 97 | 99 | 108 | 127 | 137 | 142 | 192 | 193 | 196 | 227 | 256 | 269 | 270 | 273 | ||
| AYE, CH15, CH30 | OXA-69 | T | H | D | A | A | H | Q | A | D | E | N | A | K | V | Q | K | D | N | P | K | S | Q |
| CIP 70.10 | OXA-64 | — | — | E | G | — | — | — | — | — | Q | — | — | — | — | P | — | — | D | — | — | — | — |
| CLA-1, MK8560, BAR | OXA-66 | — | — | V | — | — | — | — | — | — | K | — | — | — | — | — | — | — | — | — | — | — | — |
| CH13 | OXA-71 | — | — | E | — | — | — | — | — | — | Q | — | — | — | — | P | — | — | D | — | — | — | — |
| SDF | OXA-75 | — | Y | E | — | — | — | H | — | — | Q | — | — | N | I | — | — | — | — | S | R | G | — |
| AMA-1 | OXA-76 | — | — | V | — | — | — | — | T | — | K | — | — | — | — | — | — | — | — | — | — | — | K |
| CIP7034T | OXA-77 | S | — | E | — | — | — | — | — | — | Q | — | P | N | — | — | — | — | — | — | — | — | — |
| 788b | OXA-51 | — | — | E | — | V | Q | — | — | — | Q | D | — | — | — | P | — | — | D | — | — | — | — |
| 884, 790c | OXA-65 | — | — | E | — | — | Q | — | — | — | K | D | — | — | — | — | — | — | — | — | — | — | — |
| 809c | OXA-68 | — | — | E | — | — | Q | — | — | — | Q | N | — | N | — | — | E | — | — | — | — | — | — |
| 812c | OXA-70 | — | — | K | — | — | Q | — | — | N | Q | D | — | N | — | P | — | H | D | — | — | — | — |
Induction experiments.
Induction experiments with cultures of A. baumannii AYE and A. baumannii SDF using imipenem or cefoxitin as β-lactam inducers did not reveal any β-lactamase induction, in accordance with previous reports (4). Actually, no carbapenemase activity was detected in our conditions from the extracts of the two strains after induction. This observation is in contrast with what has been found for the naturally occurring oxacillinases OXA-22 and OXA-60 of R. pickettii or those identified in Aeromonas sp. (2, 12, 23, 32) but is similar to the lack of inducibility of naturally occurring OXA-50-like oxacillinases of P. aeruginosa (11).
Expression of the blaOXA-69 gene in A. baumannii.
In order to evaluate the role of OXA-69 in providing the natural β-lactam resistance profile in A. baumannii, cloning of its gene on a broad-host-range plasmid was performed and the resulting recombinant plasmid was transformed into a wild-type A. baumannii reference strain and in an AdeABC efflux hyperproducer. Our results did not show an overall increase of the β-lactam resistance level in A. baumannii CIP 70.10 (pAT-OXA-69) (Table 2), suggesting that OXA-69 may not play a significant role even when expressed in A. baumannii even though a slight increase of the MIC of ticarcillin was noticed (Table 2). In addition, once expressed in the A. baumannii BM4547 reference strain, no significant MIC increase was detected, indicating that, despite overproduction of efflux that could have a synergistic effect on β-lactam resistance, production of OXA-69 in A. baumannii had a marginal effect (Table 2). By contrast, we have shown previously that production of OXA-40, once expressed from the same vector in A. baumannii, led to an increase of MICs for penicillins and carbapenems (Table 2) (14).
Conclusion.
This study showed that A. baumannii possesses, in addition to an AmpC-type cephalosporinase, another chromosomally carried gene coding for a class D β-lactamase. Since OXA-69 had a weak catalytic efficiency, it likely does not contribute to the intrinsic resistance of A. baumannii to β-lactams. It remains to be determined whether expression of this gene may be sometimes enhanced in its natural host due to mutation(s) or presence of an insertion sequence, therefore modifying the efficacy of its promoter sequences, as reported for the naturally occurring blaAmpC genes of A. baumannii (8, 15a, 34). Interestingly, the OXA-69-like enzymes were identified in all the tested strains, whatever their geographical or clinical origin was. In addition, an OXA-69 variant was even identified in strain SDF, which had been isolated from a human body louse (16). Nevertheless, no homologue of the blaOXA-69 gene was identified in the genome of Acinetobacter sp. strain ADP1, which was recently characterized (3), suggesting that such a gene may be absent in other members of the Acinetobacter species.
The OXA-69-like enzymes reported in this study are closely related to the OXA-51-like enzymes that have been recently described and claimed to be responsible for acquired carbapenem resistance (6, 7). We believe that OXA-69-like enzymes and OXA-51-like enzymes belong to the same group of naturally occurring oxacillinases in A. baumannii with low-level carbapenemase activity.
It seems now that a series of environmental gram-negative species such as P. aeruginosa, Burkholderia pseudomallei, Shewanella sp., Aeromonas sp., and R. pickettii have oxacillinase genes. Wide distribution of those types of β-lactamase genes in environmental gram-negative species may be the source for acquired oxacillinase genes in class 1 integron structures (20, 21) and also their identifications on phages recovered from bacteria present in sewage (19).
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA 3539), Université Paris XI, Paris, France, and by the European Community (6th PCRD, LSHM-CT-2003-503-335). L.P. is a researcher from the INSERM, Paris, France.
We are very grateful to Valérie Barbe and Jean Weissenbach from the Genoscope (CNRS-UMR8030), Evry, France, for their valuable contribution in the determination of the genome sequence. We thank T. Naas for valuable advice on β-lactamase purification and T. Lambert for the gift of the A. baumannii BM4547 reference strain. We thank S. Brisse and M. Gniadkowski for the gift of several A. baumannii isolates.
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alksne, L. E., and B. A. Rasmussen. 1997. Expression of the AsbA1, OXA-12, and AsbM1 β-lactamases in Aeromonas jandei AER14 is coordinated by a two-component regulon. J. Bacteriol. 179:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin, E., M. L. Joly-Guillou, and K. J. Towner. 1996. Epidemiology of Acinetobacter spp. p. 71-100. In K. J. Towner et al. (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, Inc., Boca Raton, Fla.
- 5.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S., and S. G. B. Amyes. 2005. The sequences of seven class D beta-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 11:326-329. [DOI] [PubMed] [Google Scholar]
- 7.Brown, S., H. K. Young, and S. G. B. Amyes. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 8.Corvec, S., N. Caroff, E. Espaze, C. Giraudeau, H. Drugeon, and A. Reynaud. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629-635. [DOI] [PubMed] [Google Scholar]
- 9.Couture, F., J. Lachapelle, and R. C. Lévesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 10.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girlich, D., T. Naas, and P. Nordmann. 2004. OXA-60, a chromosomal, inducible, and imipenem-hydrolyzing class D β-lactamase from Rasltonia pickettii. Antimicrob. Agents Chemother. 48:4217-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Héritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Héritier, C., L. Poirel, and P. Nordmann. 2004. Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing Ambler class D β-lactamase from Shewanella algae. Antimicrob. Agents Chemother. 48:1670-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Héritier, C., L. Poirel, and P. Nordmann. Cephalosporinase overexpression as a result of insertion of ISAba1 in Acinetobacter baumannii. Clin. Microb. Infect., in press. [DOI] [PubMed]
- 16.La Scola, B., and D. Raoult. 2004. Acinetobacter baumannii in human body louse. Emerg. Infect. Dis. 10:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maveyraud, L., D. Golemi, L. P. Kotra, S. Tranier, S. Vakulenko, S. Mobashery, and J. P. Samama. 2000. Insights into class D β-lactamases are revealed by the crystal structure of the OXA-10 enzyme from Pseudomonas aeruginosa. Structure 8:1289-1298. [DOI] [PubMed] [Google Scholar]
- 19.Muniesa, M., A. Garcia, E. Miro, B. Mirelis, G. Prats, J. Jofre, and F. Navarro. 2004. Bacteriophages and diffusion of β-lactamase genes. Emerg. Infect. Dis. 10:1134-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 21.Naas, T., W. Sougakoff, A. Casetta, and P. Nordmann. 1998. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, p. 1. Approved standard M7-A6. NCCLS, Wayne, Pa.
- 23.Nordmann, P., L. Poirel, M. Kubina, A. Casetta, and T. Naas. 2000. Biochemical-genetic characterization and distribution of OXA-22, a chromosomal and inducible class D β-lactamase from Ralstonia (Pseudomonas) pickettii. Antimicrob. Agents Chemother. 44:2201-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paetzel, M., F. Danel, L. De Castro, S. C. Mosimann, M. G. Page, and N. C. Strynadka. 2000. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 7:919-925. [DOI] [PubMed] [Google Scholar]
- 25.Paton, R., R. S. Miles, J. Hood, and S. G. B. Amyes. 1993. ARI-1: β-lactamase mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2:81-88. [DOI] [PubMed] [Google Scholar]
- 26.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic-acid inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., C. Héritier, and P. Nordmann. 2004. Chromosome-encoded Ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., C. Héritier, V. Tolün, and P. Nordmann. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., A. Karim, A. Mercat, I. Le Thomas, H. Vahaboglu, C. Richard, and P. Nordmann. 1999. Extended-spectrum beta-lactamase-producing strain of Acinetobacter baumannii isolated from a patient in France. J. Antimicrob Chemother. 43:157-158. [PubMed] [Google Scholar]
- 30.Poirel, L., S. Marqué, C. Héritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen, B. A., D. Keeney, Y. Yang, and K. Bush. 1994. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob. Agents Chemother. 38:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 34.Segal, H., E. C. Nelson, and B. G. Elisha. 2004. Genetic environment of ampC in Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1989. PAUP (version 3.0): phylogenetic analysis using parsimony. Illinois Natural History Survey, Champaign.